Rosenbergiella meliponini D21B Isolated from Pollen Pots of the Australian Stingless Bee Tetragonula carbonaria
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Media Components
2.2. Microorganisms
2.3. Deposition of Rosenbergiella meliponini D21B
2.4. Isolation of Microorganisms from T. carbonaria
2.5. Cultivation of Microorganisms from T. carbonaria
2.6. Genome Sequencing and Genome Assembly
2.7. Annotation of the R. meliponini D21B Genome
2.8. Genomic DNA Isolation
2.9. 16S rDNA Amplification by PCR
2.10. Phylogenetic Analysis
2.11. Physiological and Biochemical Characterization of R. meliponini D21B
2.12. Electron Microscopy of R. meliponini D21B
2.13. Fatty Acid Methyl Ester (FAME) Profile of Hydrolyzed Lipids from R. meliponini D21B
2.14. Fatty Acid Trimethylsilyl Ester Profile of Hydrolyzed Lipids from R. meliponini D21B
2.15. Spent Liquid Medium Extractions for the Detection of Lipophilic Secondary Metabolites
2.16. Collection of Volatiles from R. meliponini D21B
2.17. Quantification of 2-Phenylethanol and 2-Phenylacetic Acid
2.18. Antibiotic Resistance of the Rosenbergiella Isolates
3. Results
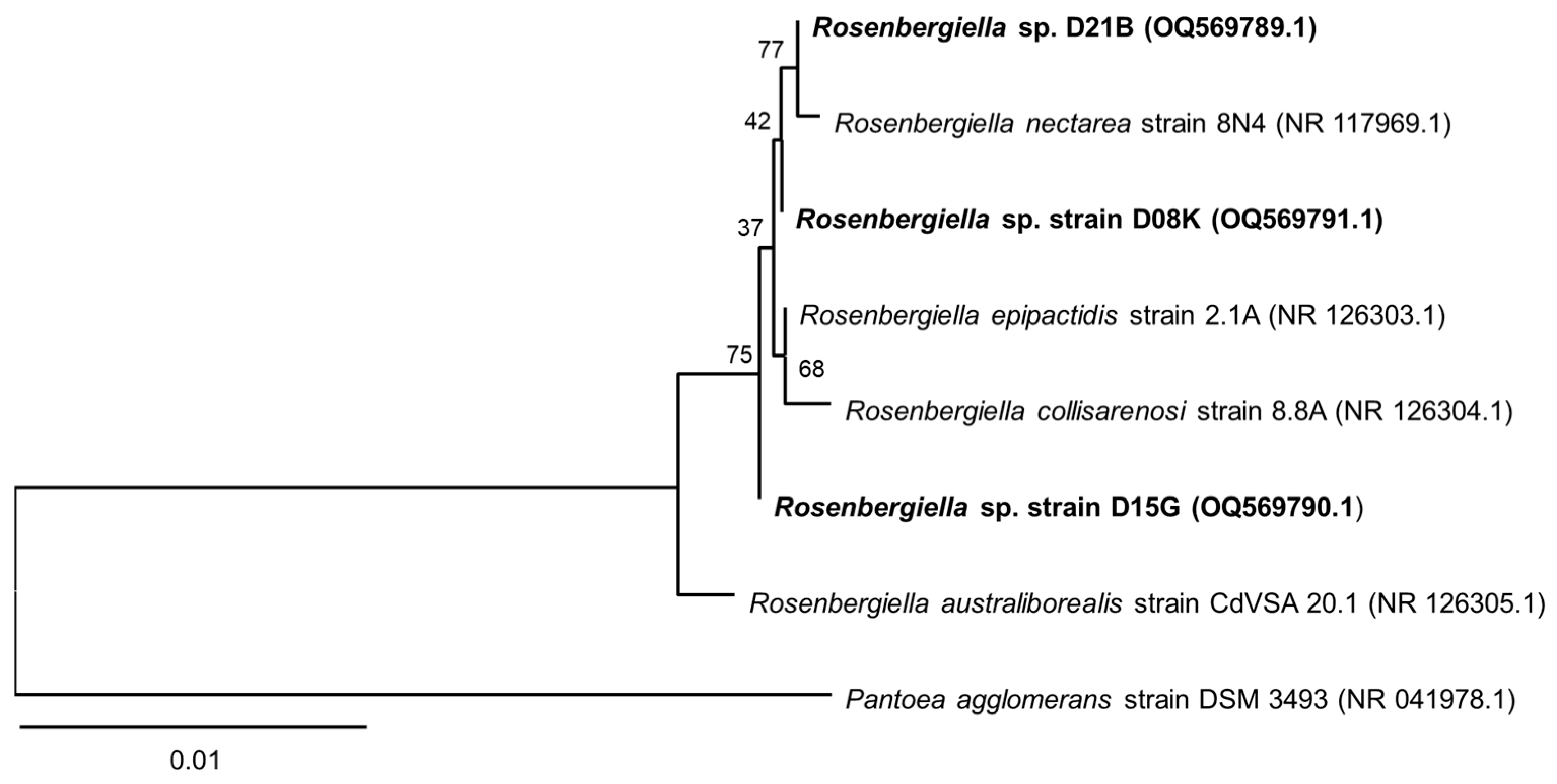
3.1. 16S rRNA Sequence Analysis of the Isolates from T. carbonaria
3.2. Genome of R. meliponini D21B
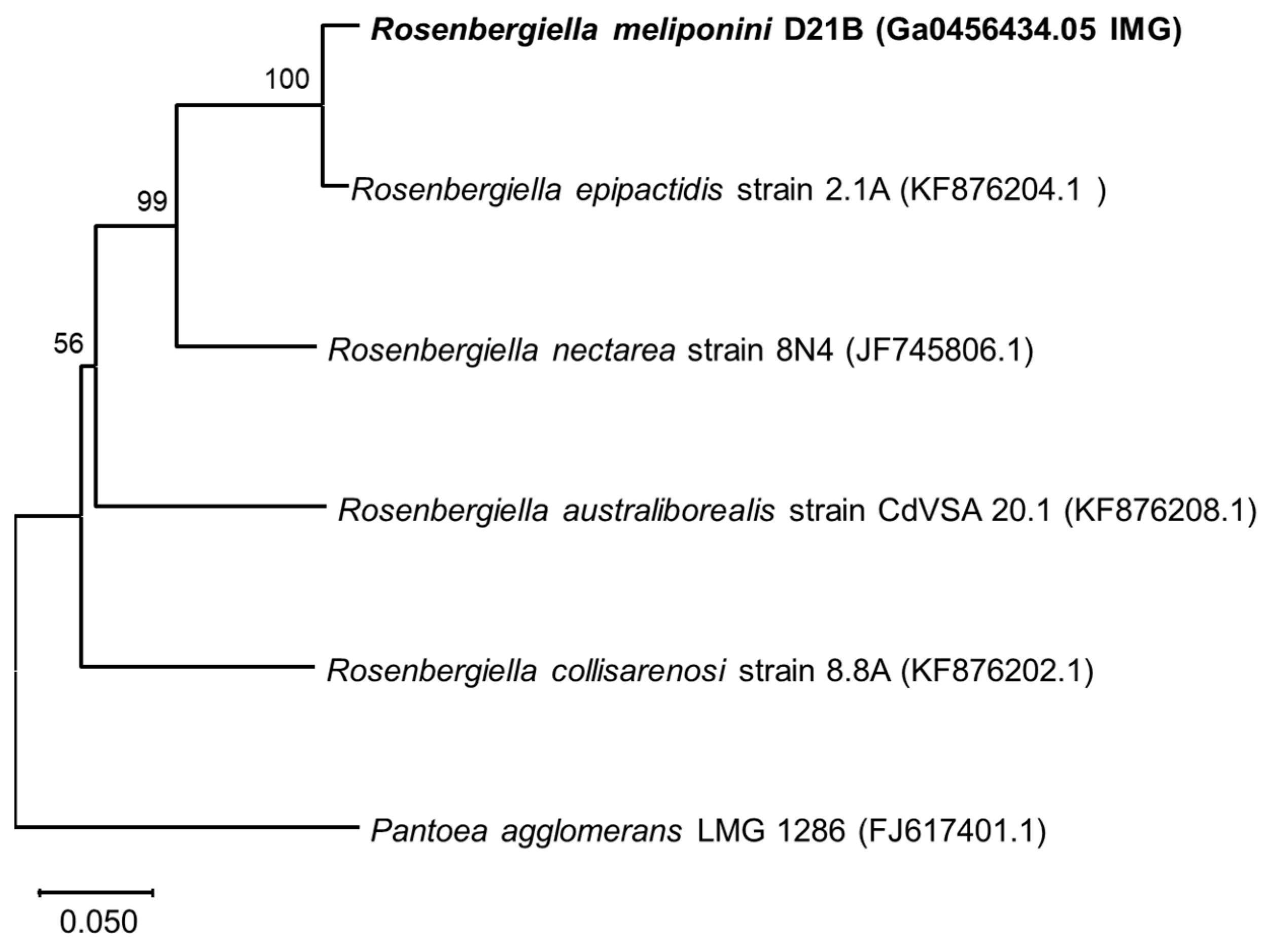
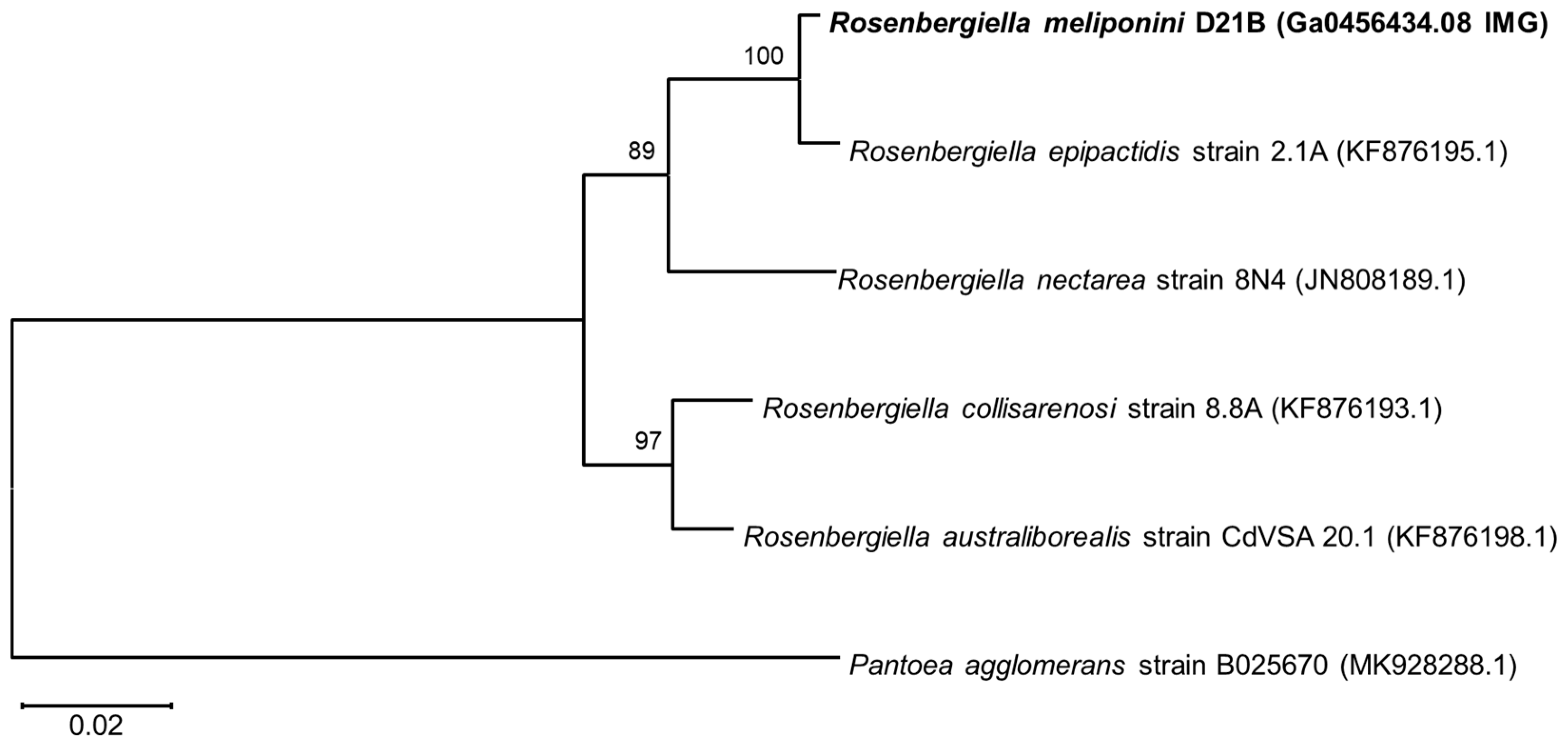
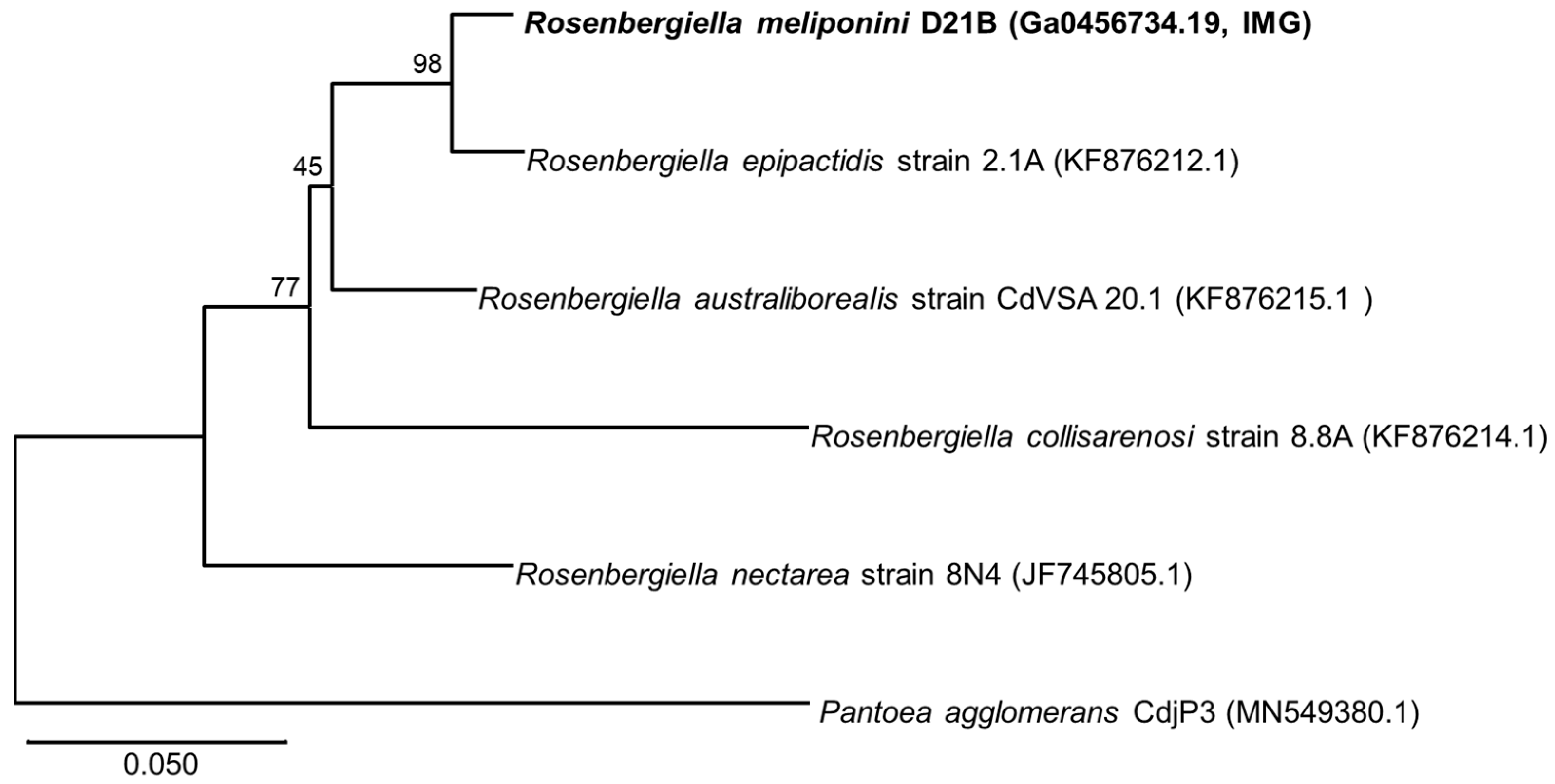
3.3. Phylogenetic Comparison of R. meliponini D21B and R. epipactidis 2.1A
3.4. Analysis of Secondary Metabolite Biosynthetic Gene Clusters from R. meliponini D21B
3.5. Physiological and Biochemical Characterization of R. meliponini D21B
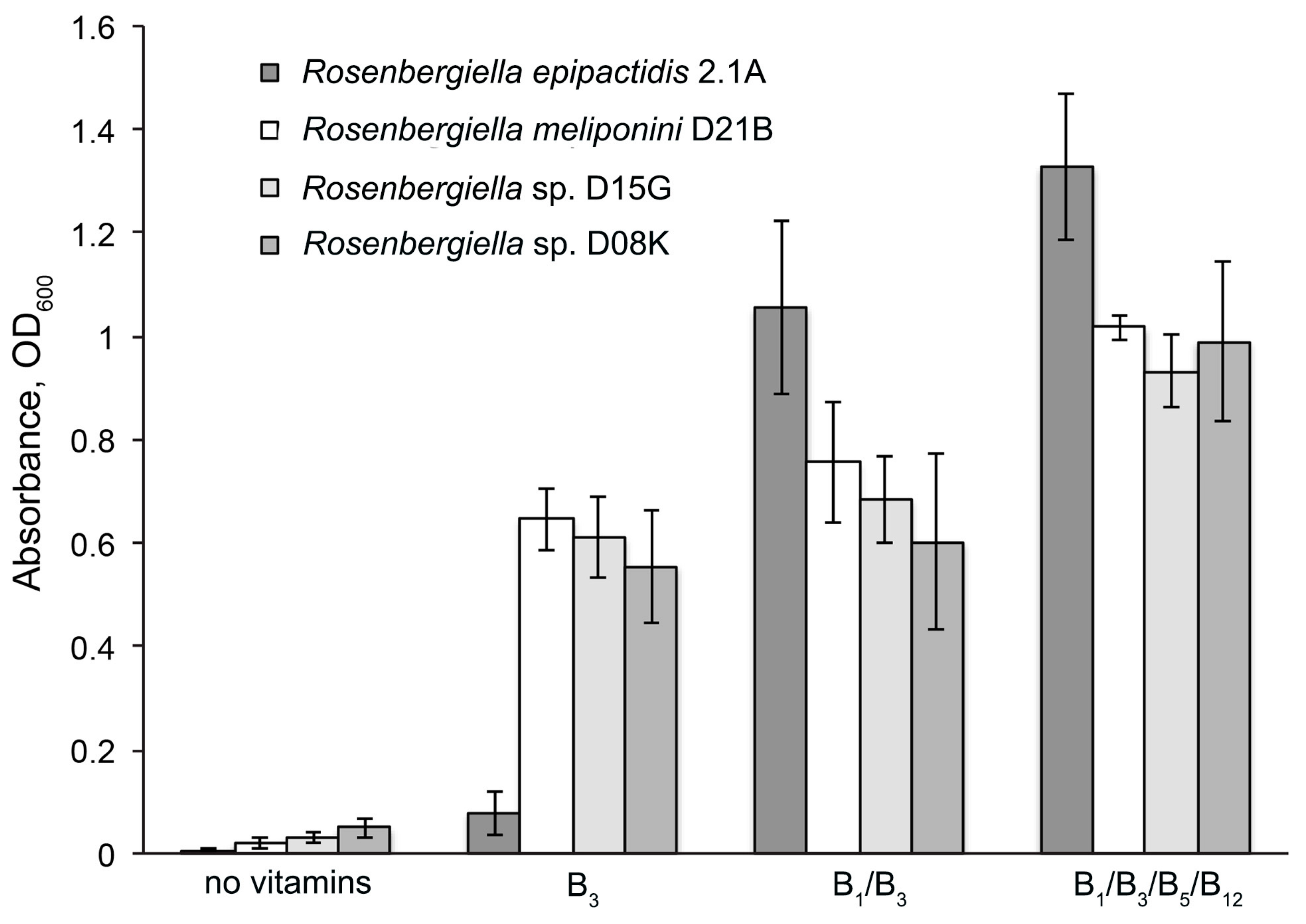
3.6. Minimal Growth Requirements of Rosenbergiella
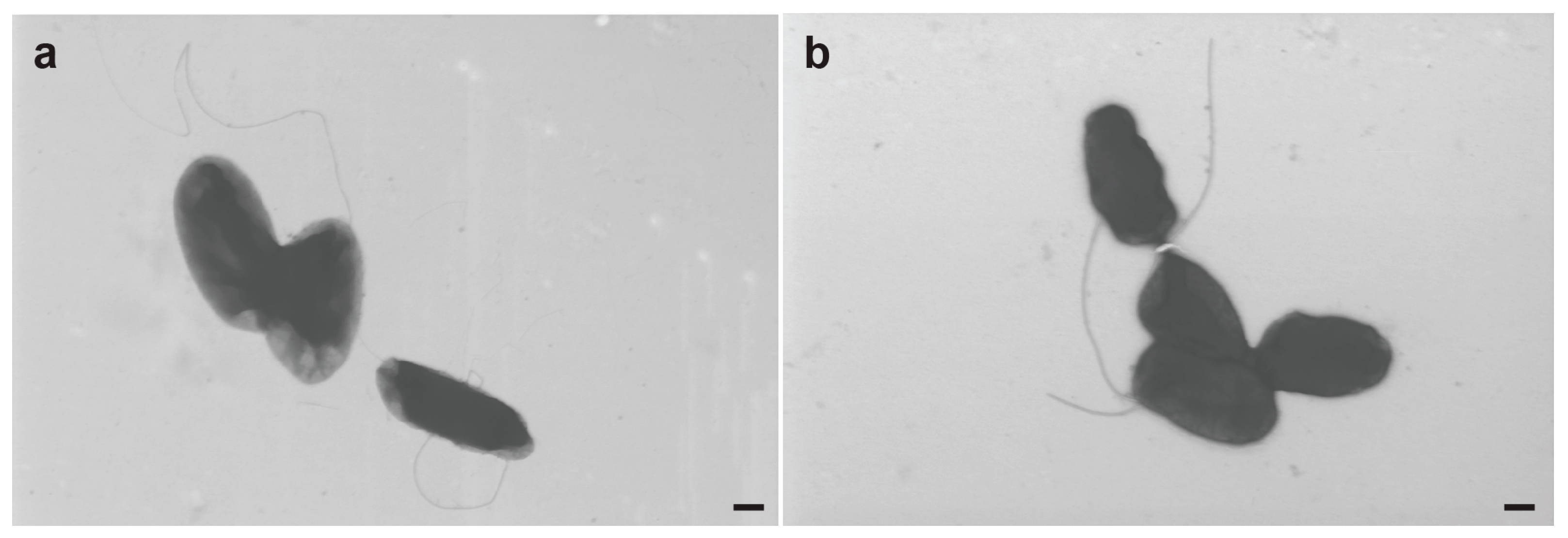
3.7. Electron Microscopy of R. meliponini D21B
3.8. Fatty Acid Profile of Hydrolyzed Lipids
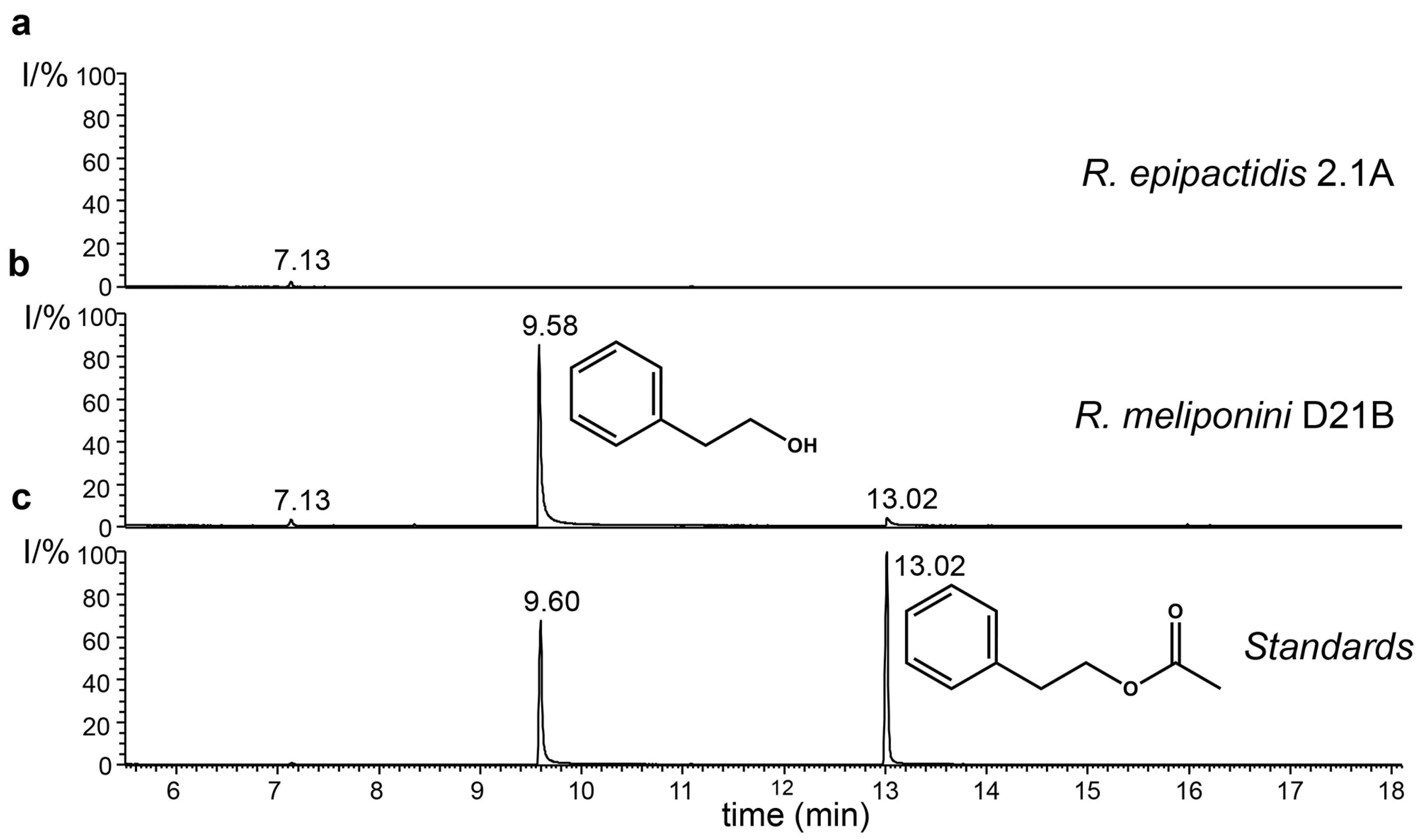
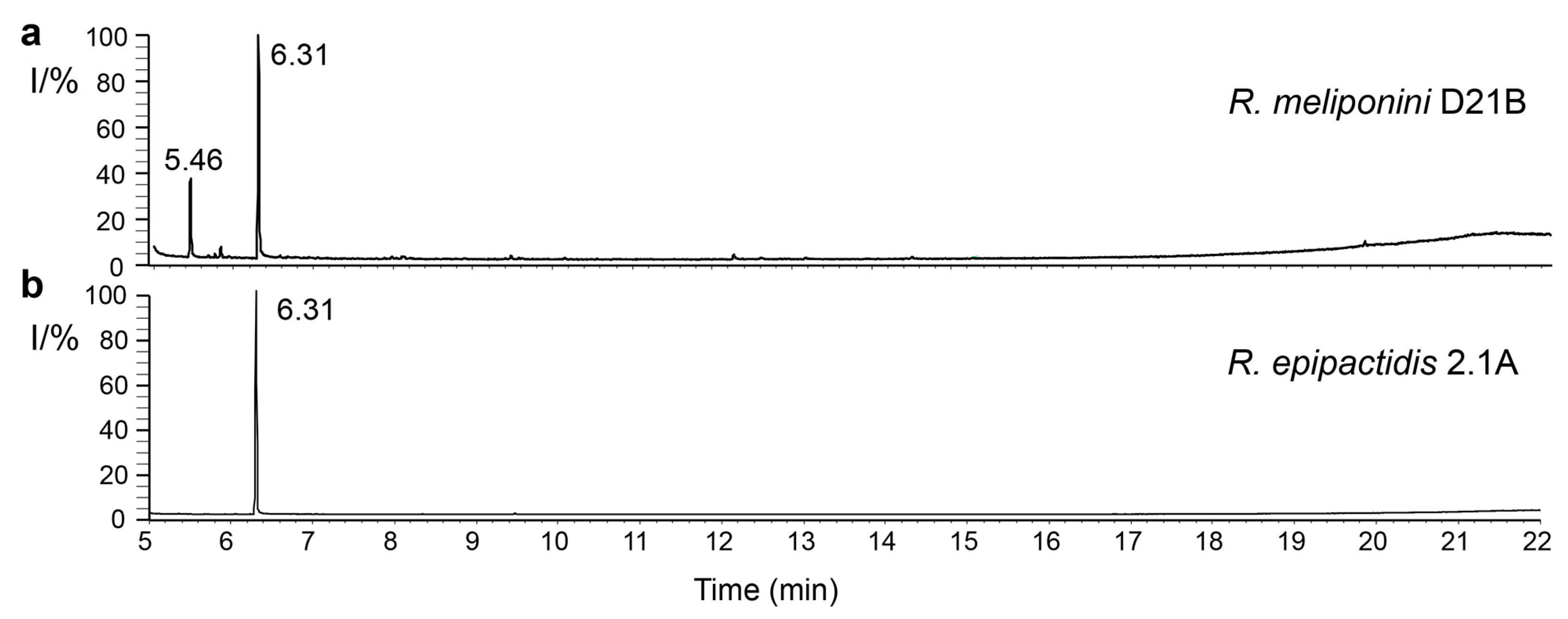
3.9. Production of 2-Phenylethanol by the Rosenbergiella Isolates from T. carbonaria
3.10. Analysis of spent medium of R. epipactidis 2.1A and R. meliponini D21B
3.11. Quantification of 2-Phenylethanol and 2-Phenylacetic Acid Production by Rosenbergiella
3.12. Antibiotic Resistance of Rosenbergiella Strains
4. Discussion
4.1. Possible Symbiotic Benefits of Rosenbergiella for Bees
4.2. Amino Acid and Vitamin Synthesis
4.3. Digestion of Food
4.4. Ecological Role of Secondary Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, Q.-H.; Miao, C.-H.; Chen, Y.-F.; Dong, Z.-X.; Cao, Z.; Liao, S.-Q.; Wang, J.-X.; Wang, Z.-W.; Guo, J. The composition of bacteria in gut and beebread of stingless bees (Apidae: Meliponini) from tropics Yunnan, China. Antonie Van Leeuwenhoek 2021, 114, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, W.-Z.; Xu, H.; Wang, Z.-W.; He, S.-Y. Bacillus in the guts of honey bees (Apis mellifera; Hymenoptera: Apidae) mediate changes in amylase values. EJE 2015, 112, 619–624. [Google Scholar] [CrossRef]
- Du Rand, E.E.; Stutzer, C.; Human, H.; Pirk, C.W.W.; Nicolson, S.W. Antibiotic treatment impairs protein digestion in the honeybee, Apis mellifera. Apidologie 2020, 51, 94–106. [Google Scholar] [CrossRef]
- Kešnerová, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef] [PubMed]
- Paludo, C.R.; Ruzzini, A.C.; Silva-Junior, E.A.; Pishchany, G.; Currie, C.R.; Nascimento, F.S.; Kolter, R.G.; Clardy, J.; Pupo, M.T. Whole-Genome Sequence of Bacillus sp. SDLI1, Isolated from the social bee Scaptotrigona depilis. Genome Announc. 2016, 4, e00174-116. [Google Scholar] [CrossRef] [PubMed]
- Vásquez, A.; Olofsson, T.C. The lactic acid bacteria involved in the production of bee pollen and bee bread. J. Apic. Res. 2009, 48, 189–195. [Google Scholar] [CrossRef]
- Motta, E.; Moran, N. Impact of Glyphosate on the Honey Bee Gut Microbiota: Effects of Intensity, Duration, and Timing of Exposure. mSystems 2020, 5, e00268-00220. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef]
- Zulkhairi Amin, F.A.; Sabri, S.; Ismail, M.; Chan, K.W.; Ismail, N.; Mohd Esa, N.; Mohd Lila, M.A.; Zawawi, N. Probiotic Properties of Bacillus Strains Isolated from Stingless Bee (Heterotrigona itama) Honey Collected across Malaysia. Int. J. Environ. Res. Public Health 2019, 17, 278. [Google Scholar] [CrossRef]
- Wang, X.; Hu, W.; Zhu, L.; Yang, Q. Bacillus subtilis and surfactin inhibit the transmissible gastroenteritis virus from entering the intestinal epithelial cells. Biosci. Rep. 2017, 37, 82. [Google Scholar] [CrossRef] [PubMed]
- Sabaté, D.C.; Audisio, M.C. Inhibitory activity of surfactin, produced by different Bacillus subtilis subsp. subtilis strains, against Listeria monocytogenes sensitive and bacteriocin-resistant strains. Microbiol. Res. 2013, 168, 125–129. [Google Scholar] [CrossRef]
- Sabaté, D.C.; Carrillo, L.; Audisio, M.C. Inhibition of Paenibacillus larvae and Ascosphaera apis by Bacillus subtilis isolated from honeybee gut and honey samples. Res. Microbiol. 2009, 160, 193–199. [Google Scholar] [CrossRef]
- Baffoni, L.; Gaggìa, F.; Alberoni, D.; Cabbri, R.; Nanetti, A.; Biavati, B.; Di Gioia, D. Effect of dietary supplementation of Bifidobacterium and Lactobacillus strains in Apis mellifera L. against Nosema ceranae. Benef. Microbes 2016, 7, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Menezes, C.; Vollet-Neto, A.; Contrera, F.A.F.L.; Venturieri, G.C.; Imperatriz-Fonseca, V.L. The Role of Useful Microorganisms to Stingless Bees and Stingless Beekeeping. In Pot-Honey: A Legacy of Stingless Bees; Vit, P., Pedro, S.R.M., Roubik, D., Eds.; Springer: New York, NY, USA, 2013; pp. 153–171. [Google Scholar]
- Brodschneider, R.; Crailsheim, K. Nutrition and health in honey bees. Apidologie 2010, 41, 278–294. [Google Scholar] [CrossRef]
- Lozo, J.; Berić, T.; Terzić-Vidojević, A.; Stanković, S.; Fira, D.; Stanisavljević, L. Microbiota associated with pollen, bee bread, larvae and adults of solitary bee Osmia cornuta (Hymenoptera: Megachilidae). Bull. Entomol. Res. 2015, 105, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Dharampal, P.S.; Diaz-Garcia, L.; Haase, M.A.B.; Zalapa, J.; Currie, C.R.; Hittinger, C.T.; Steffan, S.A. Microbial Diversity Associated with the Pollen Stores of Captive-Bred Bumble Bee Colonies. Insects 2020, 11, 250. [Google Scholar] [CrossRef]
- Gilliam, M.; Roubik, D.W.; Lorenz, B.J. Microorganisms associated with pollen, honey, and brood provisions in the nest of a stingless bee, Melipona fasciata. Apidologie 1990, 21, 89–97. [Google Scholar] [CrossRef]
- Halcroft, M.; Spooner-Hart, R.; Dollin, L.A. Australian Stingless Bees. In Pot-Honey: A Legacy of Stingless Bees; Vit, P., Pedro, S.R.M., Roubik, D., Eds.; Springer: New York, NY, USA, 2013; pp. 35–72. [Google Scholar]
- Shanks, J.L.; Haigh, A.M.; Riegler, M.; Spooner-Hart, R.N. First confirmed report of a bacterial brood disease in stingless bees. J. Invertebr. Pathol. 2017, 144, 7–10. [Google Scholar] [CrossRef]
- Forsgren, E.; Locke, B.; Sircoulomb, F.; Schäfer, M.O. Bacterial Diseases in Honeybees. Curr. Clin. Microbiol. Rep. 2018, 5, 18–25. [Google Scholar] [CrossRef]
- McMenamin, A.J.; Genersch, E. Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 2015, 8, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Jensen, A.B.; Aronstein, K.; Flores, J.M.; Vojvodic, S.; Palacio, M.A.; Spivak, M. Standard methods for fungal brood disease research. J. Apic. Res. 2013, 52, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Fries, I. Nosema ceranae in European honey bees (Apis mellifera). J. Invertebrate Pathol. 2010, 103, S73–S79. [Google Scholar] [CrossRef] [PubMed]
- Chantawannakul, P.; de Guzman, L.I.; Li, J.; Williams, G.R. Parasites, pathogens, and pests of honeybees in Asia. Apidologie 2016, 47, 301–324. [Google Scholar] [CrossRef]
- Utkina, I. Declining Bee Populations Pose Threat to Global Food Security and Nutrition. Available online: https://www.fao.org/news/story/en/item/1194910/icode/#:~:text=%22The%20absence%20of%20bees%20and,sustainable%20food%20policies%20and%20systems.%22 (accessed on 20 March 2020).
- Gordon, R.E.; Smith, N.R.; Pang, C.H.-N.; Haynes, W.C. The Genus Bacillus; Agricultural Research Service, U.S. Dept. of Agriculture: Washington, DC, USA, 1973; p. 283. [Google Scholar]
- Thurnheer, T.; Cook, A.M.; Leisinger, T. Co-culture of defined bacteria to degrade seven sulfonated aromatic compounds: Efficiency, rates adn phenotypic variations. Appl. Microbiol. Biotechnol. 1988, 29, 605–609. [Google Scholar] [CrossRef]
- Pfennig, N. Rhodocyclus purpureus gen. nov. and sp. nov., a Ring-Shaped, Vitamin B12-Requiring Member of the Family Rhodospirillaceae. Int. J. Syst. Evol. Microbiol. 1978, 28, 283–288. [Google Scholar] [CrossRef]
- Marmont, L.S.; Whitfield, G.B.; Rich, J.D.; Yip, P.; Giesbrecht, L.B.; Stremick, C.A.; Whitney, J.C.; Parsek, M.R.; Harrison, J.J.; Howell, P.L. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J. Biol. Chem. 2017, 292, 19411–19422. [Google Scholar] [CrossRef]
- Markowitz, V.M.; Mavromatis, K.; Ivanova, N.N.; Chen, I.M.; Chu, K.; Kyrpides, N.C. IMG ER: A system for microbial genome annotation expert review and curation. Bioinformatics 2009, 25, 2271–2278. [Google Scholar] [CrossRef]
- Wright, M.H.; Adelskov, J.; Greene, A.C. Bacterial DNA Extraction Using Individual Enzymes and Phenol/Chloroform Separation. J. Microbiol. Biol. Educ. 2017, 18, 18.12.48. [Google Scholar] [CrossRef]
- Eden, P.A.; Schmidt, T.M.; Blakemore, R.P.; Pace, N.R. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 1991, 41, 324–325. [Google Scholar] [CrossRef]
- Jiang, H.; Dong, H.; Zhang, G.; Yu, B.; Chapman, L.R.; Fields, M.W. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl. Environ. Microbiol. 2006, 72, 3832–3845. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef]
- Shields, P.; Cathcart, L. Oxidase Test Protocol; American Society for Microbiology. Available online: http://www.microbelibrary.org/library/laboratory-test/3229-oxidase-test-protocol (accessed on 2 June 2019).
- Taylor, W.I.; Achanzar, D. Catalase test as an aid to the identification of Enterobacteriaceae. Appl. Microbiol. 1972, 24, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Laviad-Shitrit, S.; Izhaki, I.; Whitman, W.B.; Shapiro, N.; Woyke, T.; Kyrpides, N.C.; Halpern, M. Draft genome of Rosenbergiella nectarea strain 8N4(T) provides insights into the potential role of this species in its plant host. PeerJ 2020, 8, e8822. [Google Scholar] [CrossRef] [PubMed]
- Halpern, M.; Fridman, S.; Atamna-Ismaeel, N.; Izhaki, I. Rosenbergiella nectarea gen. nov., sp. nov., in the family Enterobacteriaceae, isolated from floral nectar. Int. J. Syst. Evol. Microbiol. 2013, 63, 4259–4265. [Google Scholar] [CrossRef]
- Sasser, M. Bacterial Identification by Gas Chromatographic Analysis of Fatty Acid Methy Esters (GC-FAME). Available online: http://midi-inc.com/pdf/MIS_Technote_101.pdf (accessed on 19 March 2021).
- Spiteller, D.; Boland, W. N-(15,16-Dihydroxylinoleoyl)-glutamine and N-(15,16-epoxylinoleoyl)-glutamine isolated from oral secretions of lepidopteran larvae. Tetrahedron 2003, 59, 135–139. [Google Scholar] [CrossRef]
- Grob, K.; Zürcher, F. Stripping of trace organic substances from water: Equipment and procedure. J. Chrom. A 1976, 117, 285–294. [Google Scholar] [CrossRef]
- Sulavik, M.C.; Houseweart, C.; Cramer, C.; Jiwani, N.; Murgolo, N.; Greene, J.; DiDomenico, B.; Shaw, K.J.; Miller, G.H.; Hare, R.; et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 2001, 45, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Tauch, A.; Kirchner, O.; Löffler, B.; Götker, S.; Pühler, A.; Kalinowski, J. Efficient Electrotransformation of Corynebacterium diphtheriae with a Mini-Replicon Derived from the Corynebacterium glutamicum Plasmid pGA1. Curr. Microbiol. 2002, 45, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C.; Wolfson, J.S.; McHugh, G.L.; Swartz, M.D.; Tung, C.; Swartz, M.N. Elimination of plasmid pMG110 from Escherichia coli by novobiocin and other inhibitors of DNA gyrase. Antimicrob. Agents Chemother. 1984, 25, 586–590. [Google Scholar] [CrossRef]
- Lenaerts, M.; Alvarez-Pérez, S.; de Vega, C.; Van Assche, A.; Johnson, S.D.; Willems, K.A.; Herrera, C.M.; Jacquemyn, H.; Lievens, B. Rosenbergiella australoborealis sp. nov., Rosenbergiella collisarenosi sp. nov. and Rosenbergiella epipactidis sp. nov., three novel bacterial species isolated from floral nectar. Syst. Appl. Microbiol. 2014, 37, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Perez, S.; de Vega, C.; Vanoirbeek, K.; Tsuji, K.; Jacquemyn, H.; Fukami, T.; Michiels, C.; Lievens, B. Phylogenomic analysis of the genus Rosenbergiella and description of Rosenbergiella gaditana sp. nov., Rosenbergiella metrosideri sp. nov., Rosenbergiella epipactidis subsp. epipactidis subsp. nov., Rosenbergiella epipactidis subsp. californiensis subsp. nov., Rosenbergiella epipactidis subsp. japonicus subsp. nov., Rosenbergiella nectarea subsp. nectarea subsp. nov. and Rosenbergiella nectarea subsp. apis subsp. nov., isolated from floral nectar and insects. Int. J. Syst. Evol. Microbiol. 2023, 73, 005777. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Choi, O.; Kang, B.; Lee, Y.; Lee, Y.; Kim, J.-H. Pantoea ananatis carotenoid production confers toxoflavin tolerance and is regulated by Hfq-controlled quorum sensing. MicrobiologyOpen 2021, 10, e1143. [Google Scholar] [CrossRef]
- Barona-Gómez, F.; Wong, U.; Giannakopulos, A.E.; Derrick, P.J.; Challis, G.L. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 2004, 126, 16282–16283. [Google Scholar] [CrossRef] [PubMed]
- Smits, T.H.; Duffy, B. Genomics of iron acquisition in the plant pathogen Erwinia amylovora: Insights in the biosynthetic pathway of the siderophore desferrioxamine E. Arch. Microbiol. 2011, 193, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Klinman, J.P. Biogenesis of the peptide-derived redox cofactor pyrroloquinoline quinone. Curr. Opin. Chem. Biol. 2020, 59, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial Peptides from Bacterial Origin: Overview of Their Biology and Their Impact against Multidrug-Resistant Bacteria. Microorganisms 2020, 8, 693. [Google Scholar] [CrossRef]
- Núñez-Montero, K.; Quezada-Solís, D.; Khalil, Z.G.; Capon, R.J.; Andreote, F.D.; Barrientos, L. Genomic and Metabolomic Analysis of Antarctic Bacteria Revealed Culture and Elicitation Conditions for the Production of Antimicrobial Compounds. Biomolecules 2020, 10, 673. [Google Scholar] [CrossRef]
- Govan, V.A.; Allsopp, M.H.; Davison, S. A PCR Detection Method for Rapid Identification of Paenibacillus larvae. Appl. Environ. Microbiol. 1999, 65, 2243–2245. [Google Scholar] [CrossRef]
- Atlas, R.M. (Ed.) Handbook of Microbiological Media; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Zucko, J.; Dunlap, W.C.; Shick, J.M.; Cullum, J.; Cercelet, F.; Amin, B.; Hammen, L.; Lau, T.; Williams, J.; Hranueli, D.; et al. Global genome analysis of the shikimic acid pathway reveals greater gene loss in host-associated than in free-living bacteria. BMC Genomics 2010, 11, 628. [Google Scholar] [CrossRef]
- Price, M.N.; Zane, G.M.; Kuehl, J.V.; Melnyk, R.A.; Wall, J.D.; Deutschbauer, A.M.; Arkin, A.P. Filling gaps in bacterial amino acid biosynthesis pathways with high-throughput genetics. PLoS Genet. 2018, 14, e1007147. [Google Scholar] [CrossRef]
- Liu, J.; Bai, Y.; Fan, T.P.; Zheng, X.; Cai, Y. Unveiling the Multipath Biosynthesis Mechanism of 2-Phenylethanol in Proteus mirabilis. J. Agric. Food Chem. 2020, 68, 7684–7690. [Google Scholar] [CrossRef]
- Ambika Manirajan, B.; Ratering, S.; Rusch, V.; Schwiertz, A.; Geissler-Plaum, R.; Cardinale, M.; Schnell, S. Bacterial microbiota associated with flower pollen is influenced by pollination type, and shows a high degree of diversity and species-specificity. Environ. Microbiol. 2016, 18, 5161–5174. [Google Scholar] [CrossRef] [PubMed]
- Manirajan, B.A.; Maisinger, C.; Ratering, S.; Rusch, V.; Schwiertz, A.; Cardinale, M.; Schnell, S. Diversity, specificity, co-occurrence and hub taxa of the bacterial–fungal pollen microbiome. FEMS Microbiol. Ecol. 2018, 94, fiy112. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.A.; Brettell, L.E.; Liu, H.; Nacko, S.; Spooner-Hart, R.; Riegler, M.; Cook, J.M. Temporal changes in the microbiome of stingless bee foragers following colony relocation. FEMS Microbiol. Ecol. 2020, 97, fiaa236. [Google Scholar] [CrossRef] [PubMed]
- Donkersley, P.; Rhodes, G.; Pickup, R.W.; Jones, K.C.; Wilson, K. Bacterial communities associated with honeybee food stores are correlated with land use. Ecol. Evol. 2018, 8, 4743–4756. [Google Scholar] [CrossRef]
- Disayathanoowat, T.; Li, H.; Supapimon, N.; Suwannarach, N.; Lumyong, S.; Chantawannakul, P.; Guo, J. Different Dynamics of Bacterial and Fungal Communities in Hive-Stored Bee Bread and Their Possible Roles: A Case Study from Two Commercial Honey Bees in China. Microorganisms 2020, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Graystock, P.; Rehan, S.M.; McFrederick, Q.S. Hunting for healthy microbiomes: Determining the core microbiomes of Ceratina, Megalopta, and Apis bees and how they associate with microbes in bee collected pollen. Conserv. Genet. 2017, 18, 701–711. [Google Scholar] [CrossRef]
- Praet, J.; Parmentier, A.; Schmid-Hempel, R.; Meeus, I.; Smagghe, G.; Vandamme, P. Large-scale cultivation of the bumblebee gut microbiota reveals an underestimated bacterial species diversity capable of pathogen inhibition. Environ. Microbiol. 2018, 20, 214–227. [Google Scholar] [CrossRef]
- Russell, K.A.; McFrederick, Q.S. Elevated Temperature May Affect Nectar Microbes, Nectar Sugars, and Bumble Bee Foraging Preference. Microb. Ecol. 2021, 84, 473–482. [Google Scholar] [CrossRef]
- Pozo, M.I.; Mariën, T.; van Kemenade, G.; Wäckers, F.; Jacquemyn, H. Effects of pollen and nectar inoculation by yeasts, bacteria or both on bumblebee colony development. Oecologia 2021, 195, 689–703. [Google Scholar] [CrossRef]
- Tahiliani, A.G.; Beinlich, C.J. Pantothenic acid in health and disease. Vitam. Horm. 1991, 46, 165–228. [Google Scholar] [CrossRef]
- Krehl, W.A. Pantothenic acid in Nutrition. Nutr. Rev. 1953, 11, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Rucker, R.B.; Zempleni, J.; Suttie, J.W.; McCormick, D.B. (Eds.) Handbook of Vitamins, 4th ed.; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Nanda, V.; Sarkar, B.C.; Sharma, H.K.; Bawa, A.S. Physico-chemical properties and estimation of mineral content in honey produced from different plants in Northern India. J. Food Compos. Anal. 2003, 16, 613–619. [Google Scholar] [CrossRef]
- Czipa, N.; Andrási, D.; Kovács, B. Determination of essential and toxic elements in Hungarian honeys. Food Chem. 2015, 175, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Denisow, B.; Denisow-Pietrzyk, M. Biological and therapeutic properties of bee pollen: A review. J. Sci. Food Agric. 2016, 96, 4303–4309. [Google Scholar] [CrossRef]
- Somerville, D.C.; Nicol, H.I. Crude protein and amino acid composition of honey bee-collected pollen pellets from south-east Australia and a note on laboratory disparity. Aust. J. Exp. Agric. 2006, 46, 141–149. [Google Scholar] [CrossRef]
- White, D.; Cribb, B.W.; Heard, T.A. Flower constancy of the stingless bee Trigona carbonaria Smith (Hymenoptera: Apidae: Meliponini). Aust. J. Entomol. 2001, 40, 61–64. [Google Scholar] [CrossRef]
- Anderson, L.M.; Dietz, A. Pyridoxine Requirement of the Honey Bee (Apis mellifera) for Brood Rearing. Apidologie 1976, 7, 67–84. [Google Scholar] [CrossRef]
- Ciulu, M.; Solinas, S.; Floris, I.; Panzanelli, A.; Pilo, M.I.; Piu, P.C.; Spano, N.; Sanna, G. RP-HPLC determination of water-soluble vitamins in honey. Talanta 2011, 83, 924–929. [Google Scholar] [CrossRef]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef]
- Barker, R.J. Some Carbohydrates Found in Pollen and Pollen Substitutes are Toxic To Honey Bees. J. Nutr. 1977, 107, 1859–1862. [Google Scholar] [CrossRef]
- Qian, X.; Yan, W.; Zhang, W.; Dong, W.; Ma, J.; Ochsenreither, K.; Jiang, M.; Xin, F. Current status and perspectives of 2-phenylethanol production through biological processes. Crit. Rev. Biotechnol. 2019, 39, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cao, M.; Jiang, X.; Zou, H.; Wang, C.; Xu, X.; Xian, M. De-novo synthesis of 2-phenylethanol by Enterobacter sp. CGMCC 5087. BMC Biotechnol. 2014, 14, 30. [Google Scholar] [CrossRef] [PubMed]
- Lingappa, B.T.; Prasad, M.; Lingappa, Y.; Hunt, D.F.; Biemann, K. Phenethyl alcohol and tryptophol: Autoantibiotics produced by the fungus Candida albicans. Science 1969, 163, 192–194. [Google Scholar] [CrossRef] [PubMed]
- Spinnler, H.-E.; Djian, A. Bioconversion of amino acids into flavouring alcohols and esters by Erwinia carotovora subsp. atroseptica. Appl. Microbiol. Biotechnol. 1991, 35, 264–269. [Google Scholar] [CrossRef]
- McNerney, R.; Mallard, K.; Okolo, P.I.; Turner, C. Production of volatile organic compounds by mycobacteria. FEMS Microbiol. Lett. 2012, 328, 150–156. [Google Scholar] [CrossRef]
- Hirano, J.; Miyamoto, K.; Ohta, H. Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated from 2-phenylethanol-assimilating Brevibacterium sp. KU 1309. J. Biosci. Bioeng. 2005, 100, 318–322. [Google Scholar] [CrossRef]
- Jollivet, N.; Bézenger, M.-C.; Vayssier, Y.; Belin, J.-M. Production of volatile compounds in liquid cultures by six strains of coryneform bacteria. Appl. Microbiol. Biotechnol. 1992, 36, 790–794. [Google Scholar] [CrossRef]
- Drężek, K.; Kozłowska, J.; Detman, A.; Mierzejewska, J. Development of a Continuous System for 2-Phenylethanol Bioproduction by Yeast on Whey Permeate-Based Medium. Molecules 2021, 26, 7388. [Google Scholar] [CrossRef]
- Zhu, Y.-J.; Zhou, H.-T.; Hu, Y.-h.; Tang, J.-Y.; Su, M.-X.; Guo, Y.-J.; Chen, Q.-X.; Liu, B. Antityrosinase and antimicrobial activities of 2-phenylethanol, 2-phenylacetaldehyde and 2-phenylacetic acid. Food Chem. 2011, 124, 298–302. [Google Scholar] [CrossRef]
- Kim, Y.; Cho, J.-Y.; Kuk, J.-H.; Moon, J.-H.; Cho, J.-I.; Kim, Y.-C.; Park, K.-H. Identification and Antimicrobial Activity of Phenylacetic Acid Produced by Bacillus licheniformis Isolated from Fermented Soybean, Chungkook-Jang. Curr. Microbiol. 2004, 48, 312–317. [Google Scholar] [CrossRef]
- Gutiérrez, L.; Escudero, A.; Batlle, R.; Nerín, C. Effect of mixed antimicrobial agents and flavors in active packaging films. J. Agric. Food Chem. 2009, 57, 8564–8571. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Cheng, Y.; Yang, M.; Liu, Y.; Chen, K.; Long, C.A.; Deng, X. Mechanisms of action for 2-phenylethanol isolated from Kloeckera apiculata in control of Penicillium molds of citrus fruits. BMC Microbiol. 2014, 14, 242. [Google Scholar] [CrossRef] [PubMed]
| Number | % of Total | |
|---|---|---|
| Total number of bases | 3,042,366 | 100.00% |
| Number of coding bases | 2,695,972 | 88.61% |
| G/C content | 1,433,972 | 47.13% |
| NG50 | 299,429 | |
| L50 | 4 | |
| DNA scaffolds | 21 | 100.00% |
| Genes (total number) | 3023 | 100.00% |
| Protein coding genes | 2924 | 96.73% |
| Regulatory and miscellaneous features | 44 | 1.46% |
| RNA genes | 55 | 1.82% |
| rRNA genes | 5 | 0.17% |
| 5S rRNA | 3 | 0.10% |
| 16S rRNA | 1 | 0.03% |
| 23S rRNA | 1 | 0.03% |
| tRNA genes | 47 | 1.55% |
| other RNA genes | 3 | 0.10% |
| Protein coding genes with predicted function | 2521 | 83.39% |
| Protein without function prediction | 403 | 13.33% |
| Protein coding genes with enzymes | 952 | 31.49% |
| Protein coding genes connected to KEGG pathways | 1085 | 35.89% |
| Protein coding genes not connected to KEGG pathways | 1839 | 60.83% |
| Protein coding genes connected to KEGG orthology (KO) | 2001 | 66.19% |
| Protein coding genes not connected to KEGG orthology (KO) | 923 | 30.53% |
| Protein coding genes connected to MetaCyc pathways | 795 | 26.30% |
| Protein coding genes not connected to MetaCyc pathways | 2129 | 70.43% |
| Protein coding genes with COGs3 | 2494 | 82.50% |
| with Pfam3 | 2564 | 84.82% |
| with TIGRfam3 | 1249 | 41.32% |
| with SMART | 560 | 18.52% |
| with SUPERFam | 2409 | 79.69% |
| with CATH FunFam | 2080 | 68.81% |
| in internal clusters | 539 | 17.83% |
| Strain | dDDH (d0, in %) | C.I. (d0, in %) | dDDH (d4, in %) | C.I. (d4, in %) | dDDH (d6, in %) | C.I. (d6, in %) | G + C Content Difference (in %) |
|---|---|---|---|---|---|---|---|
| Rosenbergiella epipactidis 2.1A | 78.8 | [74.8–82.3] | 61.4 | [58.6–64.2] | 78 | [74.6–81.1] | 0.46 |
| Rosenbergiella nectarea 8N4 | 73.9 | [69.9–77.5] | 31.1 | [28.7–33.6] | 61.6 | [58.3–64.8] | 0.31 |
| Rosenbergiella collisarenosi 8.8A | 51.3 | [47.9–54.8] | 20.7 | [18.5–23.1] | 39.8 | [36.9–42.9] | 1.04 |
| Rosenbergiella australiborealis CdVSA20.1 | 47 | [43.6–50.4] | 20.2 | [18.0–22.6] | 37 | [34.1–40.1] | 1.82 |
| Pantoea cypripedii LMG 2657 | 13.2 | [10.5–16.5] | 20 | [17.8–22.4] | 13.6 | [11.2–16.3] | 6.92 |
| Characteristic | R. meliponini D21B1 | R. epipactidis 2.1A |
|---|---|---|
| Urease activity | Yes | No |
| Utilization of citrate | Yes | No |
| Growth at 37 °C | Poor | No |
| NRPS/PKS gene cluster | Present | Absent |
| Production of 2-phenylethanol | Yes | No |
| Thiamine dependency | Independent | Dependent |
| Pyocin encoding genes | No | Yes |
| Hemolysin encoding genes | No | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farlow, A.J.; Rupasinghe, D.B.; Naji, K.M.; Capon, R.J.; Spiteller, D. Rosenbergiella meliponini D21B Isolated from Pollen Pots of the Australian Stingless Bee Tetragonula carbonaria. Microorganisms 2023, 11, 1005. https://doi.org/10.3390/microorganisms11041005
Farlow AJ, Rupasinghe DB, Naji KM, Capon RJ, Spiteller D. Rosenbergiella meliponini D21B Isolated from Pollen Pots of the Australian Stingless Bee Tetragonula carbonaria. Microorganisms. 2023; 11(4):1005. https://doi.org/10.3390/microorganisms11041005
Chicago/Turabian StyleFarlow, Anthony J., Darshani B. Rupasinghe, Khalid M. Naji, Robert J. Capon, and Dieter Spiteller. 2023. "Rosenbergiella meliponini D21B Isolated from Pollen Pots of the Australian Stingless Bee Tetragonula carbonaria" Microorganisms 11, no. 4: 1005. https://doi.org/10.3390/microorganisms11041005
APA StyleFarlow, A. J., Rupasinghe, D. B., Naji, K. M., Capon, R. J., & Spiteller, D. (2023). Rosenbergiella meliponini D21B Isolated from Pollen Pots of the Australian Stingless Bee Tetragonula carbonaria. Microorganisms, 11(4), 1005. https://doi.org/10.3390/microorganisms11041005








