Selected Gut Bacteria from Water Monitor Lizard Exhibit Effects against Pathogenic Acanthamoeba castellanii Belonging to the T4 Genotype
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dissection of Water Monitor Lizard and Isolation of Gut Bacteria
2.2. DNA Extraction, Amplification and Identification of Isolated Bacteria
2.3. Preparation of Bacterial Conditioned Media
2.4. Acanthamoeba castellanii Cultures
2.5. Amoebicidal Assays
2.6. HeLa Cell Cultivation
2.7. Adhesion Assays
2.8. Encystation Assays
2.9. Excystation Assays
2.10. Host Cell Cytotoxicity Assays
2.11. Cytopathogenicity Assays
2.12. Ultra-Performance Liquid Chromatography Tandem Mass Spectrometry (HPLC-MS/MS)
3. Results
3.1. Isolation and Identifcation of Bacterial Isolates Derived from the Gut of Water Monitor Lizard
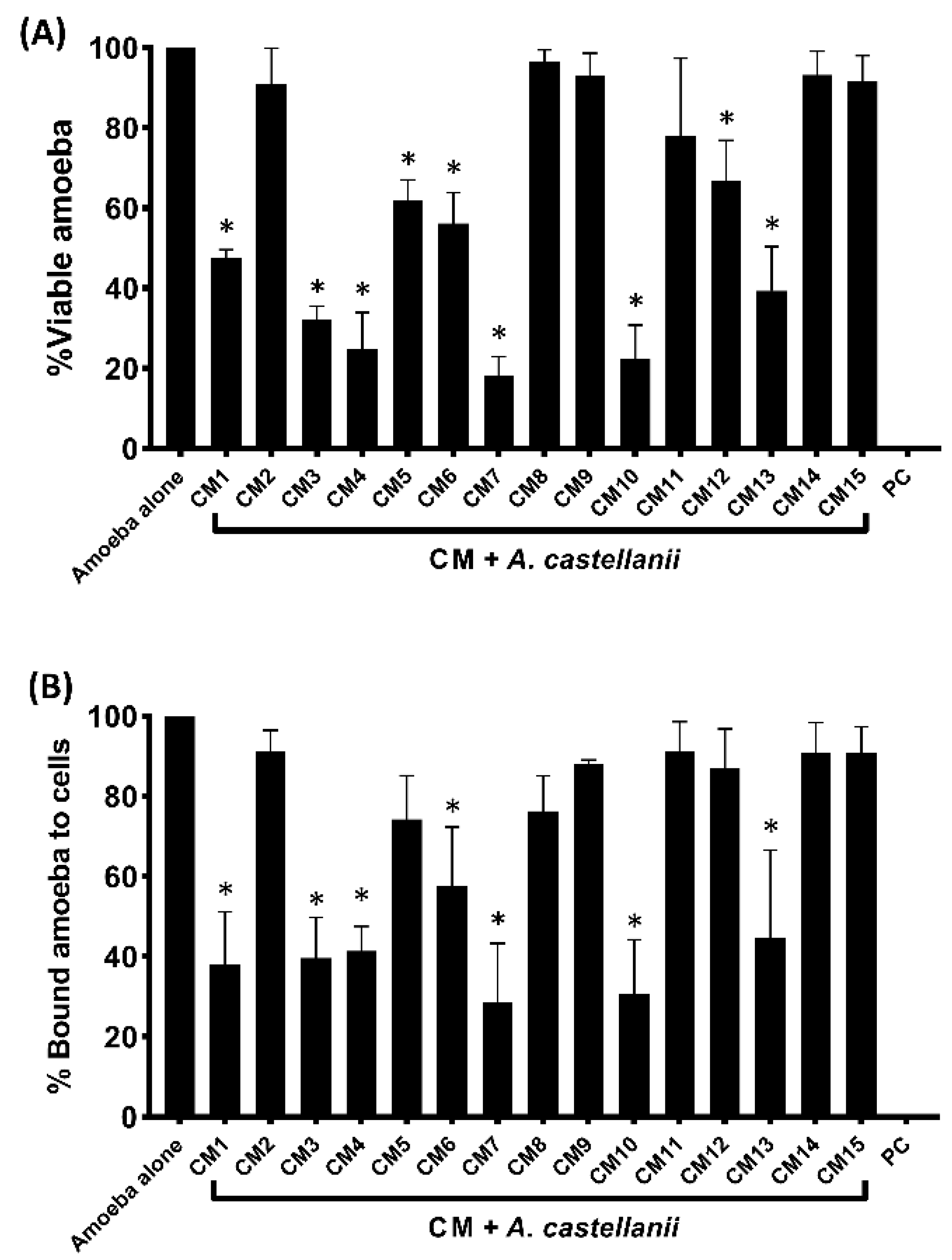
3.2. Conditioned Media Isolated from Selected Gut Bacteria of Water Monitor Lizard Showed Amoebicidal Activity
3.3. Bacterial Conditioned Media Inhibited the Binding of Amoebae to Human Cells
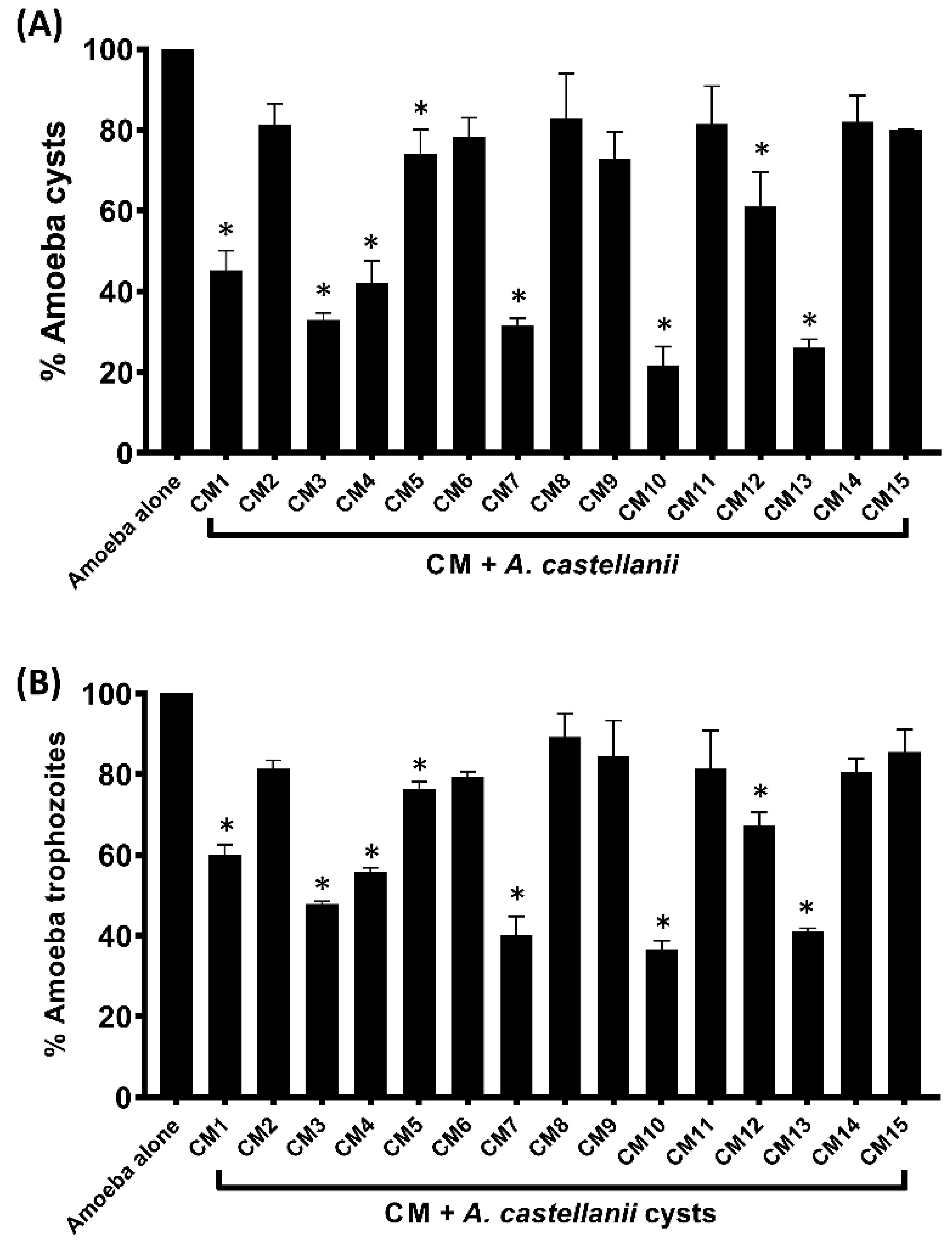
3.4. Encystation and Excystation of Amoebae were Inhibited
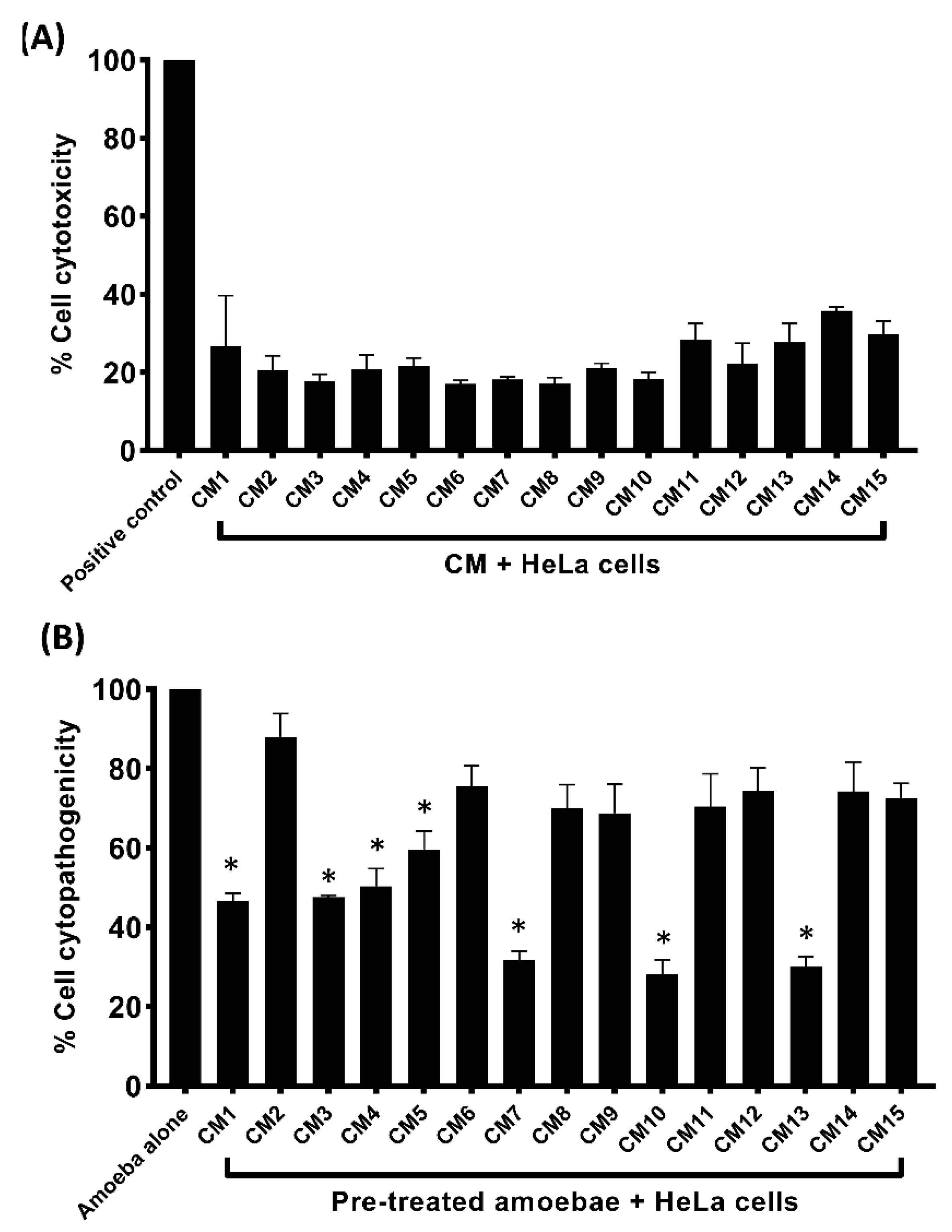
3.5. Conditioned Media Exhibited Limited Cytotoxicity and Inhibited Amoebae–Mediated Host Cell Damage
3.6. Mass Spectrometry Revealed Numerous Metabolites
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Visvesvara, G.S.; Moura, H.; Schuster, F.L. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 2007, 50, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Niederkorn, J.Y.; Alizadeh, H.; Leher, H.; McCulley, J.P. The pathogenesis of Acanthamoeba keratitis. Microbes Infect. 1999, 1, 437–443. [Google Scholar] [CrossRef]
- Marciano-Cabral, F.; Cabral, G. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 2003, 16, 273–307. [Google Scholar] [CrossRef]
- Khan, N.A. Acanthamoeba: Biology and increasing importance in human health. FEMS Microbiol. Rev. 2006, 30, 564–595. [Google Scholar] [CrossRef]
- Anwar, A.; Mungroo, M.R.; Anwar, A.; Sullivan, W.J., Jr.; Khan, N.A.; Siddiqui, R. Repositioning of guanabenz in conjugation with gold and silver nanoparticles against pathogenic amoebae Acanthamoeba castellanii and Naegleria fowleri. ACS Infect. Dis. 2019, 5, 2039–2046. [Google Scholar] [CrossRef]
- Akbar, N.; Kawish, M.; Jabri, T.; Khan, N.A.; Shah, M.R.; Siddiqui, R. Cinnamic acid and lactobionic acid based nanoformulations as a potential antiamoebic therapeutics. Exp. Parasitol. 2023, 246, 108474. [Google Scholar] [CrossRef] [PubMed]
- Chin, Y.W.; Balunas, M.J.; Chai, H.B.; Kinghorn, A.D. Drug discovery from natural sources. AAPS J. 2006, 8, E239–E253. [Google Scholar] [CrossRef]
- Simmons, T.L.; Coates, R.C.; Clark, B.R.; Engene, N.; Gonzalez, D.; Esquenazi, E.; Dorrestein, P.C.; Gerwick, W.H. Biosynthetic origin of natural products isolated from marine microorganism–invertebrate assemblages. Proc. Natl. Acad. Sci. USA 2008, 105, 4587–4594. [Google Scholar] [CrossRef]
- Show, P.L.; Oladele, K.O.; Siew, Q.Y.; Aziz Zakry, F.A.; Lan, J.C.W.; Ling, T.C. Overview of citric acid production from Aspergillus niger. Front. Life Sci. 2015, 8, 271–283. [Google Scholar] [CrossRef]
- Zida, A.; Bamba, S.; Yacouba, A.; Ouedraogo-Traore, R.; Guiguemdé, R. Anti-Candida albicans natural products, sources of new antifungal drugs: A review. J. Mycol. Med. 2017, 27, 1–19. [Google Scholar] [CrossRef]
- Akbar, N.; Khan, N.A.; Sagathevan, K.; Iqbal, M.; Tawab, A.; Siddiqui, R. Gut bacteria of Cuora amboinensis (turtle) produce broad-spectrum antibacterial molecules. Sci. Rep. 2019, 9, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Akbar, N.; Siddiqui, R.; Iqbal, M.; Khan, N.A. Antibacterial activities of selected pure compounds isolated from gut bacteria of animals living in polluted environments. Antibiotics 2020, 9, 190. [Google Scholar] [CrossRef]
- Tan, S.Y.; Tatsumura, Y. Alexander Fleming (1881–1955): Discoverer of penicillin. Singap. Med. J. 2015, 56, 366. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.; Guzmán-Trampe, S.; Ávalos, M.; Ruiz, B.; Rodríguez-Sanoja, R.; Jiménez-Estrada, M. “Microbial Natural Products,” in Natural Products in Chemical Biology; Civjan, N., Ed.; Wiley: Hoboken, NJ, USA, 2012; pp. 65–108. [Google Scholar]
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.S.; Lee, S.K.; Hong, J.S.J.; Park, S.R.; Jeong, S.J.; Han, A.R.; Sohng, J.K.; Kim, B.G.; Choi, C.Y.; Sherman, D.H.; et al. Heterologous expression of tylosin polyketide synthase and production of a hybrid bioactive macrolide in Streptomyces venezuelae. Appl. Microbiol. Biotechnol. 2006, 72, 763–769. [Google Scholar] [CrossRef]
- Stanley, V.C.; English, M.P. Some effects of nystatin on the growth of four Aspergillus species. Microbiology 1965, 40, 107–118.x. [Google Scholar] [CrossRef]
- Tevyashova, A.N.; Olsufyeva, E.N.; Solovieva, S.E.; Printsevskaya, S.S.; Reznikova, M.I.; Trenin, A.S.; Galatenko, O.A.; Treshalin, I.D.; Pereverzeva, E.R.; Mirchink, E.P.; et al. Structure-antifungal activity relationships of polyene antibiotics of the amphotericin B group. Antimicrob. Agents Chemother. 2013, 57, 3815–3822. [Google Scholar] [CrossRef]
- Silva, T.R.; Duarte, A.W.; Passarini, M.R.; Ruiz, A.L.T.; Franco, C.H.; Moraes, C.B.; de Melo, I.S.; Rodrigues, R.A.; Fantinatti-Garboggini, F.; Oliveira, V.M. Bacteria from Antarctic environments: Diversity and detection of antimicrobial, antiproliferative, and antiparasitic activities. Polar Biol. 2018, 41, 1505–1519. [Google Scholar] [CrossRef]
- Shiomi, K. Antiparasitic antibiotics from Japan. Parasitol. Int. 2021, 82, 102298. [Google Scholar] [CrossRef]
- Akbar, N.; Siddiqui, R.; Iqbal, M.; Sagathevan, K.; Kim, K.S.; Habib, F.; Khan, N.A. Gut Bacteria of Rattus rattus (Rat) Produce Broad-Spectrum Antibacterial Lipopeptides. ACS Omega 2021, 6, 12261–12273. [Google Scholar] [CrossRef]
- Akbar, N.; Siddiqui, R.; Iqbal, M.; Sagathevan, K.; Khan, N.A. Gut bacteria of cockroaches are a potential source of antibacterial compound (s). Lett. Appl. Microbiol. 2018, 66, 416–426. [Google Scholar] [CrossRef]
- Akbar, N.; Siddiqui, R.; Khamis, M.; Ibrahim, T.; Khan, N.A. Cationic surfactant-natural clay complex as a novel agent against Acanthamoeba castellanii belonging to the T4 genotype. Eye Contact Lens. 2021, 47, 592–597. [Google Scholar] [CrossRef]
- Anwar, A.; Ting, E.L.S.; Anwar, A.; ul Ain, N.; Faizi, S.; Shah, M.R.; Khan, N.A.; Siddiqui, R. Antiamoebic activity of plant-based natural products and their conjugated silver nanoparticles against Acanthamoeba castellanii (ATCC 50492). AMB Express 2020, 10, 24. [Google Scholar] [CrossRef]
- Ali, S.M.; Khan, N.A.; Sagathevan, K.; Anwar, A.; Siddiqui, R. Biologically active metabolite (s) from haemolymph of redheaded centipede Scolopendra subspinipes possess broad-spectrum antibacterial activity. AMB Express 2019, 9, 95. [Google Scholar] [CrossRef]
- Faris, M.; Madkour, M.; Al Bustanji, Y.; Semreen, M.; Giddey, A.D.; Soares, N.C.; Zeb, F. Ramadan diurnal intermittent fasting is associated with significant plasma metabolomics changes in subjects with overweight and obese: A prospective cohort study. Front. Nutr. 2022, 1–17. [Google Scholar]
- Bouyahya, A.; Mechchate, H.; Benali, T.; Ghchime, R.; Charfi, S.; Balahbib, A.; Burkov, P.; Shariati, M.A.; Lorenzo, J.M.; Omari, N.E. Health benefits and pharmacological properties of carvone. Biomolecules 2021, 11, 1803. [Google Scholar] [CrossRef]
- De Sousa, D.P.; De Farias Nóbrega, F.F.; De Almeida, R.N. Influence of the chirality of (R)-(−)-and (S)-(+)-carvone in the central nervous system: A comparative study. Chirality Pharmacol. Biol. Chem. Conseq. Mol. Asymmetry 2007, 19, 264–268. [Google Scholar] [CrossRef]
- Gibka, J.; Kunicka-Styczyńska, A.; Gliński, M. Experimental immunology antimicrobial activity of undecan-3-one, undecan-3-ol and undec-3-yl acetate. Cent. Eur. J. Immunol. 2009, 34, 154–157. [Google Scholar]
- Jarboe, L.R.; Royce, L.A.; Liu, P. Understanding biocatalyst inhibition by carboxylic acids. Front. Microbiol. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed]
- Santamaría, L.; Reverón, I.; de Felipe, F.L.; de Las Rivas, B.; Muñoz, R. Ethylphenol formation by Lactobacillus plantarum: Identification of the enzyme involved in the reduction of vinylphenols. Appl. Environ. Microbiol. 2018, 84, e01064-18. [Google Scholar] [CrossRef] [PubMed]
- Ge, T.; Gao, W.; Liang, C.; Han, C.; Wang, Y.; Xu, Q.; Wang, Q. 4-Ethylphenol, A Volatile Organic Compound Produced by Disease-Resistant Soybean, Is a Potential Botanical Agrochemical Against Oomycetes. Front. Plant Sci. 2021, 12, 717258. [Google Scholar] [CrossRef]
- Cho, J.Y.; Moon, J.H.; Seong, K.Y.; Park, K.H. Antimicrobial activity of 4-hydroxybenzoic acid and trans 4-hydroxycinnamic acid isolated and identified from rice hull. Biosci. Biotechnol. Biochem. 1998, 62, 2273–2276. [Google Scholar] [CrossRef]
- Melliou, E.; Chinou, I. Chemical analysis and antimicrobial activity of Greek propolis. Planta Med. 2004, 70, 515–519. [Google Scholar] [CrossRef]
- Kong, W.; Zhao, Y.; Shan, L.; Xiao, X.; Guo, W. Thermochemical studies on the quantity–antibacterial effect relationship of four organic acids from Radix Isatidis on Escherichia coli growth. Biol. Pharm. Bull. 2008, 31, 1301–1305. [Google Scholar] [CrossRef]
- Alvesalo, J.; Vuorela, H.; Tammela, P.; Leinonen, M.; Saikku, P.; Vuorela, P. Inhibitory effect of dietary phenolic compounds on Chlamydia pneumoniae in cell cultures. Biochem. Pharmacol. 2006, 71, 735–741. [Google Scholar] [CrossRef]
- Kubo, I.; Xiao, P.; Nihei, K.I.; Fujita, K.I.; Yamagiwa, Y.; Kamikawa, T. Molecular design of antifungal agents. J. Agric. Food Chem. 2002, 50, 3992–3998. [Google Scholar] [CrossRef]
- Innocenti, A.; Hall, R.A.; Schlicker, C.; Mühlschlegel, F.A.; Supuran, C.T. Carbonic anhydrase inhibitors. Inhibition of the β-class enzymes from the fungal pathogens Candida albicans and Cryptococcus neoformans with aliphatic and aromatic carboxylates. Bioorg. Med. Chem. 2009, 17, 2654–2657. [Google Scholar] [CrossRef]
- Ngurah, B.I.G.M.; Nyoman, Y.N.; Dafroyati, Y.; Gunadi, I.G.A.; Taneo, M. Antibacterial Evaluation of 2, 4-Dihidroxy Benzoic Acid on Escherichia coli and Vibrio Alginolyticus. J. Phys. Conf. Ser. 2020, 1503, 012027. [Google Scholar] [CrossRef]
- Korneev, S.M. Hydrocinnamic acids: Application and strategy of synthesis. Synthesis 2013, 45, 1000–1015. [Google Scholar] [CrossRef]
- He, Y.H.; Friesen, M.D.; Ruch, R.J.; Schut, H.A.J. Indole-3-carbinol as a chemopreventive agent in 2-amino-1-methyl-6-phenylimidazo [4, 5-b] pyridine (PhIP) carcinogenesis: Inhibition of PhIP–DNA adduct formation, acceleration of PhIP metabolism, and induction of cytochrome P450 in female F344 rats. Food Chem. Toxicol. 2000, 38, 15–23. [Google Scholar] [CrossRef]
- Nachshon-Kedmi, M.; Yannai, S.; Haj, A.; Fares, F.A. Indole-3-carbinol and 3, 3′-diindolylmethane induce apoptosis in human prostate cancer cells. Food Chem. Toxicol. 2003, 41, 745–752. [Google Scholar] [CrossRef]
- Sarkar, F.H.; Li, Y. Indole-3-carbinol and prostate cancer. J. Nutr. 2004, 134, 3493S–3498S. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, T.K.; Rao, G.R. Beta-indoleethanol and beta-indolelactic acid production by Candida species: Their antibacterial and autoantibiotic action. Antimicrob. Agents Chemother. 1976, 9, 375–380. [Google Scholar] [CrossRef]
- Lee, C.H.; Lee, H.S. Growth inhibiting activity of quinaldic acid isolated from Ephedra pachyclada against intestinal bacteria. J. Korean Soc. Appl. Biol. 2009, 52, 331–335. [Google Scholar] [CrossRef]
- Kumar, R.; Chandar, B.; Parani, M. Use of succinic & oxalic acid in reducing the dosage of colistin against New Delhi metallo-β-lactamase-1 bacteria. Indian J. Med. Res. 2018, 147, 97. [Google Scholar]
- De Jonckheere, J.F. Ecology of acanthamoeba. Rev. Infect. Dis. 1991, 13 (Suppl. S5), S385–S387. [Google Scholar] [CrossRef]
- Akbar, N.; Siddiqui, R.; Sagathevan, K.; Iqbal, M.; Khan, N.A. Gut bacteria of water monitor lizard (Varanus salvator) are a potential source of antibacterial compound (s). Antibiotics 2019, 8, 164. [Google Scholar] [CrossRef] [PubMed]
- Soopramanien, M.; Khan, N.A.; Sagathevan, K.; Siddiqui, R. Gut bacteria of Varanus salvator possess potential antitumour molecules. Int. Microbiol. 2021, 24, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Shen, B. A new golden age of natural products drug discovery. Cell 2015, 163, 1297–1300. [Google Scholar]
- Iqbal, J.; Siddiqui, R.; Khan, N.A. Acanthamoeba and bacteria produce antimicrobials to target their counterpart. Parasit. Vectors 2014, 7, 56. [Google Scholar] [CrossRef]
- Huang, S.; Chen, X.; Yan, R.; Huang, M.; Chen, D. Isolation, identification and antibacterial mechanism of the main antibacterial component from pickled and dried mustard (Brassica juncea Coss. Var. foliosa Bailey). Molecules 2022, 27, 2418. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Nisani, S.; Chamovitz, D.A. Indole-3-carbinol: A plant hormone combatting cancer. F1000Research 2018, 7, 689. [Google Scholar] [CrossRef]
- Warth, A.D. Mechanism of action of benzoic acid on Zygosaccharomyces bailii: Effects on glycolytic metabolite levels, energy production, and intracellular pH. Appl. Environ. Microbiol. 1991, 57, 3410–3414. [Google Scholar] [CrossRef]
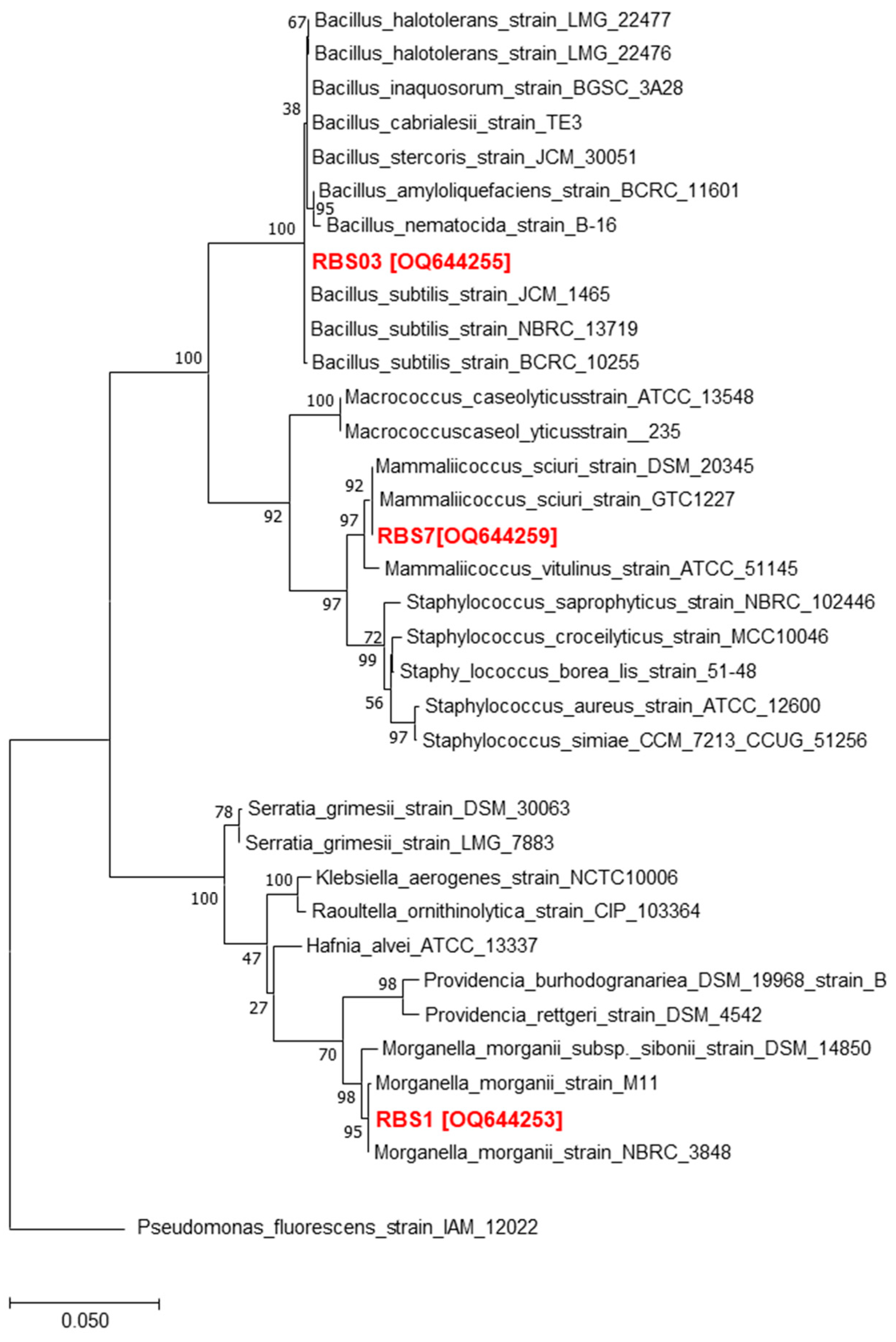
| S. No. | Conditioned Media Code | Bacteria Species | Accession Number |
|---|---|---|---|
| 1. | CM1 | Morganella morganii | OQ644253 |
| 2. | CM2 | Empedobacter brevis | OQ644254 |
| 3. | CM3 | Bacillus subtilis | OQ644255 |
| 4. | CM4 | Pseudomonas otitidis | OQ644256 |
| 5. | CM5 | Pseudoclavibacter alba | OQ644257 |
| 6. | CM6 | Pseudomonas aeruginosa | OQ644258 |
| 7. | CM7 | Mammaliicoccus sciuri | OQ644259 |
| 8. | CM8 | Proteus terrae | OQ644260 |
| 9. | CM9 | Chryseobacterium taklimakanense | OQ644261 |
| 10. | CM10 | Bacillus aerius | OQ644262 |
| 11. | CM11 | Morganella morganii | OQ644263 |
| 12. | CM12 | Corynebacterium atrinae | OQ644264 |
| 13. | CM13 | Mammaliicoccus sciuri | OQ644265 |
| 14. | CM14 | Chryseobacterium taklimakanense | OQ644266 |
| 15. | CM15 | Corynebacterium freneyi | OQ644267 |
| Compounds Names | RT [min] | m/z Meas. | Chemical Formula | Structure | Activity |
|---|---|---|---|---|---|
| (+)-(S)-Carvone | 11.9 | 151.113 | C10H14O |  | Antimicrobial activity (Antibacterial, antifungal, antiparasitic) by damaging plasma membrane [27], neurological activity [28], anti-neuraminidase, antioxidant, anti-inflammatory and anticancer activities [27]. |
| 1,11-Undecanedicarboxylic acid | 10.47 | 245.175 | C13H24O4 | 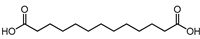 | The analogues exhibited antimicrobial activity [29,30]. |
| 4-Ethylphenol | 9.88 | 123.0804 | C8H10O | 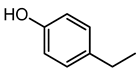 | This is a strong odorant compound produced by Lactobacillus plantarum [31]. This is an agrochemical agent produced by the disease-resistant soybean eliminating fungal infections caused by P. sojae and Phytophthora nicotianae. The volatile compound has potent anti-fungal effects against soil-borne fungi, i.e., Rhizoctonia solani, Gaeumannomyces graminis var tritici and Fusarium graminearum [32]. |
| Benzoic acid | 3.41 | 123.0484 | C7H6O2 | 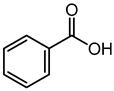 | Benzoic acid alone is considered to be a general-purpose antimicrobial agent with a broad spectrum of actions towards human pathogenic bacteria and fungi with varying minimum inhibitory concentrations (MIC) values [33,34,35,36,37]. Additionally, it was being tested as a potential inhibitor of -carbonic anhydrase, a novel molecular target found in Cryptococcus neoformans and C. albicans [38]. 2,4-dihidroxy benzoic acid showed significant antibacterial activity against Vibrio alginolyticus and E. coli [39]. |
| Hydrocinnamic acid | 5.95 | 151.0728 | C9H10O2 | 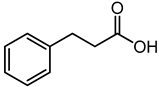 | It belongs to the phenylpropanoids class, having an important role in pharmaceuticals, cosmetics and as a food additive [40]. |
| Indole-3-carbinol | 9.52 | 148.076 | C9H9NO | 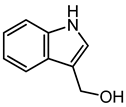 | It is used to inhibit several types of cancers in human and prevent carcinogenesis in animal models [41,42]. The dimeric counterpart of indole-3-carbinol, i.e., 3,3′-diindolylmethane, is a very crucial anti-cancer agent [43]. |
| Indolelactic acid | 7.31 | 206.084 | C11H11NO3 | 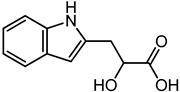 | Candida species produce Indolelactic acid that inhibit several Gram-negative and Gram-positive bacteria [44]. |
| Quinaldic acid | 5.98 | 174.0551 | C10H7NO2 | 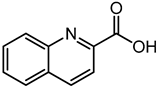 | The Quinaldic acid inhibit the growth of several intestinal bacteria [45]. |
| Succinic acid | 8.36 | 119.0351 | C4H6O4 | 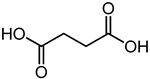 | Succinic acid has a strong antibacterial effect against E. coli [46]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbar, N.; Khan, N.A.; Giddey, A.D.; Soares, N.C.; Alharbi, A.M.; Alfahemi, H.; Siddiqui, R. Selected Gut Bacteria from Water Monitor Lizard Exhibit Effects against Pathogenic Acanthamoeba castellanii Belonging to the T4 Genotype. Microorganisms 2023, 11, 1072. https://doi.org/10.3390/microorganisms11041072
Akbar N, Khan NA, Giddey AD, Soares NC, Alharbi AM, Alfahemi H, Siddiqui R. Selected Gut Bacteria from Water Monitor Lizard Exhibit Effects against Pathogenic Acanthamoeba castellanii Belonging to the T4 Genotype. Microorganisms. 2023; 11(4):1072. https://doi.org/10.3390/microorganisms11041072
Chicago/Turabian StyleAkbar, Noor, Naveed Ahmed Khan, Alexander D. Giddey, Nelson C. Soares, Ahmad M. Alharbi, Hasan Alfahemi, and Ruqaiyyah Siddiqui. 2023. "Selected Gut Bacteria from Water Monitor Lizard Exhibit Effects against Pathogenic Acanthamoeba castellanii Belonging to the T4 Genotype" Microorganisms 11, no. 4: 1072. https://doi.org/10.3390/microorganisms11041072
APA StyleAkbar, N., Khan, N. A., Giddey, A. D., Soares, N. C., Alharbi, A. M., Alfahemi, H., & Siddiqui, R. (2023). Selected Gut Bacteria from Water Monitor Lizard Exhibit Effects against Pathogenic Acanthamoeba castellanii Belonging to the T4 Genotype. Microorganisms, 11(4), 1072. https://doi.org/10.3390/microorganisms11041072








