First Detection and Genetic Identification of Wolbachia Endosymbiont in Field-Caught Aedes aegypti (Diptera: Culicidae) Mosquitoes Collected from Southern Taiwan
Abstract
:1. Introduction
2. Materials and Methods
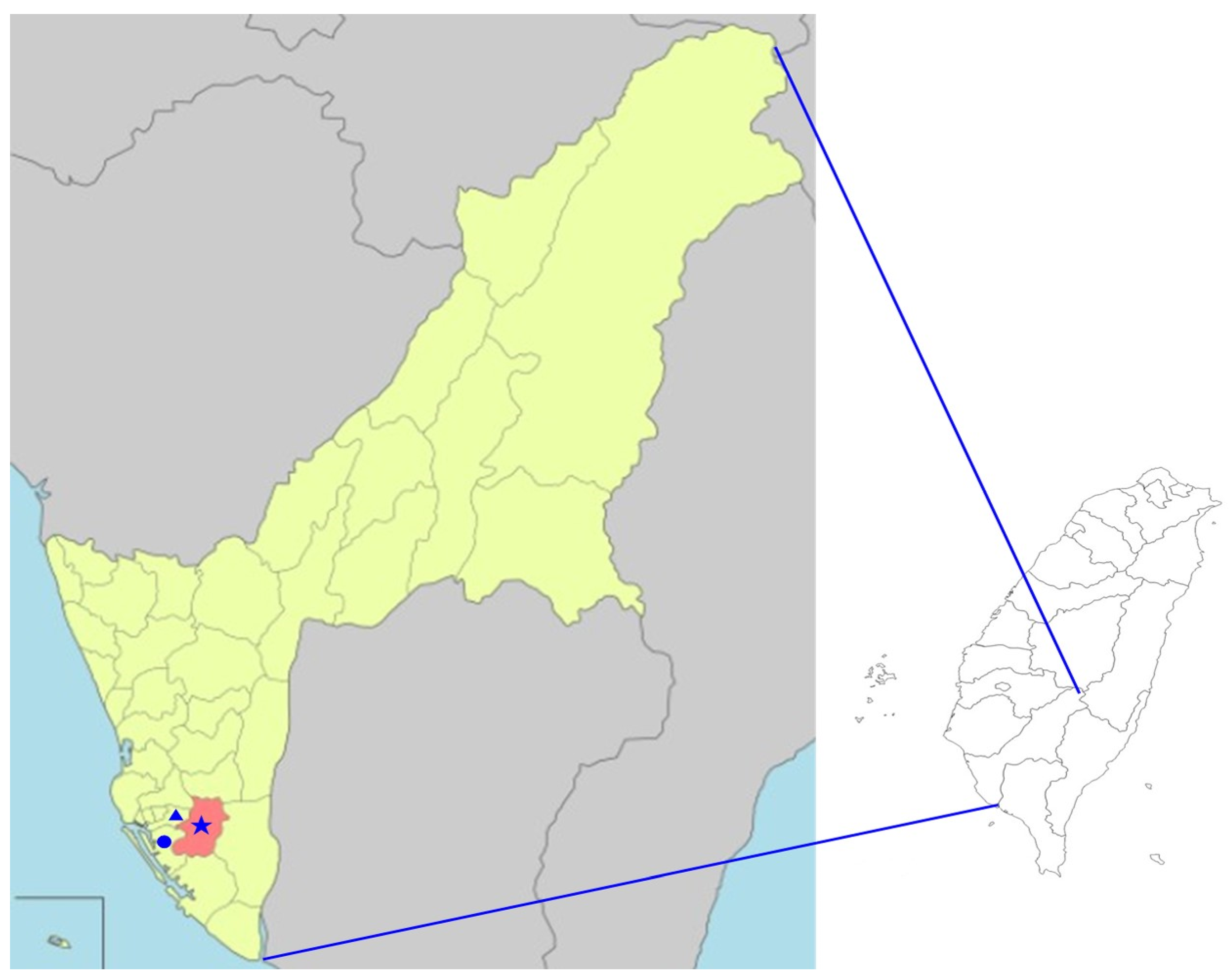
2.1. Field Collection and Genetic Identification of Mosquito Specimens
2.2. DNA Extraction from Mosquito Specimens
2.3. Wolbachia DNA Amplification via Nested Polymerase Chain Reaction (nPCR)
2.4. Genetic Identification of Mosquito Species
2.5. Phylogenetic Analysis Based on Wolbachia wsp Gene
2.6. Nucleotide Sequence Accession Numbers
3. Results
3.1. Detection of Wolbachia in Field-Caught Ae. aegypti Mosquitoes
3.2. Genetic Analysis of Wolbachia Detected in Field-Caught Ae. aegypti Mosquitoes
3.3. Phylogenetic Analysis of Wolbachia Detected in Field-Caught Ae. aegypti Mosquitoes
3.4. Molecular Identification of Field-Caught Ae. aegypti Mosquitoes of Taiwan
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Werren, J.H. Biology of Wolbachia. Ann. Rev. Entomol. 1997, 42, 587–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tram, U.; Sullivan, W. Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 2002, 296, 1124–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandi, C.; Anderson, T.J.; Genchi, C.; Blaxter, M.L. Phylogeny of Wolbachia in filarial nematodes. Proc. R. Soc. Lond. Biol. Sci. 1998, 265, 2407–2413. [Google Scholar] [CrossRef] [PubMed]
- Fischer, P.; Schmetz, C.; Bandi, C.; Bonow, I.; Mand, S.; Fischer, K.; Buttner, D.W. Tunga penetrans: Molecular identification of Wolbachia endobacteria and their recognition by antibodies against proteins of endobacteria from filarial parasites. Exp. Parasitol. 2002, 102, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Bazzocchi, C.; Jamnongluk, W.; O’Neill, S.L.; Anderson, T.J.C.; Genchi, C.; Bandi, C. Wsp gene sequences from the Wolbachia of filarial nematodes. Cur. Microbiol. 2000, 41, 96–100. [Google Scholar] [CrossRef]
- Hertig, M.; Wolbach, S.B. Studies on Rickettsia-Like Micro-Organisms in Insects. J. Med. Res. 1924, 44, 329–374.7. [Google Scholar]
- Hilgenboecker, K.; Hammerstein, P.; Schlattmann, P.; Telschow, A.; Werren, J.H. How many species are infected with Wolbachia?—A statistical analysis of current data. FEMS Microbiol. Lett. 2008, 281, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Jeyaprakash, A.; Hoy, M.A. Long PCR improves Wolbachia DNA amplification: Wsp sequences found in 76% of sixty-three arthropod species. Insect Mol. Biol. 2000, 9, 393–405. [Google Scholar] [CrossRef]
- Rasgon, J.L. Wolbachia induces male-specific mortality in the mosquito Culex pipiens (LIN strain). PLoS ONE 2012, 7, e30381. [Google Scholar] [CrossRef]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Laven, H. Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature 1967, 216, 383–384. [Google Scholar] [CrossRef]
- Aliota, M.T.; Peinado, S.A.; Velez, I.D.; Osorio, J.E. The wMel strain of Wolbachia reduces transmission of zika virus by Aedes aegypti. Sci. Rep. 2016, 6, 28792. [Google Scholar] [CrossRef] [Green Version]
- Aliota, M.T.; Walker, E.C.; Yepes, A.; Velez, I.D.; Christensen, B.M.; Osorio, J.E. The wMel Strain of Wolbachia reduces transmission of chikungunya virus in Aedes aegypti. PLoS Negl. Trop. Dis. 2016, 10, e0004677. [Google Scholar] [CrossRef] [Green Version]
- Nazni, W.A.; Hoffmann, A.A.; NoorAfizah, A.; Cheong, Y.L.; Mancini, M.V.; Golding, N.; Kamarul, G.M.; Arif, M.A.; Thohir, H.; NurSyamimi, H.; et al. Establishment of Wolbachia strain wAlbB in Malaysian populations of Aedes aegypti for dengue control. Curr. Biol. 2019, 29, 4241–4248. [Google Scholar] [CrossRef] [Green Version]
- Frentiu, F.D.; Zakir, T.; Walker, T.; Popovici, J.; Pyke, A.T.; van den Hurk, A.; McGraw, E.A.; O’Neill, S.C. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Negl. Trop. Dis. 2014, 8, e2688. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, A.A.; Montgomery, B.L.; Popovici, J.; Iturbe-Ormaetxe, I.; Johnson, P.H.; Muzzi, F.; Greenfield, M.; Durkan, M.; Leong, Y.S.; Dong, Y.; et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 2011, 476, 454–457. [Google Scholar] [CrossRef]
- McMenimen, C.J.; Lane, R.V.; Cass, B.N.; Fong, A.W.; Sidhu, M.; Wang, Y.F.; O’Neill, S.L. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 2009, 323, 141–144. [Google Scholar] [CrossRef] [Green Version]
- Moreira, L.A.; Iturbe-Ormaetxe, I.; Jeffery, J.A.; Lu, G.; Pyke, A.T.; Hedges, L.M.; Rocha, B.C.; Hall-Mendelin, S.; Day, A.; Riegler, M.; et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 2009, 139, 1268–1278. [Google Scholar] [CrossRef] [Green Version]
- Bian, G.; Xu, Y.; Lu, P.; Xie, Y.; Xi, Z. The endosymbiotic bacterium Wolbachia induces resistence to dengue virus in Aedes aegypti. PLoS Path. 2010, 6, e1000833. [Google Scholar] [CrossRef] [Green Version]
- Hancock, P.A.; Sinkins, S.P.; Godfray, H.C. Strategies for introducing Wolbachia to reduce transmission of mosquito-borne diseases. PLoS Negl. Trop. Dis. 2011, 5, e1024. [Google Scholar] [CrossRef]
- Walker, T.J.P.H.; Johnson, P.H.; Moreira, L.A.; Iturbe-Ormaetxe, I.; Frentiu, F.D.; McMeniman, C.J.; Leong, Y.S.; Dong, Y.; Axford, J.; Kriesner, P.; et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 2011, 476, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Turelli, M. Wolbachia versus dengue: Evolutionary forecasts. Evol. Med. Public Health 2013, 2013, 197–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benelli, G.; Mehlhom, H. Declining malaria, rising of dengue and zika virus: Insights for mosquito vector control. Parasitol. Res. 2016, 115, 1747–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kittayapong, P.; Baisley, K.J.; Baimai, V.; O’Neill, S.L. Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae). J. Med. Entomol. 2000, 37, 340–345. [Google Scholar] [CrossRef]
- Ricci, I.; Cancrini, G.; Gabrielli, S.; D’amelio, S.; Favia, G. Searching for Wolbachia (Rickettsiales, Rickettsiaceae) in mosquitoes (Diptera: Culicidae): Large polymerase chain reaction survey and new identifications. J. Med. Entomol. 2002, 39, 562–567. [Google Scholar] [CrossRef]
- Gloria-Soria, A.; Chiodo, T.G.; Powell, J.R. Lack of evidence for natural Wolbachia infections in Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 2018, 55, 1354–1356. [Google Scholar] [CrossRef]
- Teo, C.H.; Lim, P.K.; Voon, K.; Mak, J.W. Detection of dengue viruses and Wolbachia in Aedes aegypti and Aedes albopictus larvae from four urban localities in Kuala Lumpur, Malaysia. Trop. Biomed. 2017, 34, 583–597. [Google Scholar]
- Carvajal, T.M.; Hashimoto, K.; Harnandika, R.K.; Amalin, D.M.; Watanabe, K. Detection of Wolbachia in field-collected Aedes aegypti mosquitoes in metropolitan Manila, Philippines. Parasit. Vectors 2019, 12, 361. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, A.; Yu, W.; Jiang, J.; Sanchez, C.; Karna, A.K.; Martinezk, K.J.L.; Hanley, K.A.; Buenemann, M.; Hansen, I.A.; Xue, R.D.; et al. Wolbachia pipientis in Aedes aegypti populations in New Mexico and Florida, USA. Ecol. Evol. 2019, 9, 6148–6156. [Google Scholar] [CrossRef] [Green Version]
- Balaji, S.; Jayachandran, S.; Prabagaran, S.R. Evidence for the natural occurrence of Wolbachia in Aedes aegypti mosquitoes. FEMS Microbiol. Lett. 2019, 366, fnz055. [Google Scholar] [CrossRef]
- Benson, M.J.; Gawronski, J.D.; Eveleigh, D.E.; Benson, D.R. Intracellular symbionts and other bacteria associated with deer ticks (Ixodes scapularis) from Nantucket and Wellfleet, Cape Cod, Massachusetts. Appl. Environ. Microbiol. 2004, 70, 616–620. [Google Scholar] [CrossRef] [Green Version]
- Bordenstein, S.; Rosengaus, R.B. Discovery of a novel Wolbachia super group in Isoptera. Curr. Microbiol. 2005, 51, 393–398. [Google Scholar] [CrossRef]
- Andreotti, R.; Perez, A.A.; Scoles, G.A. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 2011, 11, 6–9. [Google Scholar] [CrossRef] [Green Version]
- Carpi, G.; Cagnacci, F.; Wittekindt, N.E.; Zhao, F.; Qi, J.; Tomsho, L.P.; Drautz, D.I.; Rizzoli, A.; Schuster, S.C. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS ONE 2011, 6, e25604. [Google Scholar] [CrossRef]
- Bing, X.L.; Xia, W.Q.; Gui, J.D.; Yan, G.H.; Wang, X.W.; Liu, S.S. Diversity and evolution of the Wolbachia endosymbionts of Bemisia (Hemiptera: Aleyrodidae) whiteflies. Ecol. Evol. 2014, 4, 2714–2737. [Google Scholar] [CrossRef]
- Zhou, W.; Rousset, F.; O’Neil, S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. Lond. Biol. Sci. 1998, 265, 509–515. [Google Scholar] [CrossRef]
- Ruang-Areerate, T.; Kittayapong, P.; Baimai, V.; O’Neill, S.L. Molecular phylogeny of Wolbachia endosymbionts in Southeast Asian mosquitoes (Diptera: Culicidae) based on wsp gene sequences. J. Med. Entomol. 2003, 40, 1–5. [Google Scholar] [CrossRef]
- Chai, H.N.; Du, Y.Z.; Qiu, B.L.; Zhai, B.P. Detection and phylogenetic analysis of Wolbachia in the Asiatic rice leafroller, Cnaphalocrocis medinalis, in Chinese populations. J. Insect Sci. 2011, 11, 123. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Norris, D.E.; Rasgon, J.L. Distribution and molecular characterization of Wolbachia endosymbionts and filarial nematodes in Maryland populations of the lone star tick (Amblyomma americanum). FEMS Microbiol. Ecol. 2011, 77, 50–56. [Google Scholar] [CrossRef]
- Wang, G.H.; Jia, L.Y.; Xiao, J.H.; Huang, D.W. Discovery of a new Wolbachia supergroup in cave spider species and the lateral transfer of phage WO among distant hosts. Infect. Genet. Evol. 2016, 41, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werren, J.H.; Zhang, W.; Guo, L.R. Evolution and phylogeny of Wolbachia: Reproductive parasites of arthropods. Proc. R. Soc. Lond. Biol. Sci. 1995, 261, 55–63. [Google Scholar]
- WHO. Pictorial Identification Key of Important Disease Vectors in the WHO South-East Asia Region; WHO: Geneva, Switzerland, 2020; ISBN 978-92-9022-758-8. [Google Scholar]
- Chan, A.; Chiang, L.P.; Hapuarachchi, H.C.; Tan, C.H.; Pang, S.C.; Lee, R.; Lee, K.S.; Ng, L.C.; Lam-Phua, S.G. DNA barcoding: Complementing morphological identification of mosquito species in Singapore. Parasit. Vectors 2014, 7, 569. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Bio. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 52, 1119–1134. [Google Scholar]
- Coon, K.L.; Brown, M.R.; Strand, M.R. Mosquitoes host communities of bacterial that are essential for development but vary greatly between local habitats. Mol. Ecol. 2016, 25, 5806–5826. [Google Scholar] [CrossRef] [Green Version]
- Thongsripong, P.; Chandler, J.A.; Green, A.B.; Kittayapong, P.; Wilcox, B.A.; Kapan, D.D.; Bennett, S.N. Mosquito vector-associated microbiota: Metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecol. Evol. 2018, 8, 1352–1368. [Google Scholar] [CrossRef]
- Atyame, C.M.; Labbe, P.; Dumas, E.; Milesi, P.; Charlat, S.; Fort, P.; Weill, M. Wolbachia divergence and the evolution of cytoplasmic incompatibility in Culex pipiens. PLoS ONE 2014, 9, e87336. [Google Scholar] [CrossRef] [Green Version]
- Ravikumar, H.; Ramachandraswamy, N.; Sampathkumar, S.; Prakash, B.M.; Huchesh, H.C.; Uday, J.; Puttaraju, H.P. A preliminary survey for Wolbachia and bacteriophage WO infection in Indian mosquitoes (Diptera: Culicidae). Trop. Biomed. 2010, 27, 384–393. [Google Scholar]
- Zheng, X.; Zhang, D.; Li, Y.; Yang, C.; Wu, Y.; Liang, X.; Liang, Y.; Pan, X.; Hu, L.; Sun, Q.; et al. Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature 2019, 572, 56–61. [Google Scholar] [CrossRef]
- Crawford, J.E.; Clarke, D.W.; Criswell, V.; Desnoyer, M.; Cornel, D.; Deegan, B.; Gong, K.; Hopkins, K.C.; Howell, P.; Hyde, J.S.; et al. Efficient production of male Wolbachia infected Aedes aegypti mosquitoes enables large-scale suppression of wild populations. Nat. Biotechnol. 2020, 38, 482–492. [Google Scholar] [CrossRef] [Green Version]
- National Environment Agency, Singapore. Project Wolbachia Singapore. Available online: https://www.nea.gov.sg/corporate-functions/resources/research/wolbachia-aedes-mosquito-suppression-strategy (accessed on 15 June 2023).
- Caputo, B.; Moretti, R.; Manica, M.; Serini, P.; Lampazzi, E.; Bonanni, M.; Fabbri, G.; Pichler, V.; della Torre, A. A bacterium against the tiger: Preliminary evidence of fertility reduction after release of Aedes albopictus males with manipulated Wolbachia infection in an Italian urban area. Pest Manag. Sci. 2020, 76, 1324–1332. [Google Scholar] [CrossRef]
- Dengue Fever in Taiwan. CDC, Taiwan. Available online: https://www.cdc.gov.tw/Disease/SubIndex/WYbKe3aE7LiY5gb-eA8PBw (accessed on 15 June 2023).
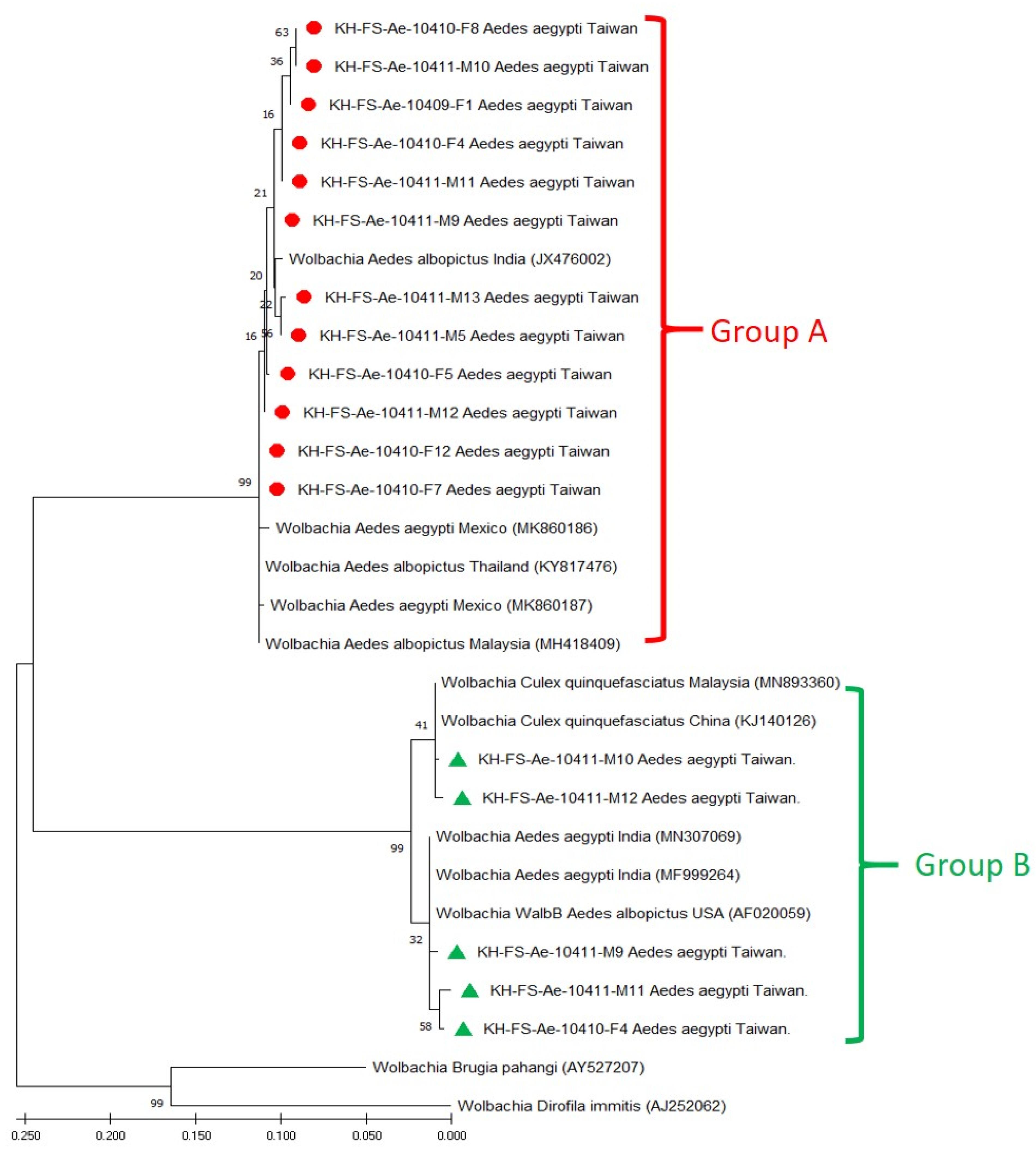
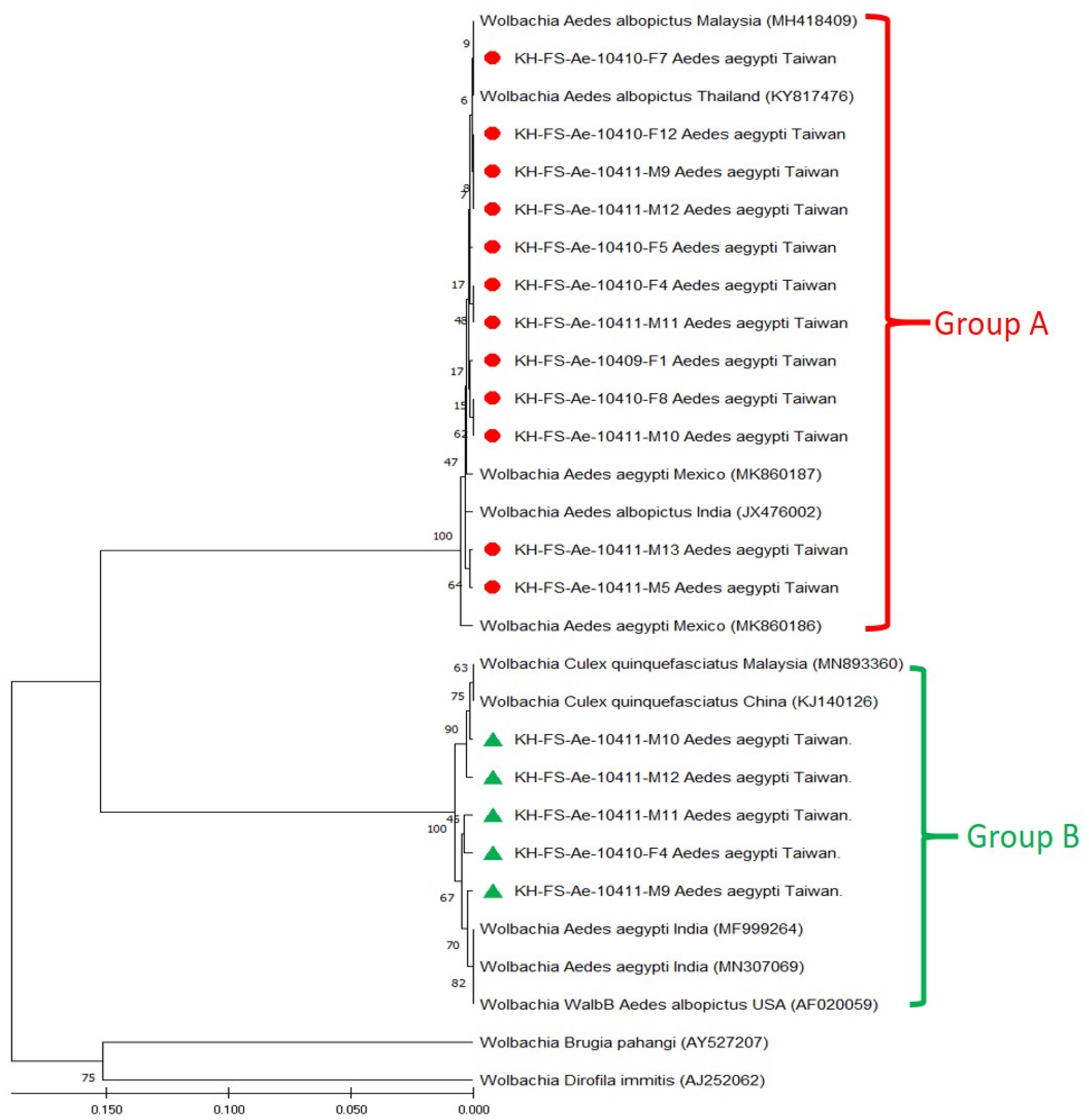

| Genogroup and Strain | Origin of Wolbachia Strain | wsp Gene | |
|---|---|---|---|
| Biological | Geographic | Accession Number a | |
| Supergroup A | |||
| KH-FS-Ae-10409-F1 | Aedes aegypti | Kaohsiung, Taiwan | OP882272 |
| KH-FS-Ae-10410-F4 | Aedes aegypti | Kaohsiung, Taiwan | OP882273 |
| KH-FS-Ae-10410-F5 | Aedes aegypti | Kaohsiung, Taiwan | OP882274 |
| KH-FS-Ae-10410-F7 | Aedes aegypti | Kaohsiung, Taiwan | OP882275 |
| KH-FS-Ae-10410-F8 | Aedes aegypti | Kaohsiung, Taiwan | OP882276 |
| KH-FS-Ae-10410-F12 | Aedes aegypti | Kaohsiung, Taiwan | OP882277 |
| KH-FS-Ae-10411-M5 | Aedes aegypti | Kaohsiung, Taiwan | OP882278 |
| KH-FS-Ae-10411-M9 | Aedes aegypti | Kaohsiung, Taiwan | OP882279 |
| KH-FS-Ae-10411-M10 | Aedes aegypti | Kaohsiung, Taiwan | OP882280 |
| KH-FS-Ae-10411-M11 | Aedes aegypti | Kaohsiung, Taiwan | OP882281 |
| KH-FS-Ae-10411-M12 | Aedes aegypti | Kaohsiung, Taiwan | OP882282 |
| KH-FS-Ae-10411-M13 | Aedes aegypti | Kaohsiung, Taiwan | OP882283 |
| wol 5/Odisha | Aedes albopictus | India | JX476002 |
| AlbA03 | Aedes aegypti | Mexico | MK860186 |
| AP66-1W | Aedes albopictus | Thailand | KY817476 |
| wAlbA04 | Aedes aegypti | Mexico | MK860187 |
| F8A | Aedes albopictus | Malaysia | MH418409 |
| Supergroup B | |||
| KH-FS-Ae-10410-F4 | Aedes aegypti | Kaohsiung, Taiwan | OP896740 |
| KH-FS-Ae-10411-M9 | Aedes aegypti | Kaohsiung, Taiwan | OP896741 |
| KH-FS-Ae-10411-M10 | Aedes aegypti | Kaohsiung, Taiwan | OP896742 |
| KH-FS-Ae-10411-M11 | Aedes aegypti | Kaohsiung, Taiwan | OP896743 |
| KH-FS-Ae-10411-M12 | Aedes aegypti | Kaohsiung, Taiwan | OP896744 |
| Kla6 | Culex quinquefasciatus | Malaysia | MN893360 |
| GD13098 | Culex quinquefasciatus | China | KJ140126 |
| wAegB | Aedes aegypti | India | MN307069 |
| wAegB | Aedes aegypti | India | MF999264 |
| WalbB | Aedes albopictus | USA | AF020059 |
| Outgroup | |||
| TRS | Brugia pahangi | USA | AY527207 |
| unknown | Dirofilaria immitis | Italy | AJ252062 |
| Sex of Mosquito | No. Examined | Wolbachia-Group Infection | Total | |||
|---|---|---|---|---|---|---|
| A | B | A&B | No. Infected | % Infection | ||
| Female | 395 | 6 | 1 | 1 | 8 | 2.0 |
| Male | 270 | 6 | 4 | 4 | 14 | 5.2 |
| Total | 665 | 12 | 5 | 5 | 22 | 3.3 |
| (%) | (1.8) | (0.8) | (0.8) | |||
| Wolbachia Strains b | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. Wolbachia Thailand (KY817476) | — | ||||||||||||||||
| 2. Wolbachia Malaysia (MH418409) | 0.000 | — | |||||||||||||||
| 3. Wolbachia India (JX476002) | 0.000 | 0.000 | — | ||||||||||||||
| 4. KH-FS-Ae-10410-F12 (Taiwan) | 0.000 | 0.000 | 0.000 | — | |||||||||||||
| 5. KH-FS-Ae-10410-F7 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | — | ||||||||||||
| 6. KH-FS-Ae-10411-M9 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | — | |||||||||||
| 7. KH-FS-Ae-10410-F8 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | — | ||||||||||
| 8. KH-FS-Ae-10410-F4 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | — | |||||||||
| 9. KH-FS-Ae-10410-F5 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | — | ||||||||
| 10. KH-FS-Ae-10411-M11 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | — | |||||||
| 11. KH-FS-Ae-10411-M12 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | — | ||||||
| 12. KH-FS-Ae-10409-F1 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | — | |||||
| 13. KH-FS-Ae-10411-M10 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | — | ||||
| 14. KH-FS-Ae-10411-M13 (Taiwan) | 0.006 | 0.000 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | — | |||
| 15. KH-FS-Ae-10411-M5 (Taiwan) | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.000 | — | ||
| 16. Wolbachia India (MF999264) | 0.518 | 0.518 | 0.518 | 0.518 | 0.518 | 0.518 | 0.498 | 0.518 | 0.518 | 0.518 | 0.518 | 0.518 | 0.518 | 0.532 | 0.532 | — | |
| 17. Wolbachia B. pahangi (AY527207) | 0.788 | 0.788 | 0.788 | 0.788 | 0.788 | 0.788 | 0.793 | 0.788 | 0.788 | 0.788 | 0.788 | 0.788 | 0.788 | 0.804 | 0.804 | 1.093 | — |
| Wolbachia Strains b | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. Wolbachia WalbB USA (AF020059) | — | ||||||||||||
| 2. Wolbachia India (MF999264) | 0.000 | — | |||||||||||
| 3. Wolbachia India (MN307069) | 0.000 | 0.000 | — | ||||||||||
| 4. KH-FS-Ae-10411-M9 (Taiwan) | 0.000 | 0.000 | 0.000 | — | |||||||||
| 5. KH-FS-Ae-10410-F4 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | — | ||||||||
| 6. KH-FS-Ae-10411-M11 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | — | |||||||
| 7. KH-FS-Ae-10411-M10 (Taiwan) | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | — | ||||||
| 8. KH-FS-Ae-10411′-M12 (Taiwan) | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.000 | — | |||||
| 9. Wolbachia China (KJ140126) | 0.018 | 0.018 | 0.018 | 0.008 | 0.008 | 0.008 | 0.000 | 0.000 | — | ||||
| 10. Wolbachia Malaysia (MN893360) | 0.018 | 0.018 | 0.018 | 0.008 | 0.008 | 0.008 | 0.000 | 0.000 | 0.000 | — | |||
| 11. Wolbachia Thailand (KY817476) | 0.417 | 0.417 | 0.417 | 0.302 | 0.302 | 0.302 | 0.286 | 0.286 | 0.415 | 0.415 | — | ||
| 12. Wolbachia B. pahangi (AY527207) | 0.768 | 0.768 | 0.768 | 0.527 | 0.527 | 0.527 | 0.515 | 0.515 | 0.677 | 0.677 | 0.559 | — | |
| 13. Wolbachia D. immitis (AJ252062) | 0.602 | 0.602 | 0.602 | 0.482 | 0.482 | 0.482 | 0.480 | 0.480 | 0.574 | 0.574 | 0.681 | 0.369 | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chao, L.-L.; Shih, C.-M. First Detection and Genetic Identification of Wolbachia Endosymbiont in Field-Caught Aedes aegypti (Diptera: Culicidae) Mosquitoes Collected from Southern Taiwan. Microorganisms 2023, 11, 1911. https://doi.org/10.3390/microorganisms11081911
Chao L-L, Shih C-M. First Detection and Genetic Identification of Wolbachia Endosymbiont in Field-Caught Aedes aegypti (Diptera: Culicidae) Mosquitoes Collected from Southern Taiwan. Microorganisms. 2023; 11(8):1911. https://doi.org/10.3390/microorganisms11081911
Chicago/Turabian StyleChao, Li-Lian, and Chien-Ming Shih. 2023. "First Detection and Genetic Identification of Wolbachia Endosymbiont in Field-Caught Aedes aegypti (Diptera: Culicidae) Mosquitoes Collected from Southern Taiwan" Microorganisms 11, no. 8: 1911. https://doi.org/10.3390/microorganisms11081911
APA StyleChao, L.-L., & Shih, C.-M. (2023). First Detection and Genetic Identification of Wolbachia Endosymbiont in Field-Caught Aedes aegypti (Diptera: Culicidae) Mosquitoes Collected from Southern Taiwan. Microorganisms, 11(8), 1911. https://doi.org/10.3390/microorganisms11081911








