Evidence of Bacterial Community Coalescence between Freshwater and Discharged tpm-Harboring Bacterial Taxa from Hospital and Domestic Wastewater Treatment Plants among Epilithic Biofilms
Abstract
:1. Introduction
2. Materials and Methods
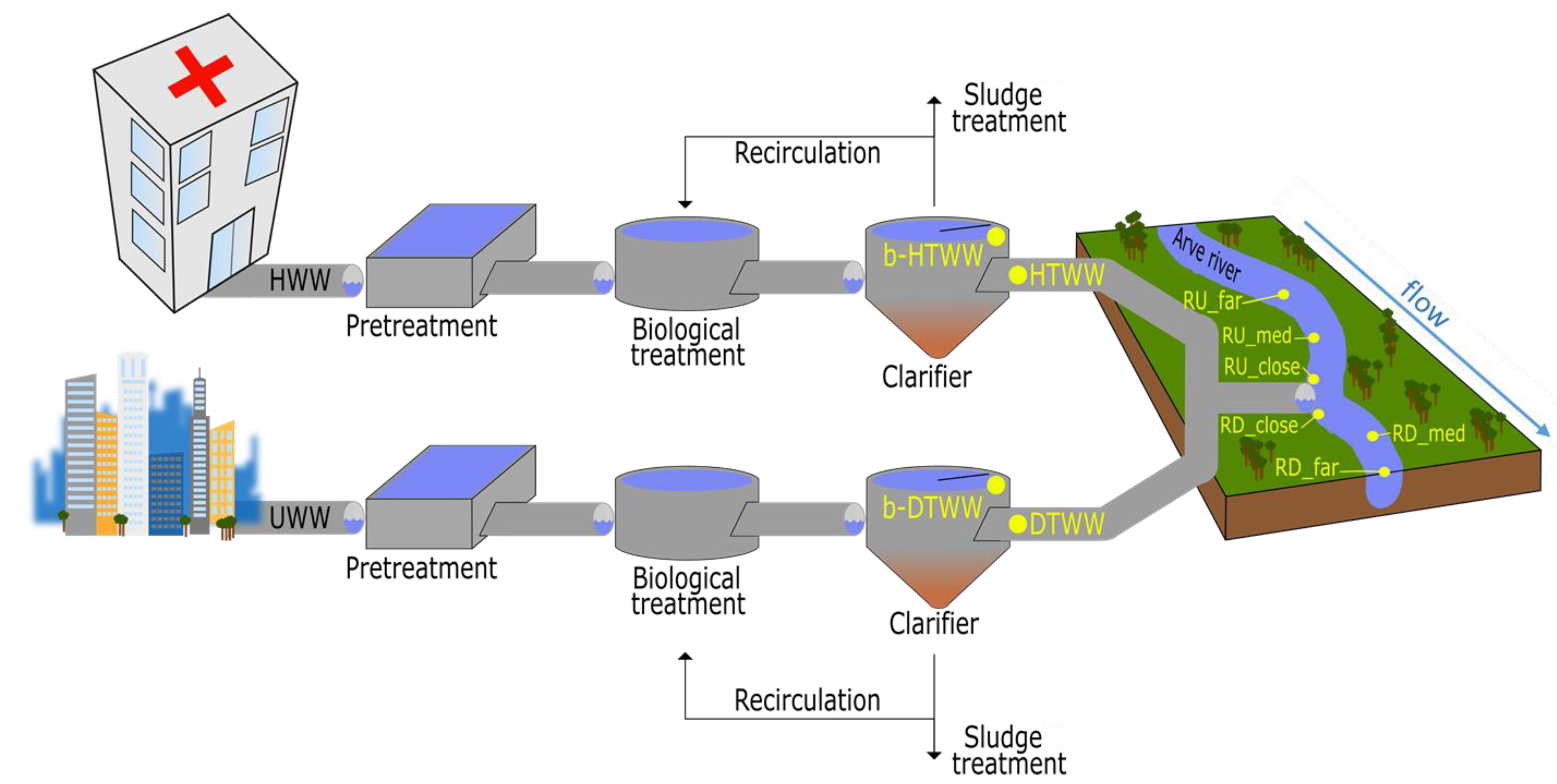
2.1. Study Site
2.2. Sampling Strategy
2.3. Physico-Chemichal Analyses
2.4. Bacterial Counts and DNA Extractions
2.5. The tpm Metabarcoding Analytical Scheme
2.6. Statistical Analyses
2.7. Pseudomonas aeruginosa Isolation and Antibiotic Resistance Assays
3. Results
3.1. Monitoring of Pharmaceutical Loads, Physico-chemical, and Classical Microbial Parameters
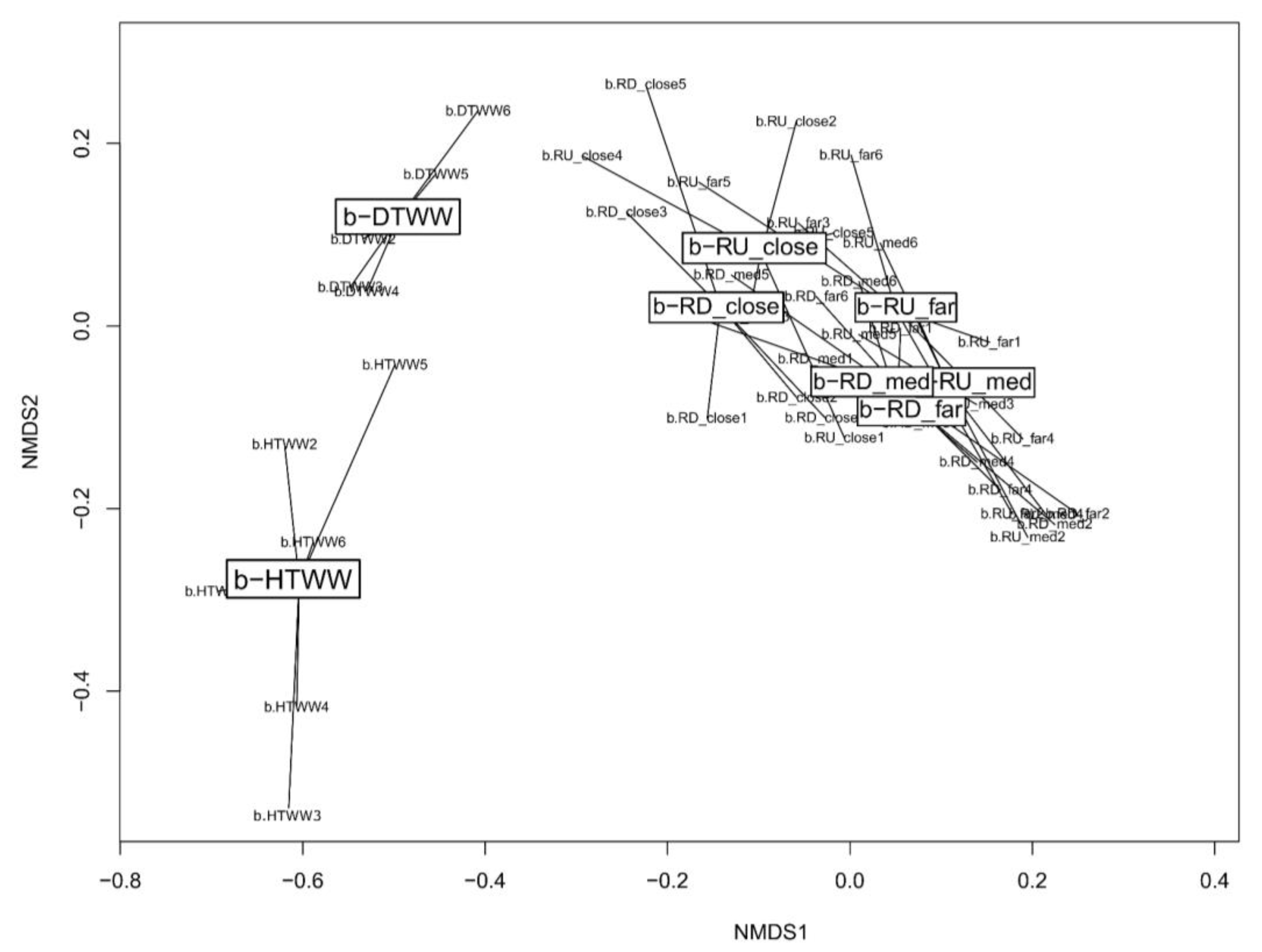
3.2. Genetic Structure of tpm Bacterial Communities among Biofilms
3.2.1. General Features
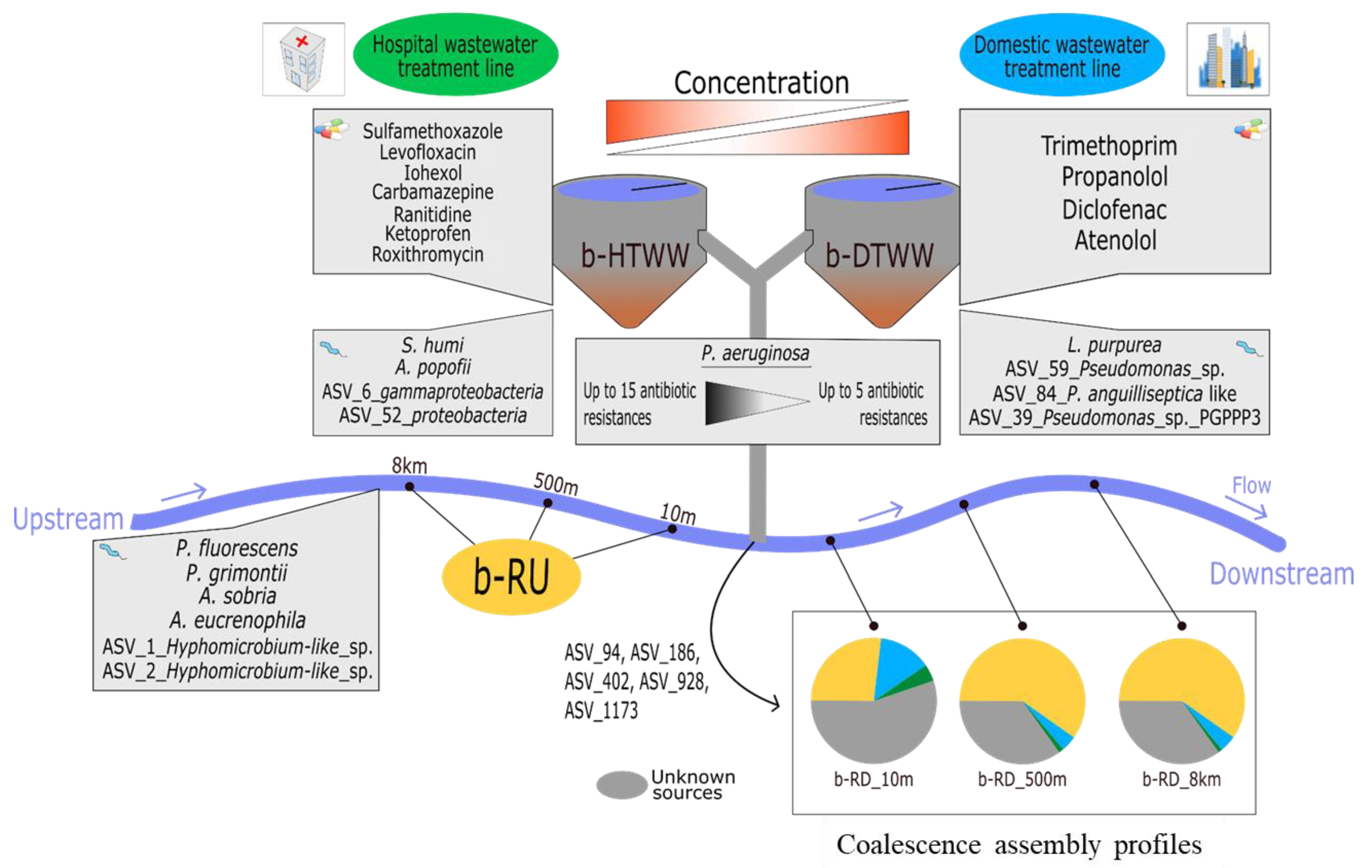
3.2.2. tpm ASV Specific of Biofilms Generated from DTWW and/or HTWW
3.2.3. Relationships between the Relative Content of tpm-Harboring Species and Pharmaceuticals among b-HTWW and b-DTWW
3.3. Coalescence of b-DTWW and b-HTWW tpm Bacterial Communities among Epilithic Biofilms
3.4. Antibiotic Resistance of P. aeruginosa Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deblonde, T.; Cossu-Leguille, C.; Hartemann, P. Emerging Pollutants in Wastewater: A Review of the Literature. Int. J. Hyg. Environ. Health 2011, 214, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Bootsma, M.J.; Morrison, H.G.; Sogin, M.L.; McLellan, S.L. A Microbial Signature Approach to Identify Fecal Pollution in the Waters Off an Urbanized Coast of Lake Michigan. Microb. Ecol. 2013, 65, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- McLellan, S.L.; Fisher, J.C.; Newton, R.J. The Microbiome of Urban Waters. Int. Microbiol. 2015, 18, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Cydzik-Kwiatkowska, A.; Zielińska, M. Bacterial Communities in Full-Scale Wastewater Treatment Systems. World J. Microbiol. Biotechnol. 2016, 32, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stalder, T.; Alrhmoun, M.; Louvet, J.-N.; Casellas, M.; Maftah, C.; Carrion, C.; Pons, M.-N.; Pahl, O.; Ploy, M.-C.; Dagot, C. Dynamic Assessment of the Floc Morphology, Bacterial Diversity, and Integron Content of an Activated Sludge Reactor Processing Hospital Effluent. Environ. Sci. Technol. 2013, 47, 7909–7917. [Google Scholar] [CrossRef] [PubMed]
- Chonova, T.; Keck, F.; Labanowski, J.; Montuelle, B.; Rimet, F.; Bouchez, A. Separate Treatment of Hospital and Urban Wastewaters: A Real Scale Comparison of Effluents and Their Effect on Microbial Communities. Sci. Total Environ. 2016, 542, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Escher, B.I.; Baumgartner, R.; Koller, M.; Treyer, K.; Lienert, J.; McArdell, C.S. Environmental Toxicology and Risk Assessment of Pharmaceuticals from Hospital Wastewater. Water Res. 2011, 45, 75–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barceló, D.; Hennion, M.-C. Trace Determination of Pesticides and Their Degradation Products in Water; Elsevier Science: Amsterdam, The Netherlands; New York, NY, USA, 1997; ISBN 978-0-08-054312-3. [Google Scholar]
- Fuchs, S.; Haritopoulou, T.; Schäfer, M.; Wilhelmi, M. Heavy Metals in Freshwater Ecosystems Introduced by Urban Rainwater Runoff—Monitoring of Suspended Solids, River Sediments and Biofilms. Water Sci. Technol. 1997, 36, 277–282. [Google Scholar] [CrossRef]
- Göbel, P.; Dierkes, C.; Coldewey, W.G. Storm Water Runoff Concentration Matrix for Urban Areas. J. Contam. Hydrol. 2007, 91, 26–42. [Google Scholar] [CrossRef]
- Revitt, D.M.; Lundy, L.; Coulon, F.; Fairley, M. The Sources, Impact and Management of Car Park Runoff Pollution: A Review. J. Environ. Manag. 2014, 146, 552–567. [Google Scholar] [CrossRef] [Green Version]
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilbäck, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. PLoS Pathog. 2011, 7, e1002158. [Google Scholar] [CrossRef] [Green Version]
- Tello, A.; Austin, B.; Telfer, T.C. Selective Pressure of Antibiotic Pollution on Bacteria of Importance to Public Health. Environ. Health Perspect. 2012, 120, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Chagas, T.P.G.; Seki, L.M.; Cury, J.C.; Oliveira, J.A.L.; Dávila, A.M.R.; Silva, D.M.; Asensi, M.D. Multiresistance, Beta-Lactamase-Encoding Genes and Bacterial Diversity in Hospital Wastewater in Rio de Janeiro, Brazil: Resistant Bacteria in Hospital Sewage. J. Appl. Microbiol. 2011, 111, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Varela, A.R.; André, S.; Nunes, O.C.; Manaia, C.M. Insights into the Relationship between Antimicrobial Residues and Bacterial Populations in a Hospital-Urban Wastewater Treatment Plant System. Water Res. 2014, 54, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of Antibiotic-Resistant Bacteria and Their Resistance Genes in Wastewater, Surface Water, and Drinking Water Biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of Antibiotics and Antibiotic Resistance Genes in Hospital and Urban Wastewaters and Their Impact on the Receiving River. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef]
- Shapiro, J. Mobile Genetic Elements; Elsevier Science: Burlington, VT, USA, 2012; ISBN 978-0-12-638680-6. [Google Scholar]
- Su, J.-Q.; An, X.-L.; Li, B.; Chen, Q.-L.; Gillings, M.R.; Chen, H.; Zhang, T.; Zhu, Y.-G. Metagenomics of Urban Sewage Identifies an Extensively Shared Antibiotic Resistome in China. Microbiome 2017, 5, 84. [Google Scholar] [CrossRef]
- Bertrand-Krajewski, J.-L.; Bournique, R.; Lecomte, V.; Pernin, N.; Wiest, L.; Bazin, C.; Bouchez, A.; Brelot, E.; Cournoyer, B.; Chonova, T.; et al. SIPIBEL Observatory: Data on Usual Pollutants (Solids, Organic Matter, Nutrients, Ions) and Micropollutants (Pharmaceuticals, Surfactants, Metals), Biological and Ecotoxicity Indicators in Hospital and Urban Wastewater, in Treated Effluent and Sludge from Wastewater Treatment Plant, and in Surface and Groundwater. Data Brief 2022, 40, 107726. [Google Scholar] [CrossRef]
- Aigle, A.; Colin, Y.; Bouchali, R.; Bourgeois, E.; Marti, R.; Ribun, S.; Marjolet, L.; Pozzi, A.C.M.; Misery, B.; Colinon, C.; et al. Spatio-Temporal Variations in Chemical Pollutants Found among Urban Deposits Match Changes in Thiopurine S-, Se-Methyltransferase-Harboring Bacteria Tracked by the tpm Metabarcoding Approach. Sci. Total Environ. 2021, 145425. [Google Scholar] [CrossRef]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian Community-Wide Culture-Independent Microbial Source Tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef] [Green Version]
- Wiest, L.; Chonova, T.; Bergé, A.; Baudot, R.; Bessueille-Barbier, F.; Ayouni-Derouiche, L.; Vulliet, E. Two-Year Survey of Specific Hospital Wastewater Treatment and Its Impact on Pharmaceutical Discharges. Environ. Sci. Pollut. Res. 2018, 25, 9207–9218. [Google Scholar] [CrossRef] [PubMed]
- AFNOR 1997; French Standard Operating Procedures, Water Quality-Analytical Methods. Afnor: Paris, France; Volume 3.
- Navratil, O.; Boukerb, M.A.; Perret, F.; Breil, P.; Caurel, C.; Schmitt, L.; Lejot, J.; Petit, S.; Marjolet, L.; Cournoyer, B. Responses of Streambed Bacterial Groups to Cycles of Low-flow and Erosive Floods in a Small Peri-urban Stream. Ecohydrology 2020, 13, e2206. [Google Scholar] [CrossRef]
- Park, J.-W.; Crowley, D.E. Dynamic Changes in nahAc Gene Copy Numbers during Degradation of Naphthalene in PAH-Contaminated Soils. Appl. Microbiol. Biotechnol. 2006, 72, 1322–1329. [Google Scholar] [CrossRef] [PubMed]
- Layton, A.; McKay, L.; Williams, D.; Garrett, V.; Gentry, R.; Sayler, G. Development of Bacteroides 16S rRNA Gene TaqMan-Based Real-Time PCR Assays for Estimation of Total, Human, and Bovine Fecal Pollution in Water. Appl. Environ. Microbiol. 2006, 72, 4214–4224. [Google Scholar] [CrossRef] [Green Version]
- Seurinck, S.; Defoirdt, T.; Verstraete, W.; Siciliano, S.D. Detection and Quantification of the Human-Specific HF183 Bacteroides 16S rRNA Genetic Marker with Real-Time PCR for Assessment of Human Faecal Pollution in Freshwater. Environ. Microbiol. 2005, 7, 249–259. [Google Scholar] [CrossRef]
- Marti, R.; Ribun, S.; Aubin, J.-B.; Colinon, C.; Petit, S.; Marjolet, L.; Gourmelon, M.; Schmitt, L.; Breil, P.; Cottet, M.; et al. Human-Driven Microbiological Contamination of Benthic and Hyporheic Sediments of an Intermittent Peri-Urban River Assessed from MST and 16S rRNA Genetic Structure Analyses. Front. Microbiol. 2017, 8, 19. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple Statistical Identification and Removal of Contaminant Sequences in Marker-Gene and Metagenomics Data. Microbiome 2018, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Pozzi, A.C.M.; Bouchali, R.; Marjolet, L.; Cournoyer, B. The tpm Metabarcoding DNA Sequence Database for Taxonomic Allocations Using the Mothur and DADA2 Bio-Informatic Tools (Version 2.0.1) [Data set]. Zenodo 2021. [Google Scholar] [CrossRef]
- Bouchali, R.; Mandon, C.; Marti, R.; Michalon, J.; Aigle, A.; Marjolet, L.; Vareilles, S.; Kouyi, G.L.; Polomé, P.; Toussaint, J.-Y.; et al. Bacterial Assemblages of Urban Microbiomes Mobilized by Runoff Waters Match Land Use Typologies and Harbor Core Species Involved in Pollutant Degradation and Opportunistic Human Infections. Sci. Total Environ. 2022, 815, 152662. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, D.T.; Jovanovic, D.; Lintern, A.; Teakle, I.; Barnes, M.; Deletic, A.; Coleman, R.; Rooney, G.; Prosser, T.; Coutts, S.; et al. Source Tracking Using Microbial Community Fingerprints: Method Comparison with Hydrodynamic Modelling. Water Res. 2017, 109, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Lavenir, R.; Jocktane, D.; Laurent, F.; Nazaret, S.; Cournoyer, B. Improved Reliability of Pseudomonas aeruginosa PCR Detection by the Use of the Species-Specific ecfX Gene Target. J. Microbiol. Methods 2007, 70, 20–29. [Google Scholar] [CrossRef]
- Li, Y.; Xie, Y.-Y.; Chen, R.-X.; Xu, H.-Z.; Zhang, G.-J.; Li, J.-Z.; Li, X.-M. Effects of Combined Treatment with Sansanmycin and Macrolides on Pseudomonas aeruginosa and Formation of Biofilm. Biomed. Environ. Sci. 2009, 22, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Maurya, N.S.; Tiwari, B. Hospital Wastewater Treatment Scenario around the Globe. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2020; pp. 549–570. ISBN 978-0-12-819722-6. [Google Scholar]
- Imhoff, J.F. Transfer of Pfennigia purpurea Tindall 1999 (Amoebobacter purpureus Eichler and Pfennig 1988) to the Genus Lamprocystis as Lamprocystis purpurea Comb. Nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 1699–1701. [Google Scholar] [CrossRef] [Green Version]
- Hua, H.T.; Bollet, C.; Tercian, S.; Drancourt, M.; Raoult, D. Aeromonas popoffii Urinary Tract Infection. J. Clin. Microbiol. 2004, 42, 5427–5428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heylen, K.; Vanparys, B.; Peirsegaele, F.; Lebbe, L.; De Vos, P. Stenotrophomonas terrae Sp. Nov. and Stenotrophomonas humi Sp. Nov., Two Nitrate-Reducing Bacteria Isolated from Soil. Int. J. Syst. Evol. Microbiol. 2007, 57, 2056–2061. [Google Scholar] [CrossRef]
- Chonova, T.; Labanowski, J.; Cournoyer, B.; Chardon, C.; Keck, F.; Laurent, É.; Mondamert, L.; Vasselon, V.; Wiest, L.; Bouchez, A. River Biofilm Community Changes Related to Pharmaceutical Loads Emitted by a Wastewater Treatment Plant. Environ. Sci. Pollut. Res. 2018, 25, 9254–9264. [Google Scholar] [CrossRef]


| Rock Biofilms | b-DTWW | b-HTWW | b-RU | Unknown | ||||
|---|---|---|---|---|---|---|---|---|
| Contribution (%) | rsd (%) | Contribution (%) | rsd (%) | Contribution (%) | rsd (%) | Contribution (%) | rsd (%) | |
| b-RD_close1 | 7.15 | 0.66 | 0.57 | 0.13 | 11.33 | 2.41 | 80.95 | 2.77 |
| b-RD_close2 | 7.3 | 1.06 | 0.85 | 0.34 | 55.92 | 1.17 | 35.93 | 0.65 |
| b-RD_close3 | 35.08 | 1.19 | 1.84 | 0.5 | 16.5 | 0.75 | 46.58 | 1.02 |
| b-RD_close4 | 6.44 | 1.07 | 2.49 | 0.64 | 49.2 | 1.17 | 41.87 | 1.15 |
| b-RD_close5 | 18.13 | 0.84 | 0.74 | 0.4 | 7.75 | 1.11 | 73.38 | 1.09 |
| b-RD_med_1 | 10.87 | 1.08 | 0.44 | 0.22 | 49.24 | 1.19 | 39.45 | 0.71 |
| b-RD_med_2 | 0.21 | 0.09 | 0.2 | 0.17 | 90.18 | 0.24 | 9.41 | 0.27 |
| b-RD_med_3 | 1.86 | 0.49 | 0.23 | 0.12 | 81.12 | 0.52 | 16.79 | 0.41 |
| b-RD_med_4 | 1.25 | 0.22 | 0.36 | 0.14 | 83.77 | 0.44 | 14.62 | 0.36 |
| b-RD_med_5 | 19.28 | 0.75 | 0.89 | 0.52 | 24.54 | 1.26 | 55.29 | 1.5 |
| b-RD_med_6 | 4.4 | 0.94 | 0.41 | 0.23 | 44.51 | 1.1 | 50.68 | 0.71 |
| b-RD_far1 | 1.36 | 0.45 | 0.42 | 0.15 | 59.32 | 0.94 | 38.9 | 0.82 |
| b-RD_far2 | 0.16 | 0.1 | 0.18 | 0.07 | 90 | 0.23 | 9.66 | 0.23 |
| b-RD_far3 | 2.15 | 0.57 | 0.27 | 0.14 | 69.64 | 0.45 | 27.94 | 0.61 |
| b-RD_far4 | 0.34 | 0.13 | 0.13 | 0.1 | 85.03 | 0.41 | 14.5 | 0.33 |
| b-RD_far5 | 23.04 | 1.22 | 0.61 | 0.3 | 26.06 | 0.94 | 50.29 | 1.12 |
| b-RD_far6 | 10.4 | 1.03 | 1.29 | 0.42 | 39.65 | 1.47 | 48.66 | 1.07 |
| Mean | 8.79 | 0.7 | 0.7 | 0.27 | 51.99 | 0.93 | 38.52 | 0.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouchali, R.; Marjolet, L.; Mondamert, L.; Chonova, T.; Ribun, S.; Laurent, E.; Bouchez, A.; Labanowski, J.; Cournoyer, B. Evidence of Bacterial Community Coalescence between Freshwater and Discharged tpm-Harboring Bacterial Taxa from Hospital and Domestic Wastewater Treatment Plants among Epilithic Biofilms. Microorganisms 2023, 11, 922. https://doi.org/10.3390/microorganisms11040922
Bouchali R, Marjolet L, Mondamert L, Chonova T, Ribun S, Laurent E, Bouchez A, Labanowski J, Cournoyer B. Evidence of Bacterial Community Coalescence between Freshwater and Discharged tpm-Harboring Bacterial Taxa from Hospital and Domestic Wastewater Treatment Plants among Epilithic Biofilms. Microorganisms. 2023; 11(4):922. https://doi.org/10.3390/microorganisms11040922
Chicago/Turabian StyleBouchali, Rayan, Laurence Marjolet, Leslie Mondamert, Teofana Chonova, Sébastien Ribun, Elodie Laurent, Agnès Bouchez, Jérôme Labanowski, and Benoit Cournoyer. 2023. "Evidence of Bacterial Community Coalescence between Freshwater and Discharged tpm-Harboring Bacterial Taxa from Hospital and Domestic Wastewater Treatment Plants among Epilithic Biofilms" Microorganisms 11, no. 4: 922. https://doi.org/10.3390/microorganisms11040922
APA StyleBouchali, R., Marjolet, L., Mondamert, L., Chonova, T., Ribun, S., Laurent, E., Bouchez, A., Labanowski, J., & Cournoyer, B. (2023). Evidence of Bacterial Community Coalescence between Freshwater and Discharged tpm-Harboring Bacterial Taxa from Hospital and Domestic Wastewater Treatment Plants among Epilithic Biofilms. Microorganisms, 11(4), 922. https://doi.org/10.3390/microorganisms11040922







