Detection of Oxacillin/Cefoxitin Resistance in Staphylococcus aureus Present in Recurrent Tonsillitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Amplification of the Coding Gene for the 16S rRNA Subunit
2.3. Sequencing the 16S rRNA Coding Region and Analyzing the Results
2.4. Sample Preparation for Obtaining Images
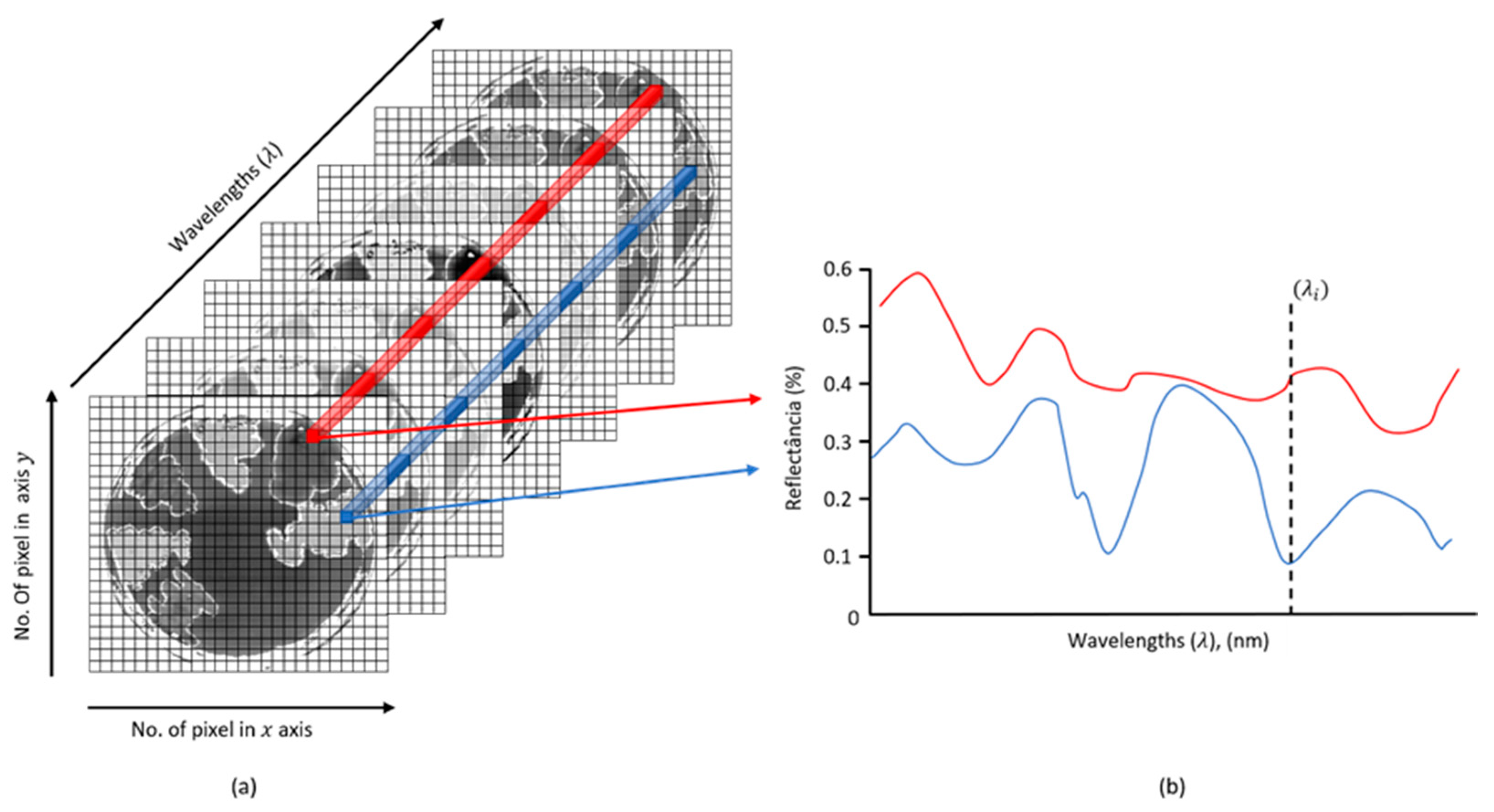
2.5. Acquisition of Hyperspectral Images
2.6. Principal Component Analysis
2.7. Partial Least Squares Discriminatory Analysis
3. Results
3.1. Sequencing the 16S rRNA
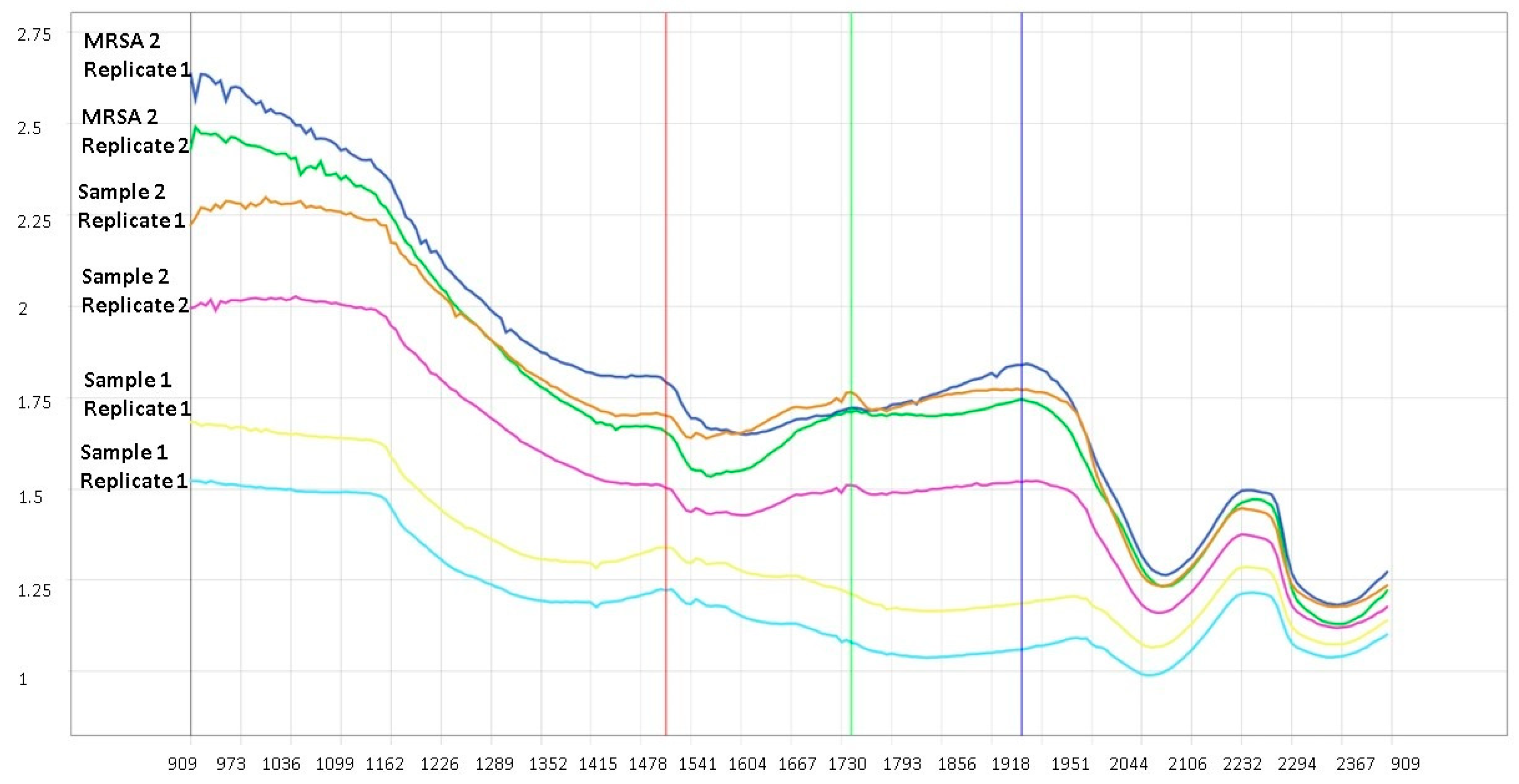
3.2. Oxacillin/Cefoxitin Resistance and Sensitivity
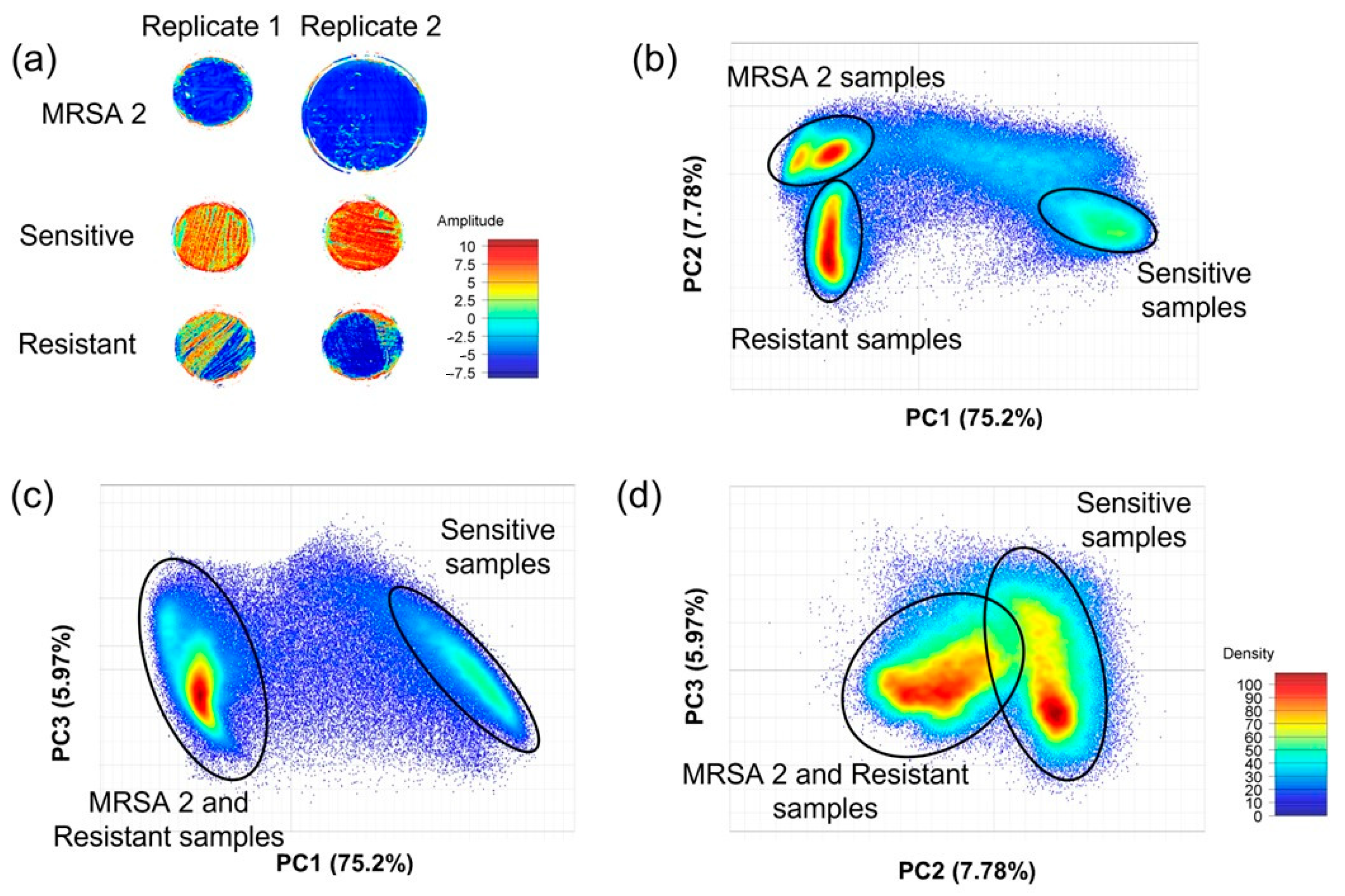
3.3. Image Analysis Using PCA
3.4. Image Analysis Using PLS-DA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prat, C.; Lacoma, A. Bacteria in the respiratory tract—how to treat? Or do not treat? Int. J. Infect. Dis. 2016, 51, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Zautner, A.E.; Krause, M.; Stropahl, G.; Holtfreter, S.; Frickmann, H.; Maletzki, C.; Kreikemeyer, B.; Pau, H.W.; Podbielski, A. Intracellular Persisting Staphylococcus aureus Is the Major Pathogen in Recurrent Tonsillitis. PloS ONE 2010, 5, e9452. [Google Scholar] [CrossRef] [Green Version]
- Weidenmaier, C.; Goerke, C.; Wolz, C. Staphylococcus aureus determinants for nasal colonization. Trends Microbiol. 2012, 20, 243–250. [Google Scholar] [CrossRef]
- Sivaraman, K.; Venkataraman, N.; Cole, A.M. Staphylococcus aureusnasal carriage and its contributing factors. Future Microbiol. 2009, 4, 999–1008. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Truque, N.; Saye, E.J.; Soper, N.; Saville, B.R.; Thomsen, I.; Edwards, K.M.; Creech, C.B. Longitudinal Assessment of Colonization With Staphylococcus aureu s in Healthy Collegiate Athletes. J. Pediatr. Infect. Dis. Soc. 2016, 5, 105–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, L.M.P.; Juman, S.; Bekele, I.; Seepersadsingh, N.; Adesiyun, A.A. Achado de bactérias selecionadas em crianças de Trinidad com doença amigdaliana crônica. Rev. Bras. De Otorrinolaringol. 2008, 74, 903–911. [Google Scholar] [CrossRef] [Green Version]
- Marquioti, C.M.J.; Lanes, L.C.; Castro, G.F.P. USO IRRACIONAL DE ANTIBACTERIANOS NA INFÂNCIA: Contribuição do profissional farmacêutico para a promoção da saúde. Rev. Transform. 2015, 7, 179–193. [Google Scholar]
- Pichichero, M.E. Group A streptococcal tonsillopharyngitis: Cost-effective diagnosis and treatment. Ann. Emerg. Med. 1995, 25, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Pinna, F.; Pádua, F.; Abdo, T. Guideline IVAS: Infecções das Vias Aéreas Superiores; Associação Brasileira de Otorrinolaringologia e Cirurgia Cérvico-Facial: São Paulo, Brazil; 196p.
- Katkowska, M.; Garbacz, K.; Stromkowski, J. Staphylococcus aureus isolated from tonsillectomized adult patients with recurrent tonsillitis. Apmis 2016, 125, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Moellering, R.C., Jr. MRSA: The first half century. J. Antimicrob. Chemother. 2012, 67, 4–11. [Google Scholar] [CrossRef] [Green Version]
- Nada, T.; Ichiyama, S.; Osada, Y.; Ohta, M.; Shimokata, K.; Kato, N.; Nakashima, N. Comparison of DNA fingerprinting by PFGE and PCR-RFLP of the coagulase gene to distinguish MRSA isolates. J. Hosp. Infect. 1996, 32, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, D.; Peters, B.M.; Li, L.; Li, B.; Xu, Z.; Shirliff, M.E. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb. Pathog. 2016, 101, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; De Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef] [Green Version]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Skov, R.L.; Han, X.; Larsen, A.R.; Larsen, J.; Sørum, M.; Wulf, M.; Voss, A.; Hiramatsu, K.; Ito, T. Novel Types of Staphylococcal Cassette Chromosome mec Elements Identified in Clonal Complex 398 Methicillin-Resistant Staphylococcus aureus Strains. Antimicrob. Agents Chemother. 2011, 55, 3046–3050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ANVISA. Agência Nacional de Vigilância Sanitária—Antimicrobial Sensitivity Teste. Available online: https://www.anvisa.gov.br/servicosaude/controle/rede_rm/cursos/boas_praticas/modulo5/sta2.htm (accessed on 19 January 2021).
- Rabelo, M.A.; Neto, A.M.B.; da Silva, E.C.B.F.; de Oliveira, W.L.M.; de Melo, F.L.; Lopes, A.C.S.; Leal, N.C.; Maciel, M.A.V. Phenotypic methods for determination of methicillin resistance in Staphylococcus spp. from health care workers. J. Bras. De Patol. E Med. Lab. 2013, 49, 91–96. [Google Scholar] [CrossRef]
- ANVISA. Agência Nacional de Vigilância Sanitária—Resistência antimicrobiana é ameaça global, diz OMS. 2019. Available online: http://portal.anvisa.gov.br/noticias/-/asset_publisher/FXrpx9qY7FbU/content/resistencia-antimicrobiana-e-ameaca-global-diz-oms/219201/pop_up?_101_INSTANCE_FXrpx9qY7FbU_viewMode=print&_101_INSTANCE_FXrpx9qY7FbU_languageId=en_US (accessed on 19 May 2020).
- Sun, Y.; Smith, E.; Wolcott, R.; Dowd, S.E. Propagation of anaerobic bacteria within an aerobic multi-species chronic wound biofilm model. J. Wound Car 2009, 18, 426–431. [Google Scholar] [CrossRef]
- Gendrin, C.; Roggo, Y.; Collet, C. Pharmaceutical applications of vibrational chemical imaging and chemometrics: A review. J. Pharm. Biomed. Anal. 2008, 48, 533–553. [Google Scholar] [CrossRef]
- Gama, A.R.; Saêta, V.P.; Gomes, R.P.; Vieira, J.D.G.; Barbosa, M.S.; Bataus, L.A.M.; Filho, A.R.G.; Coelho, C.J.; Carneiro, L.C. Differentiation of Environmental Bacteria Using Hyperspectral Imaging Technology And Multivariate Analysis. Int. J. Biotech Trends Technol. 2020, 10, 1–5. [Google Scholar] [CrossRef]
- Naumann, D. Infrared Spectroscopy in Microbiology. In MEYERS, Robert A. (Org.), Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Chichester, UK, 2006; p. a0117. [Google Scholar]
- Gowen, A.A.; Feng, Y.; Gaston, E.; Valdramidis, V. Recent applications of hyperspectral imaging in microbiology. Talanta 2015, 137, 43–54. [Google Scholar] [CrossRef]
- Sandasi, M.; Vermaak, I.; Chen, W.; Viljoen, A.M. Hyperspectral Imaging and Chemometric Modeling of Echinacea—A Novel Approach in the Quality Control of Herbal Medicines. Molecules 2014, 19, 13104–13121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, G.; Halig, L.; Wang, D.; Qin, X.; Chen, Z.G.; Fei, B. Spectral-spatial classification for noninvasive cancer detection using hyperspectral imaging. J. Biomed. Opt. 2014, 19, 106004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manley, M.; Du Toit, G.; Geladi, P. Tracking diffusion of conditioning water in single wheat kernels of different hardnesses by near infrared hyperspectral imaging. Anal. Chim. Acta 2011, 686, 64–75. [Google Scholar] [CrossRef]
- Williams, P.; Geladi, P.; Fox, G.; Manley, M. Maize kernel hardness classification by near infrared (NIR) hyperspectral imaging and multivariate data analysis. Anal. Chim. Acta 2009, 653, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Noiphung, J.; Laiwattanapaisal, W. Multifunctional Paper-Based Analytical Device for In Situ Cultivation and Screening of Escherichia coli Infections. Sci. Rep. 2019, 9, 1555. [Google Scholar] [CrossRef] [Green Version]
- van Soolingen, D.; De Haas, P.E.; Hermans, P.W.; Van Embden, J.D. [15] DNA Fingerprinting of mycobacterium tuberculosis. Methods Enzymol. 1994, 235, 196–205. [Google Scholar] [PubMed]
- Geladi, P. Chemometrics in spectroscopy. Part 1. Classical chemometrics. Spectrochim. Acta Part B At. Spectrosc. 2003, 58, 767–782. [Google Scholar] [CrossRef]
- Vidal, R.; Ma, Y.; Sastry, S.S.; Vidal, R.; Ma, Y.; Sastry, S.S. Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Reich, G. Near-infrared spectroscopy and imaging: Basic principles and pharnmaceutical applications. Adv. Drug Deliv. Rev. 2005, 57, 1109–1143. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard Normal Variate Transformation and De-Trending of Near-Infrared Diffuse Reflectance Spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Neto, E.D.A.; Guerrero, J.; Snyder, R.E.; Pereira, R.F.A.; de Freitas, M.D.F.N.; Silva-Santana, G.; Riley, L.W.; Aguiar-Alves, F. Genotypic distribution of Staphylococcus aureus colonizing children and adolescents in daycare centers, an outpatient clinic, and hospitals in a major Brazilian urban setting. Diagn. Microbiol. Infect. Dis. 2020, 97, 115058. [Google Scholar] [CrossRef]
- Esposito, S.; Terranova, L.; Macchini, F.; Bianchini, S.; Biffi, G.; Viganò, M.; Pelucchi, C.; Leva, E.; Principi, N. Staphylococcus aureus colonization and risk of surgical site infection in children undergoing clean elective surgery: A cohort study. Medicine 2018, 97, e11097. [Google Scholar] [CrossRef] [PubMed]
- Mostafavizadeh, K.; Davoodabadi, F.; Mobasherizadeh, S.; Shojaei, H.; Havaei, S.A.; Koushki, A.M.; Moghadasizadeh, Z.; Meidani, M.; Shirani, K. Nasal colonization in children with community acquired methicillin-resistant Staphylococcus aureus. Adv. Biomed. Res. 2016, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Chmielowiec-Korzeniowska, A.; Tymczyna, L.; Wlazło, Ł.; Nowakowicz-Dębek, B.; Trawińska, B. Staphylococcus aureus carriage state in healthy adult population and phenotypic and genotypic properties of isolated strains. Postep. Dermatol. I Alergol. 2020, 37, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Tan, J.; Eells, S.; Rieg, G.; Tagudar, G.; Miller, L. Body site colonization in patients with community-associated methicillin-resistant Staphylococcus aureus and other types of S. aureus skin infections. Clin. Microbiol. Infect. 2010, 16, 425–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brook, I. Role of methicillin-resistant Staphylococcus aureus in head and neck infections. J. Laryngol. Otol. 2009, 123, 1301–1307. [Google Scholar] [CrossRef]


| Microorganisms | Deposit Code |
|---|---|
| 1 Staphylococcus aureus | * |
| 2 Staphylococcus aureus | MW480543 |
| NCBI Sample | GenBank Register | Sample Test 1 Identity % | Sample Test 2 Identity % |
|---|---|---|---|
| Staphylococcus aureus strain S33 R 16S ribosomal RNA | NR_037007 | 92.44% | 99.69% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Messias, A.C.M.C.; Gama, A.R.; de Almeida Prado, L.S.; da Silva, P.A.N.; Galvão Filho, A.R.; Coelho, C.J.; Avelino, M.A.G.; Vieira, J.D.G.; Carneiro, L.C. Detection of Oxacillin/Cefoxitin Resistance in Staphylococcus aureus Present in Recurrent Tonsillitis. Microorganisms 2023, 11, 615. https://doi.org/10.3390/microorganisms11030615
Messias ACMC, Gama AR, de Almeida Prado LS, da Silva PAN, Galvão Filho AR, Coelho CJ, Avelino MAG, Vieira JDG, Carneiro LC. Detection of Oxacillin/Cefoxitin Resistance in Staphylococcus aureus Present in Recurrent Tonsillitis. Microorganisms. 2023; 11(3):615. https://doi.org/10.3390/microorganisms11030615
Chicago/Turabian StyleMessias, Aline Cristine Magalhães Costa, Aline Rodrigues Gama, Letícia Suriano de Almeida Prado, Paulo Alex Neves da Silva, Arlindo Rodrigues Galvão Filho, Clarimar José Coelho, Melissa Ameloti Gomes Avelino, José Daniel Gonçalves Vieira, and Lilian Carla Carneiro. 2023. "Detection of Oxacillin/Cefoxitin Resistance in Staphylococcus aureus Present in Recurrent Tonsillitis" Microorganisms 11, no. 3: 615. https://doi.org/10.3390/microorganisms11030615
APA StyleMessias, A. C. M. C., Gama, A. R., de Almeida Prado, L. S., da Silva, P. A. N., Galvão Filho, A. R., Coelho, C. J., Avelino, M. A. G., Vieira, J. D. G., & Carneiro, L. C. (2023). Detection of Oxacillin/Cefoxitin Resistance in Staphylococcus aureus Present in Recurrent Tonsillitis. Microorganisms, 11(3), 615. https://doi.org/10.3390/microorganisms11030615






