MicroRNAs as Biomarkers of Active Pulmonary TB Course
Abstract
:1. Introduction
2. Materials and Methods
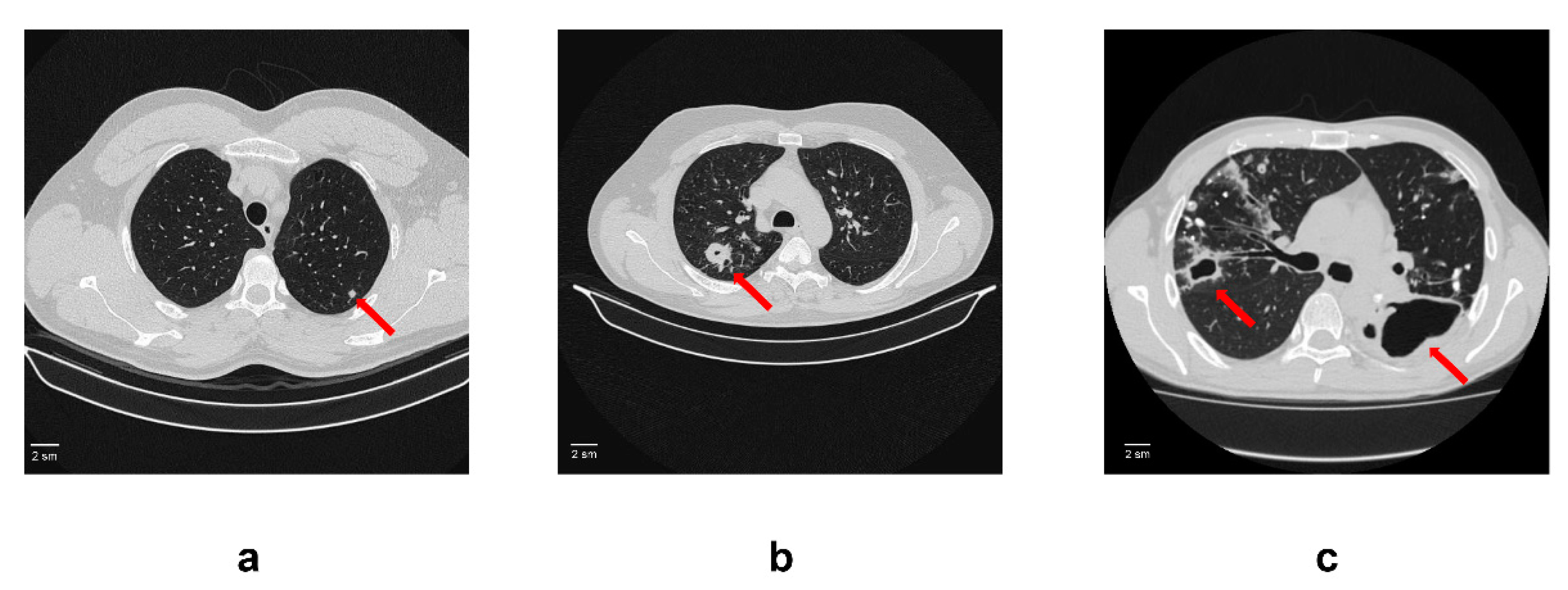
2.1. Patients and Serum Samples
2.2. Total RNA Extraction
2.3. miRNA PCR Array
2.4. Quantitative Real Time PCR
2.5. Statistical Analysis
2.6. Receiver Operating Characteristics (ROC) Curves
3. Results
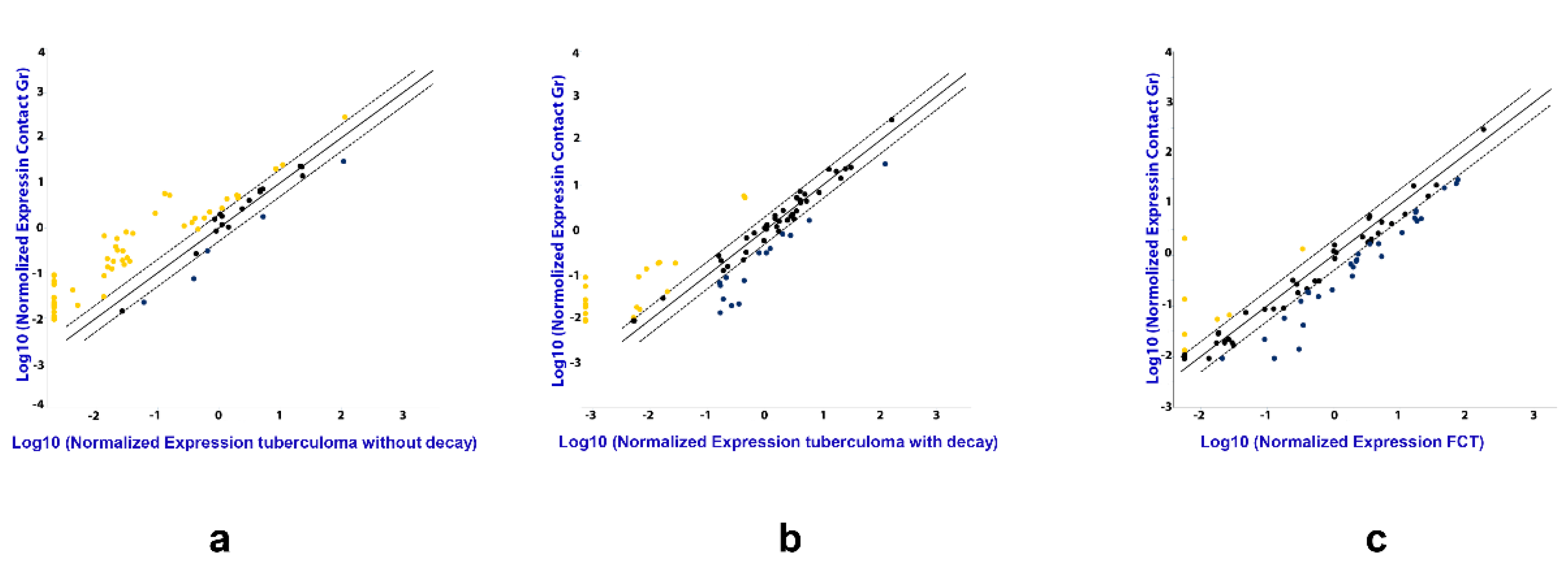
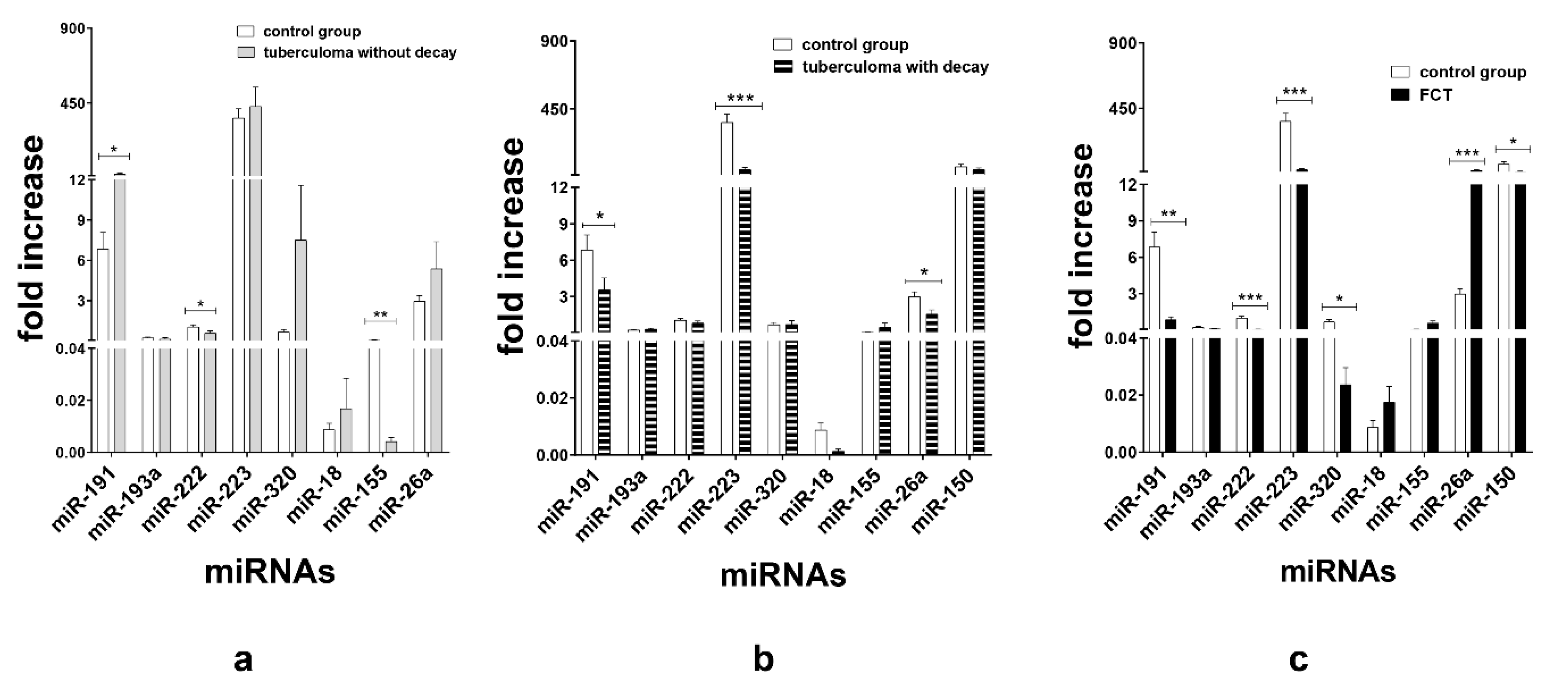
3.1. miRNA Expression Analysis
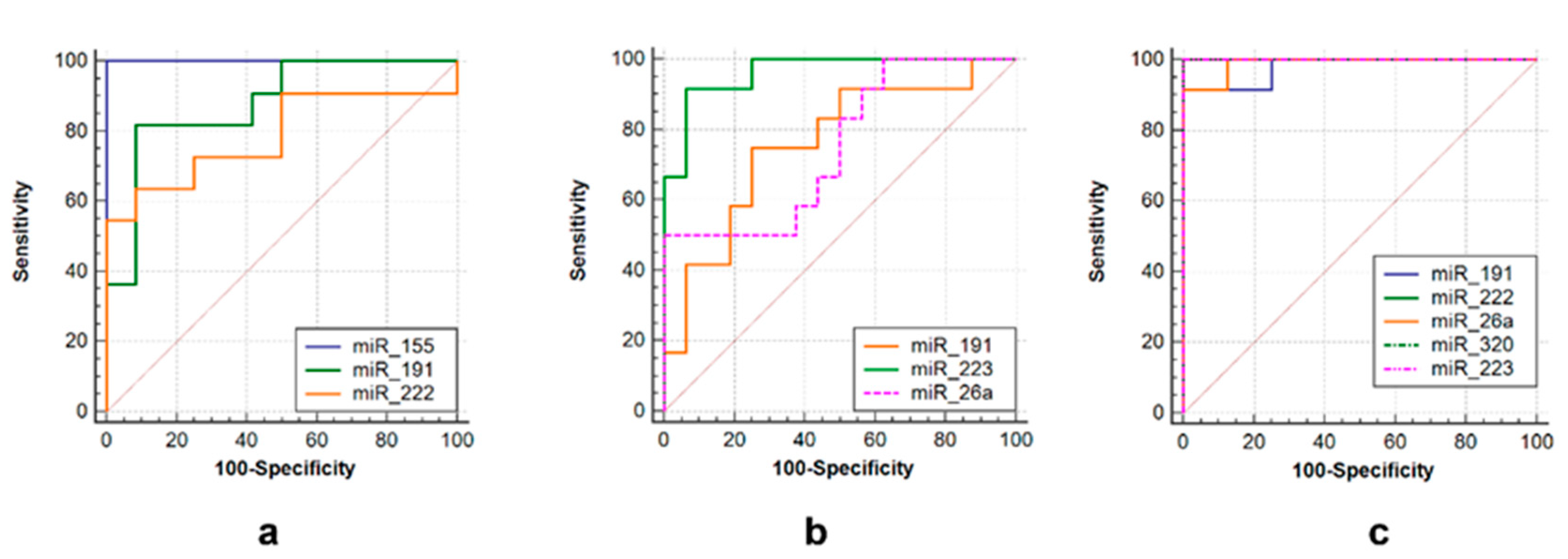
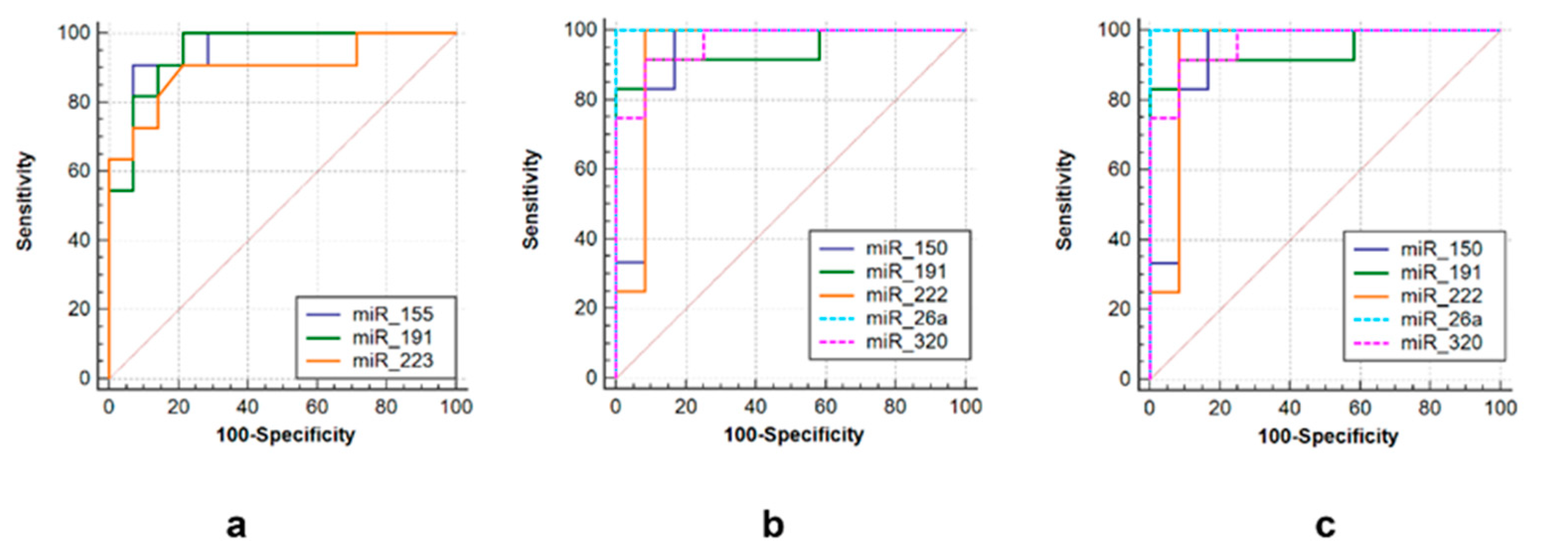
3.2. ROC Curve Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gengenbacher, M.; Kaufmann, S.H.E. Mycobacterium tuberculosis: Success through dormancy. FEMS Microbiol. Rev. 2012, 36, 514–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shilova, M.V. Tuberculosis classification. In Phthisiology: National Guidelines; Perelman, M.I., Ed.; GEOTAR-Media: Moscow, Russia, 2007; pp. 239–249. [Google Scholar]
- Bogadelnikova, I.V. Pulmonary tuberculosis. In Phthisiology: National Guidelines; Perelman, M.I., Ed.; GEOTAR-Media: Moscow, Russia, 2007; pp. 250–317. [Google Scholar]
- Subotic, D.; Yablonskiy, P.; Sulis, G.; Cordos, I.; Petrov, D.; Centis, R.; D′Ambrosio, L.; Sotgiu, G.; Migliori, G.B. Surgery and pleuropulmonary tuberculosis: A scientific literature review. J. Thorac. Dis. 2016, 8, E474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giller, D.B.; Giller, B.D.; Giller, G.V.; Shcherbakova, G.V.; Bizhanov, A.B.; Enilenis, I.I.; Glotov, A.A. Treatment of pulmonary tuberculosis: Past and present. Eur. J. Cardio-Thorac. Surg. 2018, 53, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Molnar, T.F. Tuberculosis: Mother of thoracic surgery then and now, past and prospectives: A review. J. Thorac. Dis. 2018, 10, S2628. [Google Scholar] [CrossRef] [PubMed]
- Harapan, H.; Fitra, F.; Ichsan, I.; Mulyadi, M.; Miotto, P.; Hasan, N.A.; Calado, M.; Cirillo, D.M. The roles of microRNAs on tuberculosis infection: Meaning or myth? Tuberculosis 2013, 93, 596–605. [Google Scholar] [CrossRef] [Green Version]
- Sinigaglia, A.; Peta, E.; Riccetti, S.; Venkateswaran, S.; Manganelli, R.; Barzon, L. Tuberculosis-associated microRNAs: From pathogenesis to disease biomarkers. Cells 2020, 9, 2160. [Google Scholar] [CrossRef]
- Kundu, M.; Basu, J. The role of microRNAs and Long non-coding RNAs in the regulation of the immune response to Mycobacterium tuberculosis Infection. Front. Immunol. 2021, 12, 687962. [Google Scholar] [CrossRef]
- Eremeev, V.V.; Evstifeev, V.V.; Shepelkova, G.S.; Ergeshova, A.E.; Bagirov, M.A. MicroRNA and tuberculosis. Russ. J. Infect. Immun. 2018, 8, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007, 17, 118–126. [Google Scholar] [CrossRef]
- de Araujo, L.S.; Ribeiro-Alves, M.; Leal-Calvo, T.; Leung, J.; Durán, V.; Samir, M.; Talbot, S.; Tallam, A.; Mello, F.C.Q.; Geffers, R.; et al. Reprogramming of small noncoding RNA populations in peripheral blood reveals host biomarkers for latent and active Mycobacterium tuberculosis infection. mBio 2019, 10, e01037-19. [Google Scholar] [CrossRef] [Green Version]
- Ndzi, E.N.; Nkenfou, C.N.; Mekue, L.M.; Zentilin, L.; Tamgue, O.; Pefura, E.W.Y.; Kuiaté, J.R.; Giacca, M.; Ndjolo, A. MicroRNA hsa-miR-29a-3p is a plasma biomarker for the differential diagnosis and monitoring of tuberculosis. Tuberculosis 2019, 114, 69–76. [Google Scholar] [CrossRef]
- Zhou, M.; Yu, G.; Yang, X.; Zhu, C.; Zhang, Z.; Zhan, X. Circulating microRNAs as biomarkers for the early diagnosis of childhood tuberculosis infection. Mol. Med. Rep. 2016, 13, 4620–4626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Yang, S.; Sun, G.; Tang, X.; Lu, S.; Neyrolles, O.; Gao, Q. Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PLoS ONE 2011, 6, e25832. [Google Scholar] [CrossRef] [PubMed]
- Slogotskaya, L.; Bogorodskaya, E.; Ivanova, D.; Sevostyanova, T. Comparative sensitivity of the test with tuberculosis recombinant allergen, containing ESAT6-CFP10 protein, and Mantoux test with 2 TU PPD-L in newly diagnosed tuberculosis children and adolescents in Moscow. PLoS ONE 2018, 13, e0208705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peltier, H.J.; Latham, G.J. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 2008, 14, 844–852. [Google Scholar] [CrossRef] [Green Version]
- Sumiya, R.; Nagasaka, S.; Ikeda, T.; Miyazaki, H. Solitary pleural tuberculoma diagnosed by thoracoscopic surgical resection. J. Surg. Case Rep. 2021, 2021, rjab408. [Google Scholar] [CrossRef]
- Iwai, H.; Funatogawa, K.; Matsumura, K.; Kato-Miyazawa, M.; Kirikae, F.; Kiga, K.; Sasakawa, C.; Miyoshi-Akiyama, T.; Kirikae, T. MicroRNA-155 knockout mice are susceptible to Mycobacterium tuberculosis infection. Tuberculosis 2015, 95, 246–250. [Google Scholar] [CrossRef]
- Rothchild, A.C.; Sissons, J.R.; Shafiani, S.; Plaisier, C.; Min, D.; Mai, D.; Gilchrist, M.; Peschon, J.; Larson, R.P.; Bergthaler, A.; et al. MiR-155–regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2016, 113, E6172–E6181. [Google Scholar] [CrossRef] [Green Version]
- Etna, M.P.; Sinigaglia, A.; Grassi, A.; Giacomini, E.; Romagnoli, A.; Pardini, M.; Severa, M.; Cruciani, M.; Rizzo, F.; Anastasiadou, E.; et al. Mycobacterium tuberculosis-induced miR-155 subverts autophagy by targeting ATG3 in human dendritic cells. PLoS Pathog. 2018, 14, e1006790. [Google Scholar] [CrossRef]
- Dorhoi, A.; Kaufmann, S.H. Perspectives on host adaptation in response to Mycobacterium tuberculosis: Modulation of inflammation. Semin. Immunol. 2014, 26, 533–542. [Google Scholar] [CrossRef]
- Hilda, J.N.; Das, S.; Tripathy, S.P.; Hanna, L.E. Role of neutrophils in tuberculosis: A bird’s eye view. Innate Immun. 2020, 26, 240–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorhoi, A.; Iannaccone, M.; Farinacci, M.; Faé, K.C.; Schreiber, J.; Moura-Alves, P.; Nouailles, G.; Mollenkopf, S.-J.; Oberbeck-Müller, D.; Jörg, S.; et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Investig. 2013, 123, 4836–4848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Morgan, M.J.; Choksi, S.; Zhang, Y.; Kim, Y.S.; Liu, Z.G. MicroRNAs modulate the noncanonical transcription factor NF-κB pathway by regulating expression of the kinase IKKα during macrophage differentiation. Nat. Immunol. 2010, 11, 799–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Wang, R.; Jiang, J.; Yang, B.; Cao, Z.; Cheng, X. MiR-223 is upregulated in monocytes from patients with tuberculosis and regulates function of monocyte-derived macrophages. Mol. Immunol. 2015, 67, 475–481. [Google Scholar] [CrossRef]
- Liu, F.; Chen, J.; Wang, P.; Li, H.; Zhou, Y.; Liu, H.; Liu, Z.; Zheng, R.; Wang, L.; Yang, H.; et al. MicroRNA-27a controls the intracellular survival of Mycobacterium tuberculosis by regulating calcium-associated autophagy. Nat. Commun. 2018, 9, 4295. [Google Scholar] [CrossRef] [Green Version]
- Ni, B.; Rajaram, M.V.; Lafuse, W.P.; Landes, M.B.; Schlesinger, L.S. Mycobacterium tuberculosis decreases human macrophage IFN-γ responsiveness through miR-132 and miR-26a. J. Immunol. 2014, 193, 4537–4547. [Google Scholar] [CrossRef] [Green Version]
- Sahu, S.K.; Kumar, M.; Chakraborty, S.; Banerjee, S.K.; Kumar, R.; Gupta, P.; Jana, K.; Gupta, U.D.; Ghosh, Z.; Kundu, M.; et al. MicroRNA26a (miR-26a)/KLF4 and CREB-C/EBP-regulate innate immune signaling, the polarization of macrophages and the tracking of Mycobacterium tuberculosis to lysosomes during infection. PLoS Pathog. 2017, 13, e1006410. [Google Scholar] [CrossRef] [Green Version]
- Xi, Y.; Formentini, A.; Chien, M.; Weir, D.B.; Russo, J.J.; Ju, J.; Kornmann, M.; Ju, J. Prognostic Values of microRNAs in Colorectal Cancer. Biomark. Insights 2006, 2, 113–121. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2134920/ (accessed on 26 February 2023). [CrossRef] [PubMed] [Green Version]
- Elyakim, E.; Sitbon, E.; Faerman, A.; Tabak, S.; Montia, E.; Belanis, L.; Dov, A.; Marcusson, A.G.; Bennett, C.F.; Chajut, A.; et al. Hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. 2010, 70, 8077–8087. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Su, S.; Long, J.; Mei, B.; Chen, Y. MicroRNA-191 targets N-deacetylase/N-sulfotransferase 1 and promotes cell growth in human gastric carcinoma cell line MGC803. Acta Biochim. Biophys. Sin. 2011, 43, 849–856. [Google Scholar] [CrossRef] [Green Version]
- Di Leva, G.; Piovan, C.; Gasparini, P.; Ngankeu, A.; Taccioli, C.; Briskin, D.; Cheung, D.G.; Bolon, B.; Anderlucci, L.; Alder, H.; et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013, 9, e1003311. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, N.; Kulshreshtha, R. miR-191: An emerging player in disease biology. Front. Genet. 2014, 5, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Cui, Y.; Wang, W.; Gu, J.; Guo, S.; Ma, K.; Luo, X. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia 2011, 13, 841–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, N.; Jinnin, M.; Kira-Etoh, T.; Makino, K.; Kajihara, I.; Makino, T.; Fukushima, S.; Inoue, Y.; Okamoto, Y.; Hasegawa, M.; et al. miR-150 down-regulation contributes to the constitutive type I collagen overexpression in scleroderma dermal fibroblasts via the induction of integrin β3. Am. J. Pathol. 2013, 182, 206–216. [Google Scholar] [CrossRef]
- Wang, D.; Sang, Y.; Sun, T.; Kong, P.; Zhang, L.; Dai, Y.; Cao, Y.; Tao, Z.; Liu, W. Emerging roles and mechanisms of microRNA-222-3p in human cancer. Int. J. Oncol. 2021, 58, 20. [Google Scholar] [CrossRef]
- Wang, X.; Liao, X.; Huang, K.; Zeng, X.; Liu, Z.; Zhou, X.; Yu, T.; Yang, C.; Yu, L.; Wang, Q.; et al. Clustered microRNAs hsa-miR-221-3p/hsa-miR-222-3p and their targeted genes might be prognostic predictors for hepatocellular carcinoma. J. Cancer 2019, 10, 2520–2533. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Zhao, Y.; Yin, Z.; Wang, D.W.; Chen, C. The role of miR-320 in glucose and lipid metabolism disorder-associated diseases. Int. J. Biol. Sci. 2021, 17, 402–416. [Google Scholar] [CrossRef]
- Zhuu, T.; Liu, H.; Su, L.; Xiong, X.; Wang, J.; Xiao, Y.; Zhu, Y.; Peng, Y.; Dawood, A.; Hu, C.; et al. MicroRNA-18b-5p Downregulation Favors Mycobacterium tuberculosis Clearance in Macrophages via HIF-1α by Promoting an Inflammatory Response. ACS Infect. Dis. 2021, 7, 800–810. [Google Scholar] [CrossRef]
- Braverman, J.; Sogi, K.M.; Benjamin, D.; Nomura, D.K.; Stanley, S.A. HIF-1α is an essential mediator of IFN-γ-Dependent immunity to Mycobacterium tuberculosis. J. Immunol. 2016, 197, 1287–1297. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shepelkova, G.S.; Evstifeev, V.V.; Tarasov, R.V.; Ergeshova, A.E.; Bagirov, M.A.; Yeremeev, V.V. MicroRNAs as Biomarkers of Active Pulmonary TB Course. Microorganisms 2023, 11, 626. https://doi.org/10.3390/microorganisms11030626
Shepelkova GS, Evstifeev VV, Tarasov RV, Ergeshova AE, Bagirov MA, Yeremeev VV. MicroRNAs as Biomarkers of Active Pulmonary TB Course. Microorganisms. 2023; 11(3):626. https://doi.org/10.3390/microorganisms11030626
Chicago/Turabian StyleShepelkova, Galina S., Vladimir V. Evstifeev, Ruslan V. Tarasov, Anush E. Ergeshova, Mamed A. Bagirov, and Vladimir V. Yeremeev. 2023. "MicroRNAs as Biomarkers of Active Pulmonary TB Course" Microorganisms 11, no. 3: 626. https://doi.org/10.3390/microorganisms11030626
APA StyleShepelkova, G. S., Evstifeev, V. V., Tarasov, R. V., Ergeshova, A. E., Bagirov, M. A., & Yeremeev, V. V. (2023). MicroRNAs as Biomarkers of Active Pulmonary TB Course. Microorganisms, 11(3), 626. https://doi.org/10.3390/microorganisms11030626






