Extraction of High-Quality RNA from S. aureus Internalized by Endothelial Cells
Abstract
:1. Introduction
2. Material and Methods
- Condition I: RNA from S. aureus in culture after 2 h and 5 h incubation
- Condition II: RNA extracted from endothelial cells infected with S. aureus for 90 min and immediately lysed or further cultured and lysed after 24 h and 48 h, respectively.
2.1. Bacteria and Human Endothelial Cell Culture
2.2. Bacterial Internalization
2.3. RNA Extraction of Endothelial RNA
2.4. Extraction of RNA from Intracellular S. aureus
2.4.1. Lysing Endothelial Cells including DNA and RNA
- Remove the cell culture medium and wash with PBS 1X.
- Add 1 mL 0.02% SDS/PBS 1X and incubate for 20 s (sec). The lysis destroys the endothelial cells including their DNA and RNA.
- Add 5 mL cold PBS 1X, vigorously flush multiple times, and transfer the sample to a 15 mL falcon tube.
- Add 1 mL of 0.02% SDS/PBS 1X again and incubate until all cells are detached. The process can be sped up by tilting the flask. At this point, the lysate becomes viscous.
- Add 7 mL of PBS 1X and vigorously flush multiple times, destroying the endothelial cells and releasing the intracellular bacteria. The use of SDS with PBS 1X avoids the bacterial lysis.
- Transfer the lysate to the falcon tube.
- Centrifuge the sample at 4000× g for 15 min at 4 °C to pellet the bacteria and further destroy the endothelial cells.
- Carefully remove the supernatant from the sample and add 1 mL of RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany)
- Vortex the sample at maximal (max) speed and incubate it for 5 min at RT.
- Centrifuge at 12,000× g for 10 min at RT.
- Remove the supernatant and store the pellet at −80 °C overnight or up to four weeks.
2.4.2. Mechanical Disruption of S. aureus from Culture and Infected Cells
- Thaw the samples and add 1 mL of RNApro solution (MP Biomedicals, Eschwege, Germany) and vigorously resuspend the pellet until no large clumps are visible.
- Transfer the mixture to a Lysing Matrix B tube (MP Biomedicals, Eschwege, Germany) and vortex at maximal speed for 10 s.
- Put the tube in a homogenizer (SpeedMill Plus, Analytikjena, Jena, Germany) and lyse for 3 min twice.
- Centrifuge the sample for 2 min at 15,000× g at 4 °C to pellet the whole bacterial debris.
- Transfer the supernatant containing bacterial RNA to a new reaction tube.
2.4.3. Purification of S. aureus RNA
2.5. DNAse Treatment
2.6. Gel Electrophoresis
2.7. Reverse Transcription (RT)-PCR
2.8. Real-Time Quantitative PCR (qPCR)
2.9. Data Analysis
3. Results and Discussion
3.1. Quality of RNA
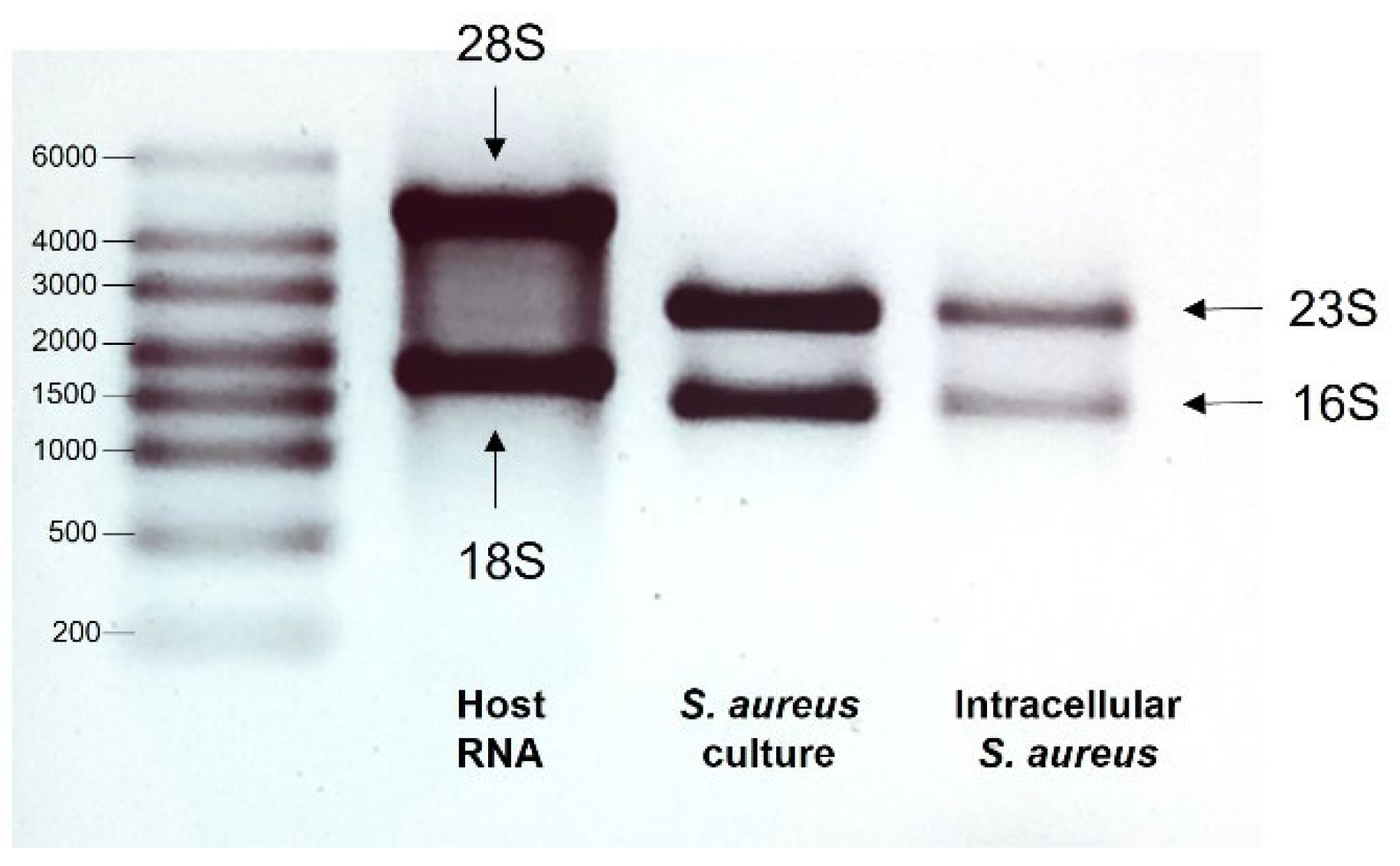
| Sample No. | Sample | Amount (ng/µL) | 260 nm/280 nm Ratio | 260 nm/230 nm Ratio |
|---|---|---|---|---|
| 1 | Bacteria from culture 2 h_1 | 234.55 | 2.21 | 2.29 |
| 2 | Bacteria from culture 2 h_2 | 255.27 | 2.22 | 2.47 |
| 3 | Bacteria from culture 2 h_3 | 371.33 | 2.19 | 2.52 |
| 4 | Bacteria from culture 2 h_4 | 411.93 | 2.22 | 2.52 |
| 5 * | Bacteria from culture 5 h_1 | 84.07 | 2.1 | 1.95 |
| 6 | Bacteria from culture 5 h_2 | 90.7 | 2.16 | 2.3 |
| 7 | Bacteria from culture 5 h_3 | 129.62 | 2.21 | 1.07 |
| 8 | Bacteria from culture 5 h_4 | 126.99 | 2.2 | 1.77 |
| 9 | Bacteria from culture 5 h_5 | 123.9 | 2.24 | 2.29 |

| Sample No. | Sample | Amount (ng/µL) | 260 nm/280 nm Ratio | 260 nm/230 nm Ratio |
|---|---|---|---|---|
| 1 | Intracellular bacteria 90 min p.i._1 | 112.01 | 2.31 | 3.14 |
| 2 | Intracellular bacteria 90 min p.i._2 | 137.13 | 2.33 | 2.66 |
| 3 | Intracellular bacteria 90 min p.i._3 | 136.16 | 2.3 | 2.21 |
| 4 | Intracellular bacteria 90 min p.i._4 | 197.18 | 2.12 | 2.18 |
| 5 * | Intracellular bacteria 90 min p.i._5 | 233.04 | 2.25 | 1.42 |
| 6 * | Intracellular bacteria 24 h p.i._1 | 79.23 | 2.33 | 2.71 |
| 7 * | Intracellular bacteria 24 h p.i._2 | 85.82 | 2.26 | 2.87 |
| 8 | Intracellular bacteria 24 h p.i._3 | 250.78 | 2.22 | 2.54 |
| 9 | Intracellular bacteria 24 h p.i._4 | 195.49 | 2.24 | 2.55 |
| 10 | Intracellular bacteria 24 h p.i._5 | 240.66 | 2.19 | 2.78 |
| 11 | Intracellular bacteria 48 h p.i._1 | 212.66 | 2.18 | 2.42 |
| 12 * | Intracellular bacteria 48 h p.i._2 | 88.21 | 2.27 | 0.4 |
| 13 | Intracellular bacteria 48 h p.i._3 | 169.4 | 2.29 | 2.99 |
| 14 | Intracellular bacteria 48 h p.i._4 | 182.99 | 2.2 | 0.88 |
| 15 | Intracellular bacteria 48 h p.i._5 | 138.57 | 2.29 | 2.66 |
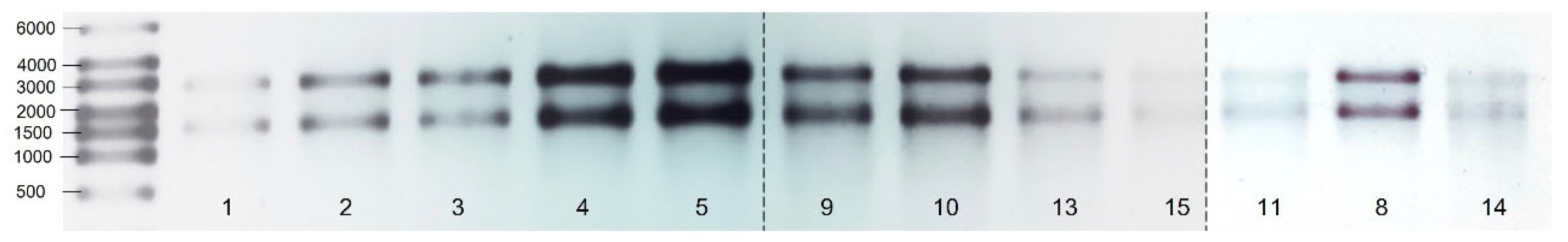
3.2. Stability of Housekeeping Genes
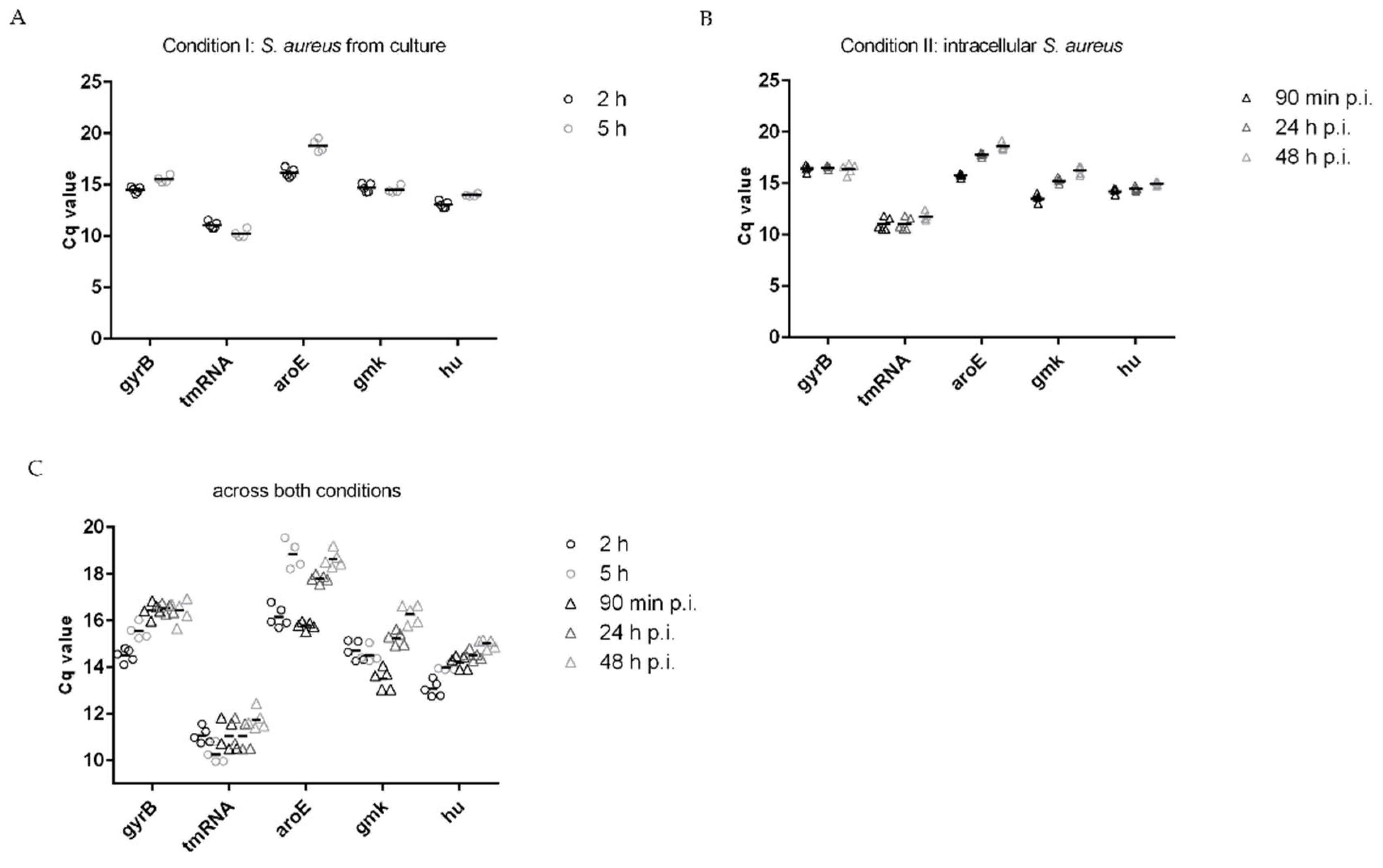
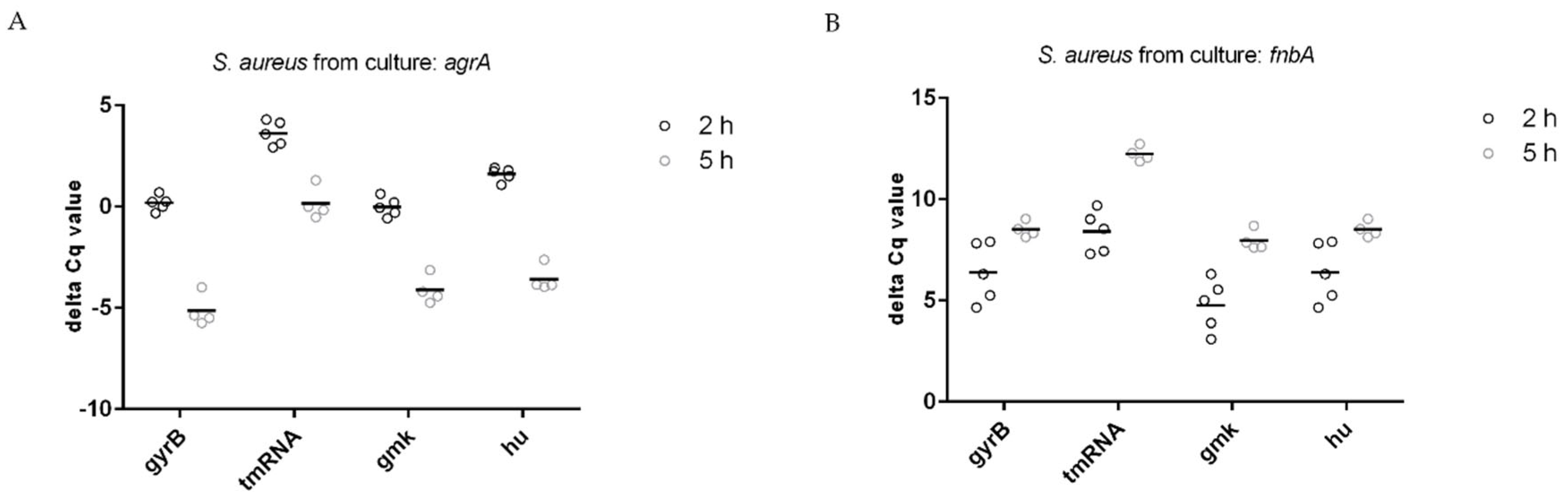
3.3. Normalization of Target Genes
3.4. Applicability and Limitations of This Protocol
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strobel, M.; Pförtner, H.; Tuchscherr, L.; Völker, U.; Schmidt, F.; Kramko, N.; Schnittler, H.J.; Fraunholz, M.J.; Löffler, B.; Peters, G.; et al. Post-invasion events after infection with Staphylococcus aureus are strongly dependent on both the host cell type and the infecting S. aureus strain. Clin. Microbiol. Infect. 2016, 22, 799–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stelzner, K.; Boyny, A.; Hertlein, T.; Sroka, A.; Moldovan, A.; Paprotka, K.; Kessie, D.; Mehling, H.; Potempa, J.; Ohlsen, K.; et al. Intracellular Staphylococcus aureus employs the cysteine protease staphopain A to induce host cell death in epithelial cells. PLoS Pathog. 2021, 17, e1009874. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.A.; von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Kreis, C.A.; Hoerr, V.; Flint, L.; Hachmeister, M.; Geraci, J.; Bremer-Streck, S.; Kiehntopf, M.; Medina, E.; Kribus, M.; et al. Staphylococcus aureus develops increased resistance to antibiotics by forming dynamic small colony variants during chronic osteomyelitis. J. Antimicrob. Chemother. 2016, 71, 438–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuchscherr, L.; Löffler, B.; Proctor, R.A. Persistence of Staphylococcus aureus: Multiple Metabolic Pathways Impact the Expression of Virulence Factors in Small-Colony Variants (SCVs). Front. Microbiol. 2020, 11, 1028. [Google Scholar] [CrossRef] [PubMed]
- Garzoni, C.; Francois, P.; Huyghe, A.; Couzinet, S.; Tapparel, C.; Charbonnier, Y.; Renzoni, A.; Lucchini, S.; Lew, D.P.; Vaudaux, P.; et al. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genom. 2007, 8, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westermann, A.J.; Barquist, L.; Vogel, J. Resolving host-pathogen interactions by dual RNA-seq. PLoS Pathog. 2017, 13, e1006033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westermann, A.J.; Gorski, S.A.; Vogel, J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 2012, 10, 618–630. [Google Scholar] [CrossRef] [Green Version]
- Stead, M.B.; Agrawal, A.; Bowden, K.E.; Nasir, R.; Mohanty, B.K.; Meagher, R.B.; Kushner, S.R. RNAsnap: A rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res. 2012, 40, e156. [Google Scholar] [CrossRef] [Green Version]
- Penaranda, C.; Hung, D.T. Single-Cell RNA Sequencing to Understand Host-Pathogen Interactions. ACS Infect. Dis. 2019, 5, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Zhao, C.; Zhang, T.; Liang, H.; Wang, X.-M.; Pan, Y.; Chen, X.; Zhao, Q.; Li, D.; Liu, F.; et al. Salmonella produce microRNA-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival. Sci. Rep. 2017, 7, 2392. [Google Scholar] [CrossRef] [PubMed]
- Raynaud, S.; Le Pabic, H.; Felden, B. Selective recovery of RNAs from bacterial pathogens after their internalization by human host cells. Methods 2018, 143, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.; Francois, P.; Liebeke, M.; Fraunholz, M.; Goerke, C.; Krismer, B.; Schrenzel, J.; Lalk, M.; Wolz, C. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 2012, 8, e1003016. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Pei, A.Y.; Oberdorf, W.E.; Nossa, C.W.; Agarwal, A.; Chokshi, P.; Gerz, E.A.; Jin, Z.; Lee, P.; Yang, L.; Poles, M.; et al. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl. Environ. Microbiol. 2010, 76, 3886–3897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-Machorro, A.L.; Hernandez, R.; Cevallos, A.M.; Lopez-Villasenor, I. Ribosomal RNA genes in eukaryotic microorganisms: Witnesses of phylogeny? FEMS Microbiol. Rev. 2010, 34, 59–86. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Theis, T.; Skurray, R.A.; Brown, M.H. Identification of suitable internal controls to study expression of a Staphylococcus aureus multidrug resistance system by quantitative real-time PCR. J. Microbiol. Methods 2007, 70, 355–362. [Google Scholar] [CrossRef]
- Ogonowska, P.; Nakonieczna, J. Validation of stable reference genes in Staphylococcus aureus to study gene expression under photodynamic treatment: A case study of SEB virulence factor analysis. Sci. Rep. 2020, 10, 16354. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [Green Version]
- Brignoli, T.; Manetti, A.G.O.; Rosini, R.; Haag, A.F.; Scarlato, V.; Bagnoli, F.; Delany, I. Absence of Protein A Expression Is Associated With Higher Capsule Production in Staphylococcal Isolates. Front. Microbiol. 2019, 10, 863. [Google Scholar] [CrossRef] [PubMed]
- Bosi, E.; Monk, J.M.; Aziz, R.K.; Fondi, M.; Nizet, V.; Palsson, B.O. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc. Natl. Acad. Sci. USA. 2016, 113, E3801–E3809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuchscherr, L.; Medina, E.; Hussain, M.; Völker, W.; Heitmann, V.; Niemann, S.; Holzinger, D.; Roth, J.; Proctor, R.A.; Becker, K.; et al. Staphylococcus aureus phenotype switching: An effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 2011, 3, 129–141. [Google Scholar] [CrossRef]
- Opitz, L.; Salinas-Riester, G.; Grade, M.; Jung, K.; Jo, P.; Emons, G.; Ghadimi, B.M.; Beißbarth, T.; Gaedcke, J. Impact of RNA degradation on gene expression profiling. BMC Med. Genom. 2010, 3, 36. [Google Scholar] [CrossRef] [Green Version]
- Traber, K.E.; Lee, E.; Benson, S.; Corrigan, R.; Cantera, M.; Shopsin, B.; Novick, R.P. agr function in clinical Staphylococcus aureus isolates. Microbiology 2008, 154 Pt 8, 2265–2274. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.; Meghji, S.; Williams, R.J.; Henderson, B.; Brock, J.H.; Nair, S.P. Staphylococcus aureus fibronectin binding proteins are essential for internalization by osteoblasts but do not account for differences in intracellular levels of bacteria. Infect. Immun. 2001, 69, 2872–2877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niemann, S.; Nguyen, M.T.; Eble, J.A.; Chasan, A.I.; Mrakovcic, M.; Böttcher, R.T.; Preissner, K.T.; Roßlenbroich, S.; Peters, G.; Herrmann, M. More Is Not Always Better-the Double-Headed Role of Fibronectin in Staphylococcus aureus Host Cell Invasion. mBio 2021, 12, e0106221. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.M.; Wiesolek, H.L.; Sumagin, R. ICAM-1: A master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. J. Leukoc. Biol. 2020, 108, 787–799. [Google Scholar] [CrossRef]
- Modesto, P.; Peletto, S.; Pisoni, G.; Cremonesi, P.; Castiglioni, B.; Colussi, S.; Caramelli, M.; Bronzo, V.; Moroni, P.; Acutis, P.L. Evaluation of internal reference genes for quantitative expression analysis by real-time reverse transcription-PCR in somatic cells from goat milk. J. Dairy Sci. 2013, 96, 7932–7944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Diao, H.; Zhao, L.; Xing, Y.; Zhang, J.; Liu, N.; Yan, Y.; Tian, X.; Sun, W.; Liu, B. Identification of suitable reference genes for real-time quantitative PCR analysis of hydrogen peroxide-treated human umbilical vein endothelial cells. BMC Mol. Biol. 2017, 18, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegmund, A.; Afzal, M.A.; Tetzlaff, F.; Keinhörster, D.; Gratani, F.; Paprotka, K.; Westermann, M.; Nietzsche, S.; Wolz, C.; Fraunholz, M.; et al. Intracellular persistence of Staphylococcus aureus in endothelial cells is promoted by the absence of phenol-soluble modulins. Virulence 2021, 12, 1186–1198. [Google Scholar] [CrossRef] [PubMed]
- Robledo, D.; Hernández-Urcera, J.; Cal, R.M.; Pardo, B.G.; Sánchez, L.; Martínez, P.; Viñas, A. Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genom. 2014, 15, 648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]


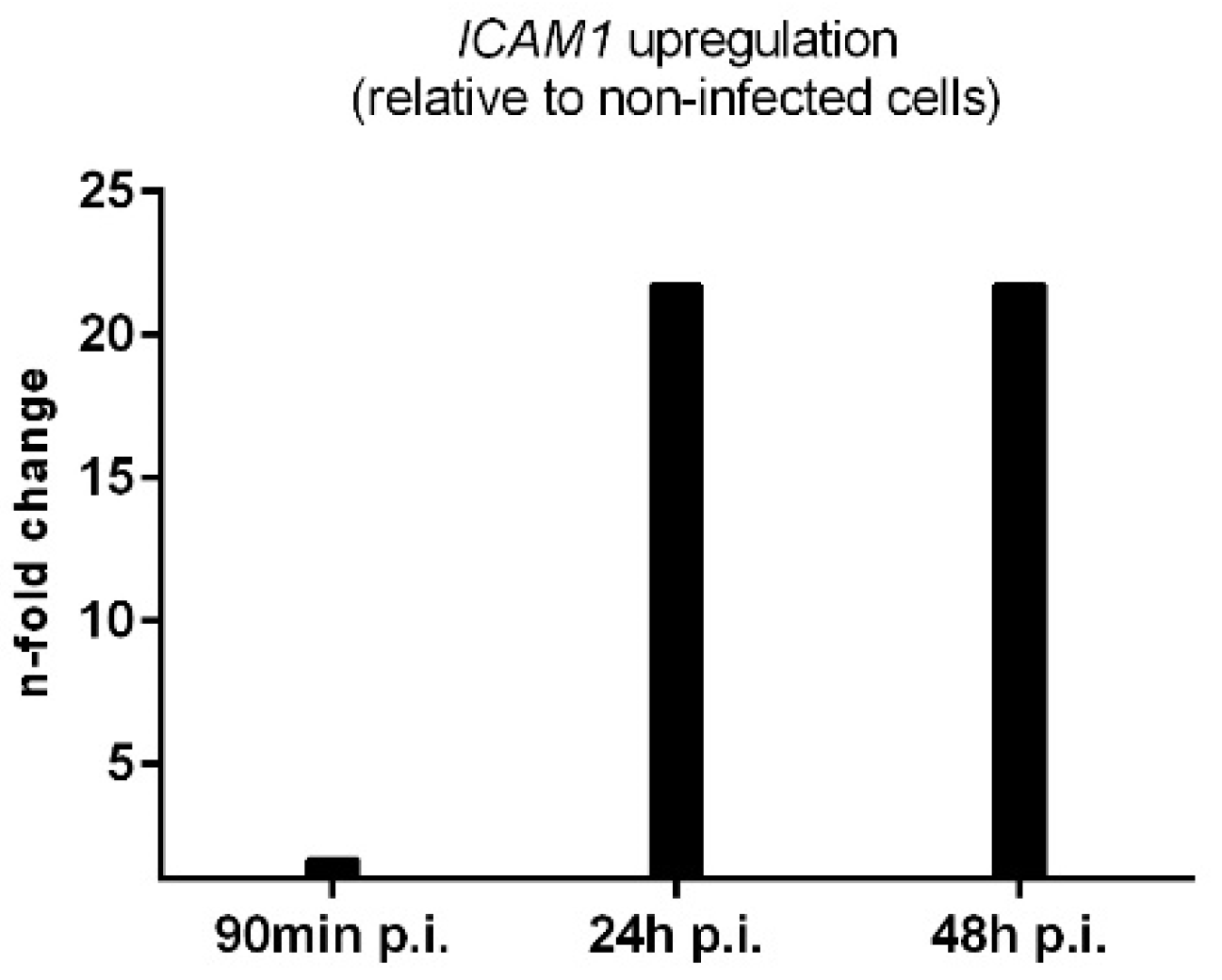
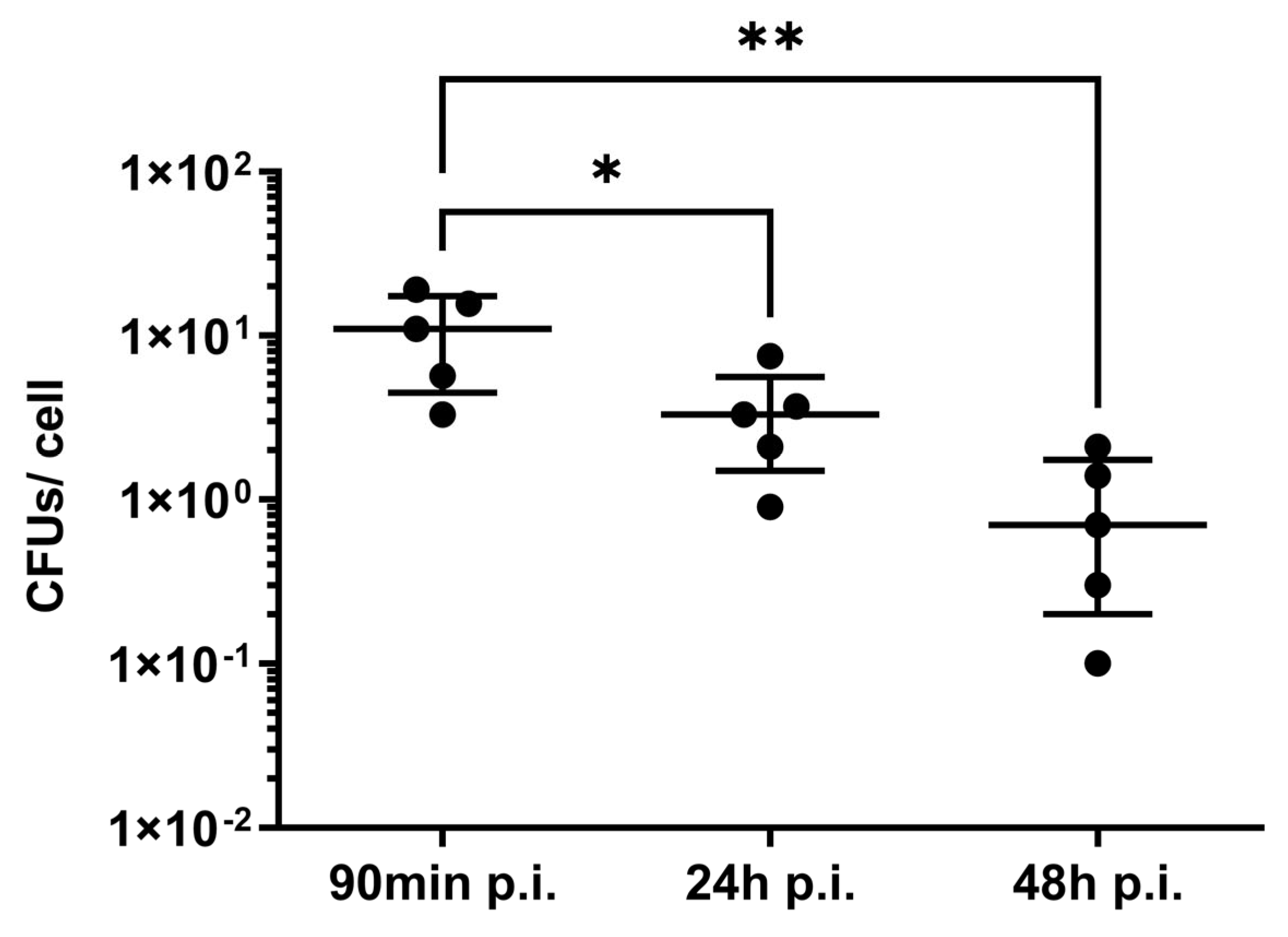
| Gene | Gene Product | Locus Tag |
|---|---|---|
| Housekeeping genes | ||
| gyrB | DNA gyrase subunit B | SAUSA300_RS00030 |
| tmRNA | tmRNA | SAUSA300_RS15205 |
| aroE | Shikimate dehydrogenase (NADP(+)) | SAUSA300_RS08475 |
| gmk | Guanylate kinase | SAUSA300_RS05970 |
| hu | Nucleotide-associated protein HU | SAUSA300_RS07430 |
| Target genes | ||
| agrA | Quorum-sensing response regulator AgrA | SAUSA300_RS10950 |
| fnbA | Fibronectin-binding protein A | SAUSA300_RS13530 |
| Ranking | Housekeeping Gene | n | std dev (±Cq) |
|---|---|---|---|
| Condition I: S. aureus from culture | |||
| 1 | gmk | 9 | 0.32 |
| 2 | tmRNA | 9 | 0.43 |
| 3 | hu | 9 | 0.46 |
| 4 | gyrB | 9 | 0.51 |
| 5 | aroE | 9 | 1.32 |
| Condition II: intracellular S. aureus | |||
| 1 | gyrB | 15 | 0.26 |
| 2 | hu | 15 | 0.32 |
| 3 | tmRNA | 15 | 0.55 |
| 4 | gmk | 15 | 1.02 |
| 5 | aroE | 15 | 1.08 |
| Across both conditions (I + II) | |||
| 1 | gyrB | 24 | 0.56 |
| 1 | tmRNA | 24 | 0.56 |
| 2 | hu | 24 | 0.73 |
| 3 | gmk | 24 | 0.81 |
| 4 | aroE | 24 | 1.18 |
| Target Gene | n | std dev (± Cq) |
|---|---|---|
| 2 h | ||
| agrA | 5 | 0.25 |
| fnbA | 5 | 0.93 |
| 5 h | ||
| agrA | 4 | 0.43 |
| fnbA | 4 | 0.35 |
| Target Gene | n | std dev (± Cq) | ||
|---|---|---|---|---|
| 90 min p.i. | 24 h p.i. | 48 h p.i. | ||
| agrA | 5 | 0.42 | 0.18 | 0.64 |
| fnbA | 5 | 0.38 | 0.95 | 0.70 |
| Reference Gene | Target Gene: agrA | |
|---|---|---|
| 2 h | 5 h | |
| gmk | −0.01 (0.46) | −4.125 (0.7) |
| tmRNA | 3.62 (0.61) | 0.15 (0.8) |
| hu | 1.61 (0.33) | −3.58 (0.64) |
| gyrB | 0.18 (0.38) | −5.15 (0.79) |
| Target gene: fnbA | ||
| gmk | 4.77 (1.29) | 7.96 (0.51) |
| tmRNA | 8.4 (1.02) | 12.24 (0.37) |
| hu | 6.39 (1.47) | 8.51 (0.39) |
| gyrB | 4.96 (0.95) | 6.94 (0.33) |
| Reference Genes | Target Gene: agrA | ||
|---|---|---|---|
| 90 min p.i. | 24 h p.i. | 48 h p.i. | |
| tmRNA | 2.32 (0.18) | 3.35 (0.45) | 4.04 (0.94) |
| hu | −0.85 (0.53) | −0.13 (0.22) | 0.79 (0.86) |
| gyrB | −3.09 (0.33) | −2.14 (0.3) | −0.63 (1.15) |
| Target gene: fnbA | |||
| 90 min p.i. | 24 h p.i. | 48 h p.i. | |
| tmRNA | 4.51 (0.48) | 7.49 (0.48) | 7.79 (0.61) |
| hu | 1.33 (0.43) | 4.02 (1.01) | 4.53 (0.91) |
| gyrB | 0.9 (0.46) | 2.0 (1.1) | 3.12 (1.26) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maurer, M.; Klassert, T.E.; Löffler, B.; Slevogt, H.; Tuchscherr, L. Extraction of High-Quality RNA from S. aureus Internalized by Endothelial Cells. Microorganisms 2023, 11, 1020. https://doi.org/10.3390/microorganisms11041020
Maurer M, Klassert TE, Löffler B, Slevogt H, Tuchscherr L. Extraction of High-Quality RNA from S. aureus Internalized by Endothelial Cells. Microorganisms. 2023; 11(4):1020. https://doi.org/10.3390/microorganisms11041020
Chicago/Turabian StyleMaurer, Michelle, Tilman E. Klassert, Bettina Löffler, Hortense Slevogt, and Lorena Tuchscherr. 2023. "Extraction of High-Quality RNA from S. aureus Internalized by Endothelial Cells" Microorganisms 11, no. 4: 1020. https://doi.org/10.3390/microorganisms11041020
APA StyleMaurer, M., Klassert, T. E., Löffler, B., Slevogt, H., & Tuchscherr, L. (2023). Extraction of High-Quality RNA from S. aureus Internalized by Endothelial Cells. Microorganisms, 11(4), 1020. https://doi.org/10.3390/microorganisms11041020








