Microbiota from Preterm Infants Who Develop Necrotizing Enterocolitis Drives the Neurodevelopment Impairment in a Humanized Mouse Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Study Design
2.2. Animals
2.3. Preparation of Human Fecal Samples for Transfaunation to GF Mice
2.4. 16S rRNA Sequencing
2.5. Immunohistochemistry
2.6. Cytokine Assay
2.7. Behavioral Studies
2.8. MRI
2.8.1. In Vivo Macromolecule Proton Fraction (MPF) Imaging
2.8.2. Ex Vivo MRI
2.8.3. MRI Data Processing
2.9. RNA Isolation and Real-Time PCR
2.10. Metabonomic Analysis
2.10.1. Data Collection
2.10.2. Untargeted Data Collection
2.10.3. Bile Acid Analysis
2.10.4. Data Processing
2.10.5. MZmine
2.10.6. The Global Natural Product Social Molecular Networking (GNPS)
2.10.7. Feature List Filtering
2.10.8. MetaboAnalyst
2.11. Statistical Methods
3. Results
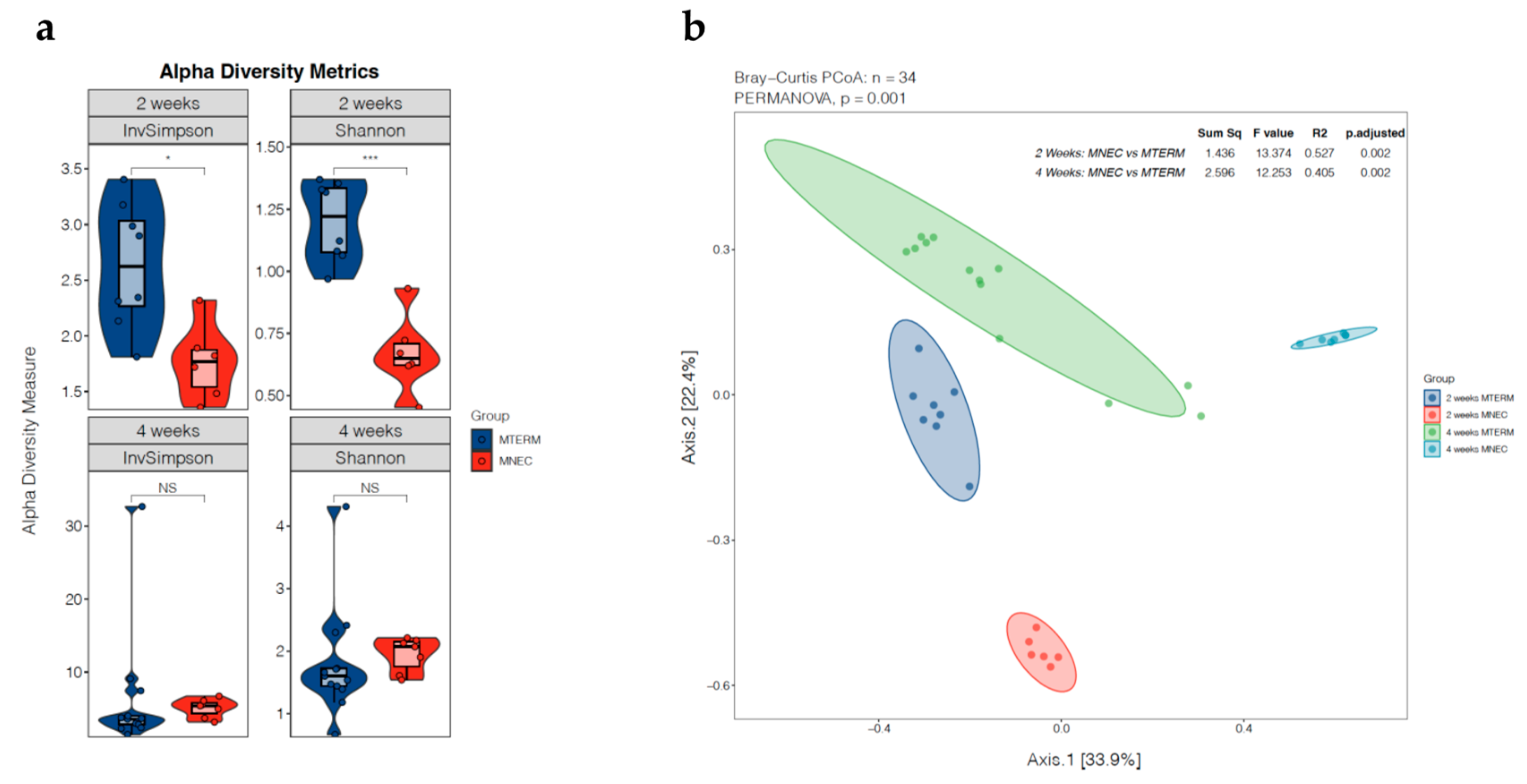
3.1. Dysbiotic Nature of Microbial Communities in Offspring Colonized with Fecal Samples from Patients Who Went on to Develop NEC
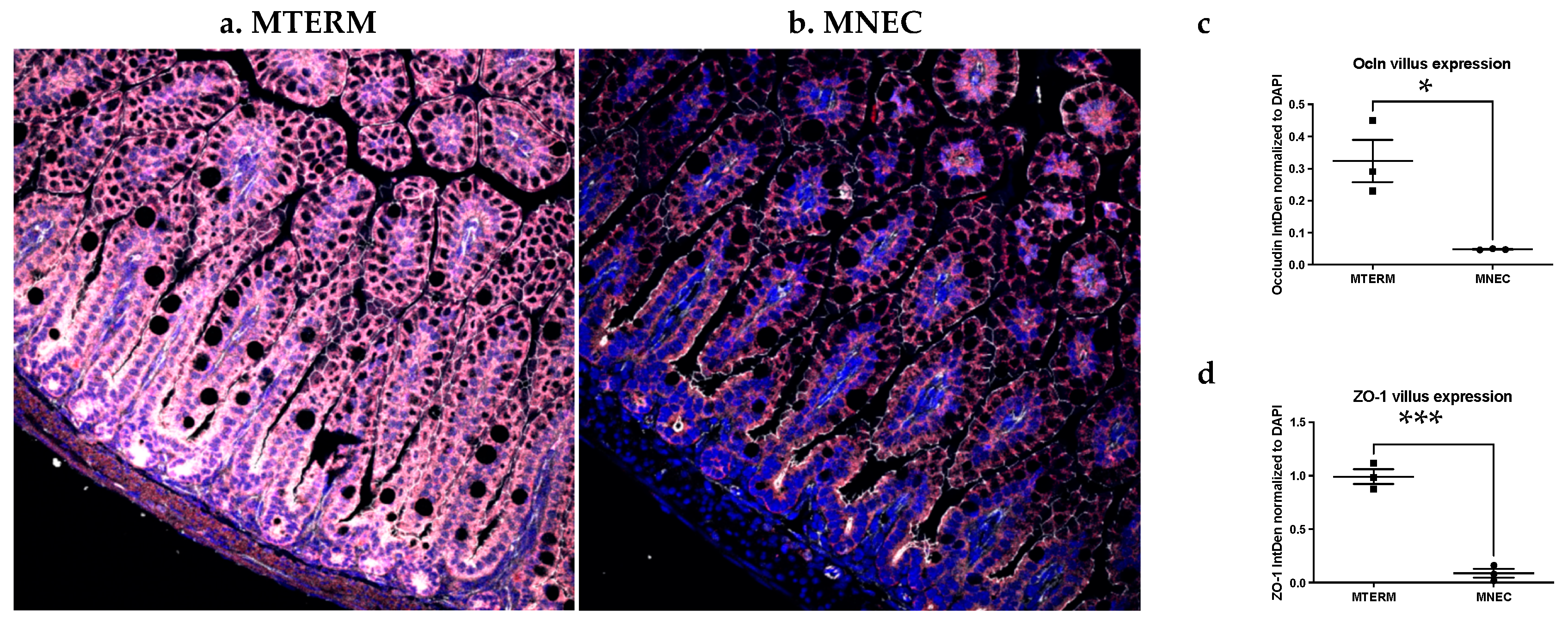
3.2. Mice Colonized with Fecal Samples from Preterm Infants Who Developed NEC Displayed Decreased Tight Junction Development and Increased Inflammation in the Small Intestine
3.3. Systemic Inflammation Was Not Different in Offspring Colonized with Fecal Samples from Preterm Infants Who Developed NEC When Compared to Those from Term Infant
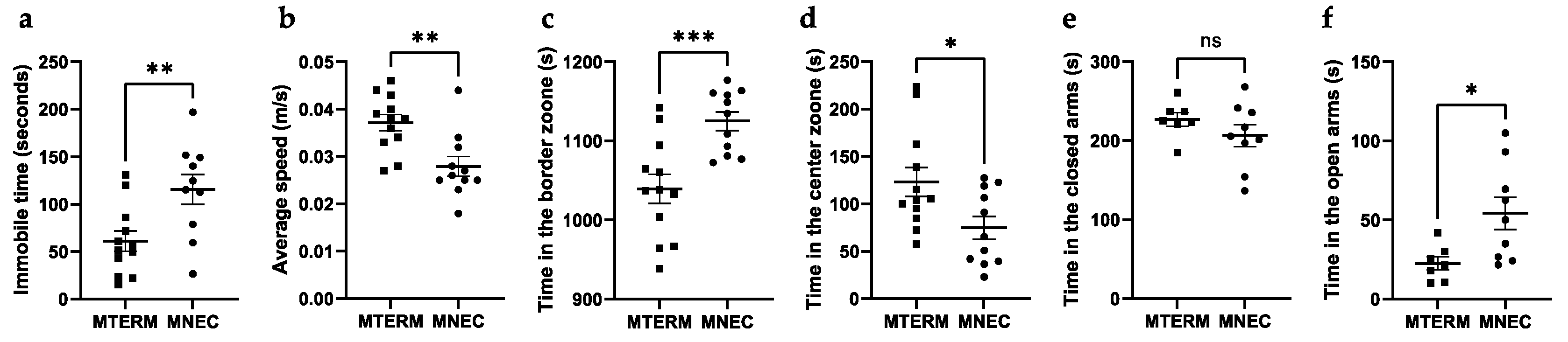
3.4. Mice Colonized with Fecal Samples from Preterm Infants Who Developed NEC Displayed Behavioral Deficits
3.4.1. MNEC Mice Exhibit Decreased Locomotion and Decreased Anxiety
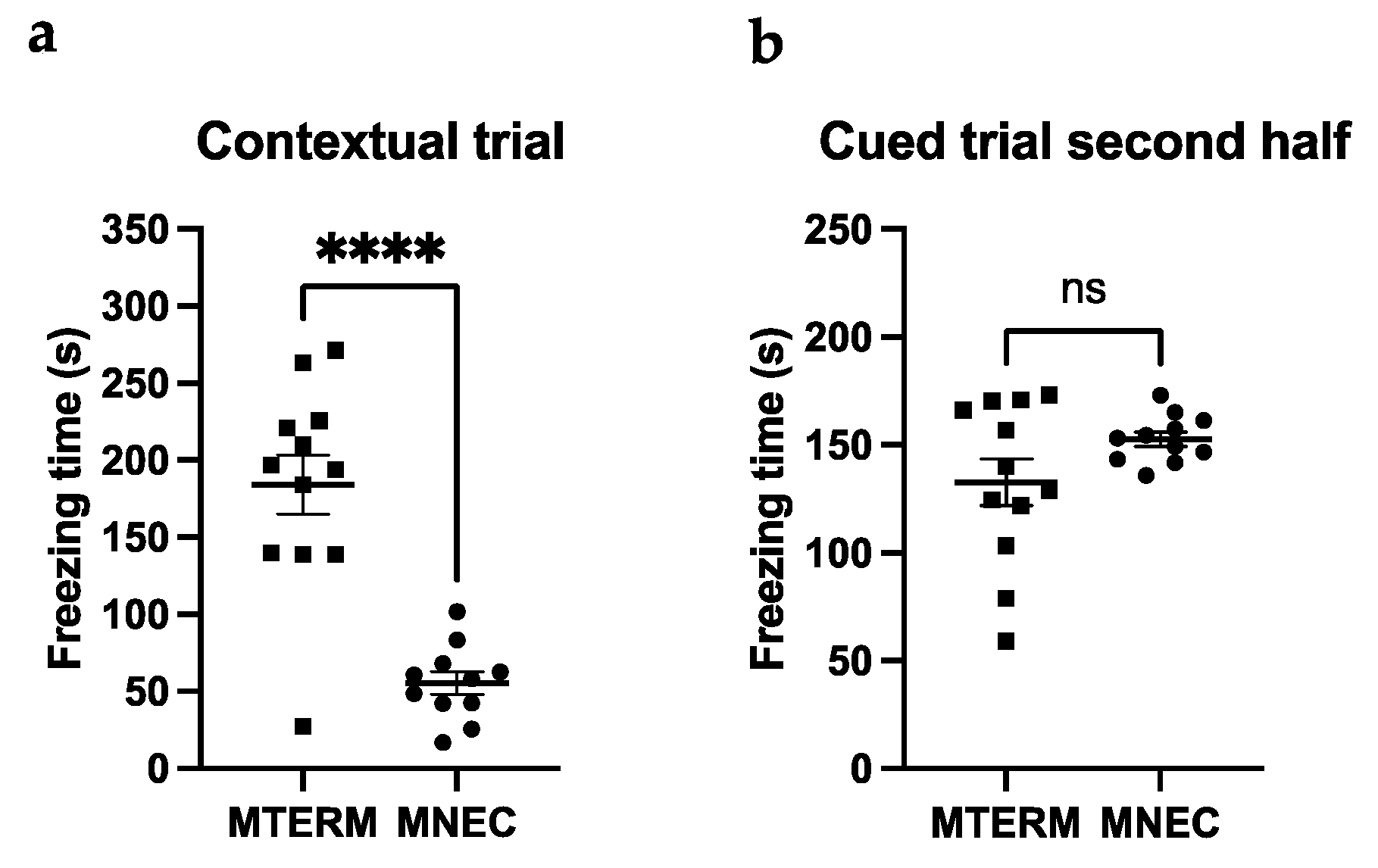
3.4.2. MNEC Mice Exhibit Impaired Contextual Memory
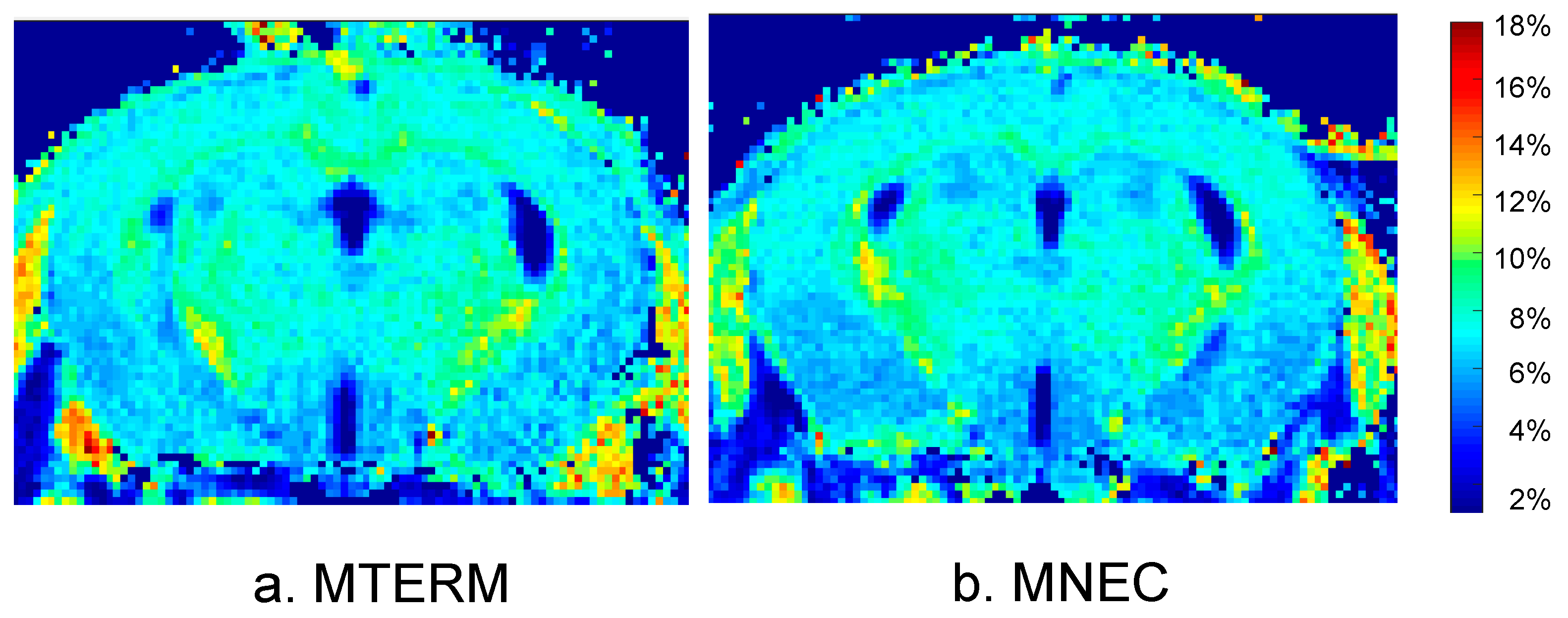
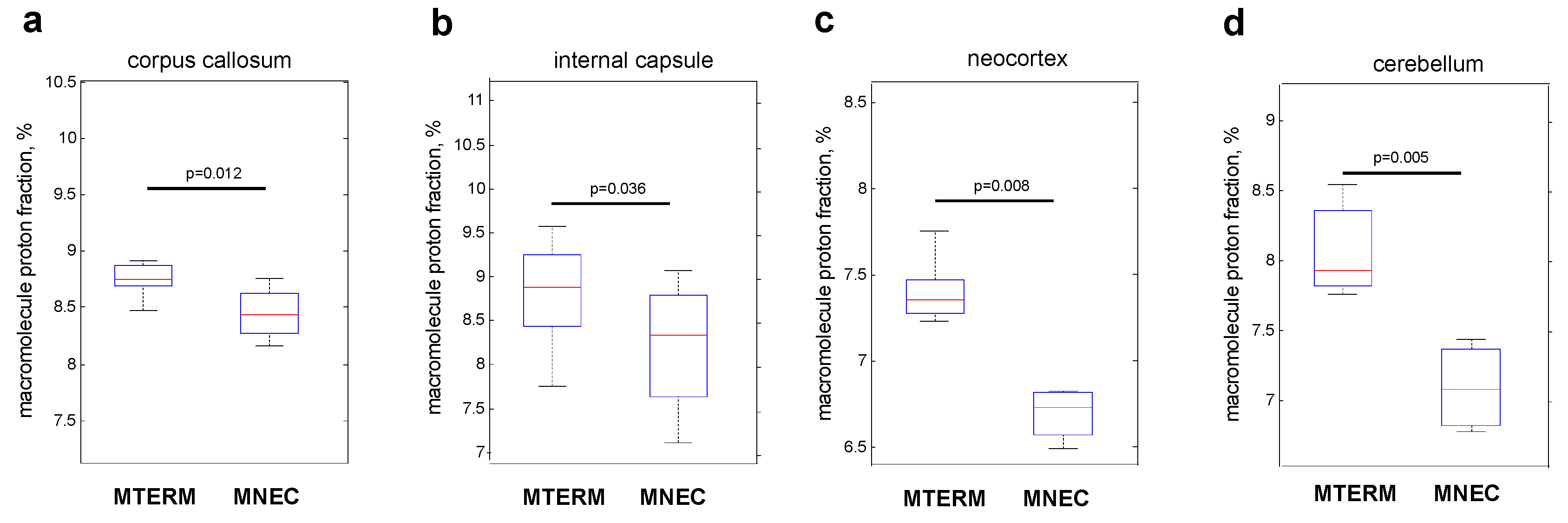
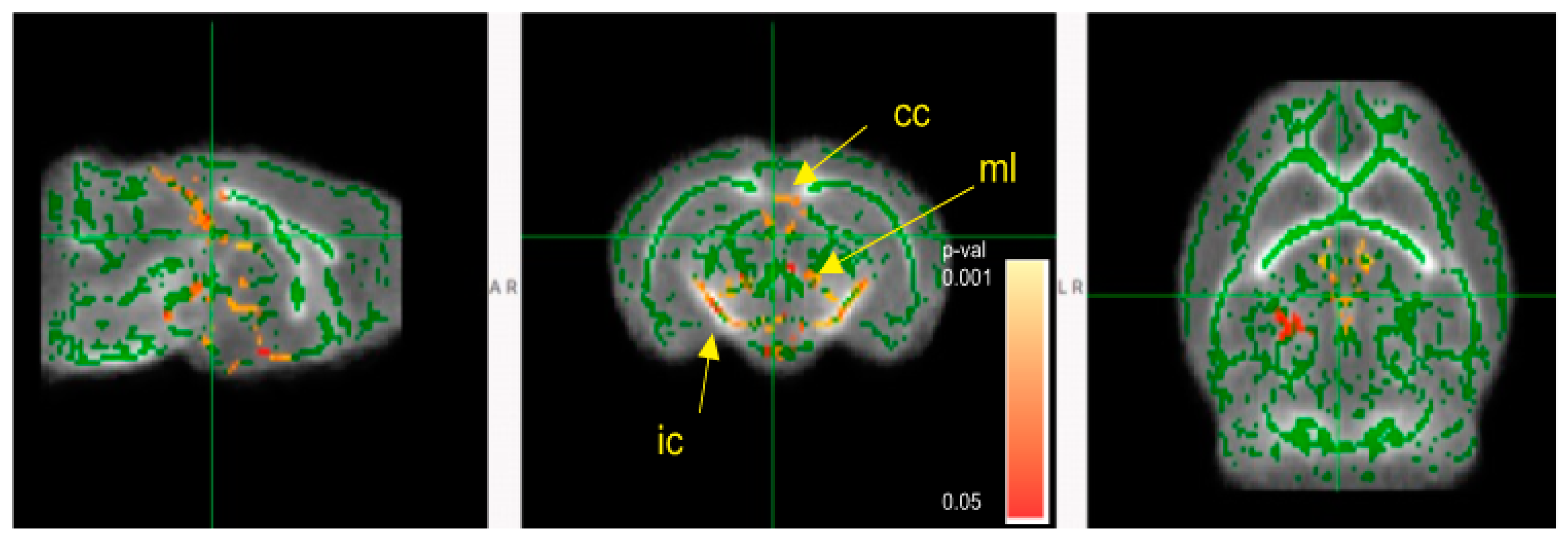
3.5. Mice Colonized with Fecal Samples from Preterm Infants Who Developed NEC Displayed Deficits in Brain Maturation
3.6. Mice Colonized with Fecal Samples from Preterm Infants Who Developed NEC Displayed Cellular Deficits in Brain Development
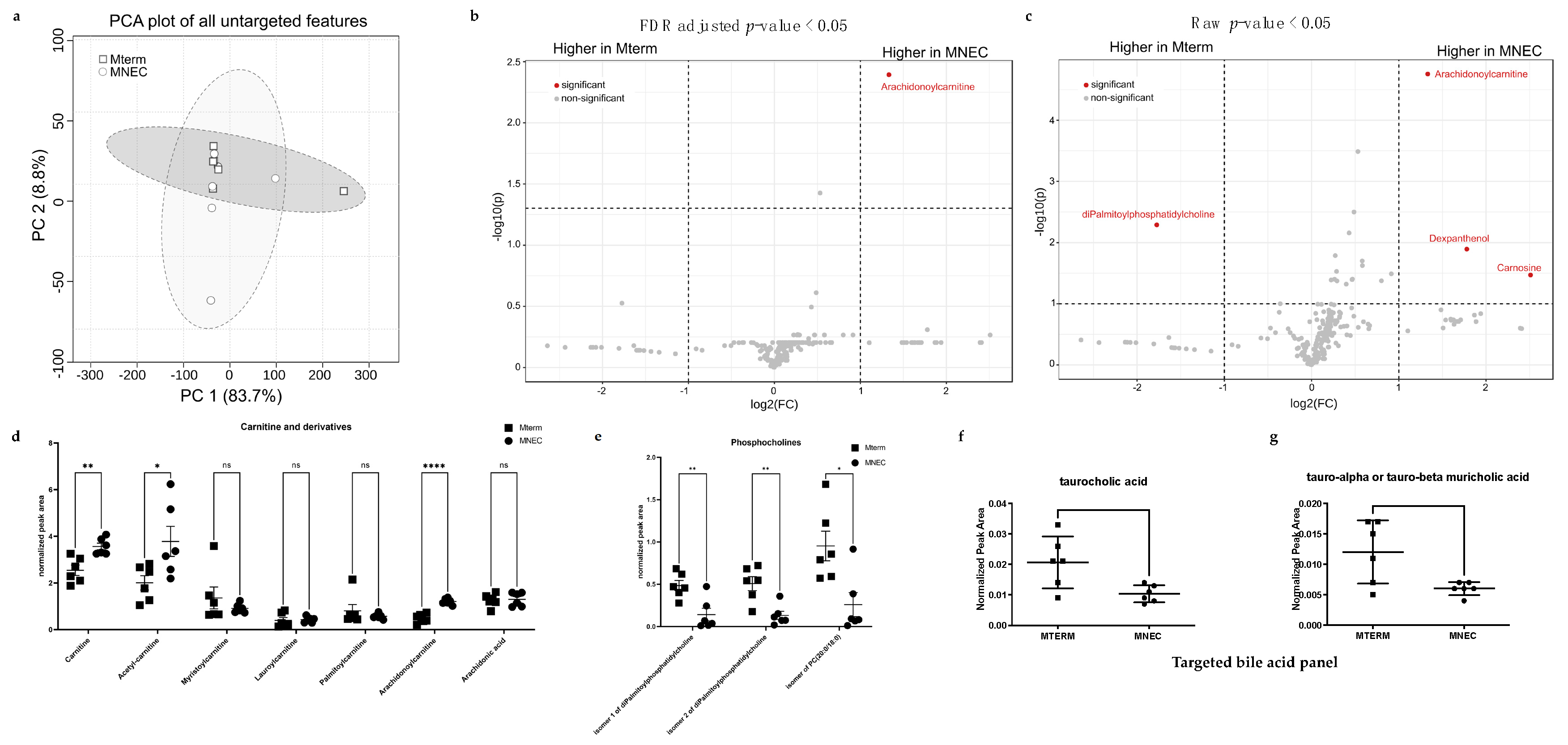
3.7. Microbial Communities from Preterm Infants Who Developed NEC Significantly Shifted the Metabolite Features in the Brain
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Federici, S.; De Biagi, L. Long Term Outcome of Infants with NEC. Curr. Pediatr. Rev. 2019, 15, 111–114. [Google Scholar] [CrossRef]
- Holmsgaard, K.W.; Petersen, S. Infants with gestational age 28 weeks or less. Dan. Med. Bull. 1996, 43, 86–91. [Google Scholar] [PubMed]
- Rees, C.M.; Pierro, A.; Eaton, S. Neurodevelopmental outcomes of neonates with medically and surgically treated necrotizing enterocolitis. Arch. Dis. Child. Fetal Neonatal Ed. 2007, 92, F193–F198. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, J.; Grimmer, I.; Scholz, T.; Metze, B.; Wit, J.; Obladen, M. Growth and neurodevelopmental outcome of very low birthweight infants with necrotizing enterocolitis. Acta Paediatr. 2000, 89, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Castro, L.; Yolton, K.; Haberman, B.; Roberto, N.; Hansen, N.I.; Ambalavanan, N.; Vohr, B.R.; Donovan, E.F. Bias in reported neurodevelopmental outcomes among extremely low birth weight survivors. Pediatrics 2004, 114, 404–410. [Google Scholar] [CrossRef]
- Hintz, S.R.; Kendrick, D.E.; Stoll, B.J.; Vohr, B.R.; Fanaroff, A.A.; Donovan, E.F.; Poole, W.K.; Blakely, M.L.; Wright, L.; Higgins, R.; et al. Neurodevelopmental and growth outcomes of extremely low birth weight infants after necrotizing enterocolitis. Pediatrics 2005, 115, 696–703. [Google Scholar] [CrossRef]
- Simon, N.P. Follow-up for infants with necrotizing enterocolitis. Clin. Perinatol. 1994, 21, 411–424. [Google Scholar] [CrossRef]
- Chacko, J.; Ford, W.D.; Haslam, R. Growth and neurodevelopmental outcome in extremely-low-birth-weight infants after laparotomy. Pediatr. Surg. Int. 1999, 15, 496–499. [Google Scholar] [CrossRef]
- Tobiansky, R.; Lui, K.; Roberts, S.; Veddovi, M. Neurodevelopmental outcome in very low birthweight infants with necrotizing enterocolitis requiring surgery. J. Paediatr. Child. Health 1995, 31, 233–236. [Google Scholar] [CrossRef]
- Mayr, J.; Fasching, G.; Hollwarth, M.E. Psychosocial and psychomotoric development of very low birthweight infants with necrotizing enterocolitis. Acta Paediatr. Suppl. 1994, 396, 96–100. [Google Scholar] [CrossRef]
- Walsh, M.C.; Kliegman, R.M.; Hack, M. Severity of necrotizing enterocolitis: Influence on outcome at 2 years of age. Pediatrics 1989, 84, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Waugh, J.; O’Callaghan, M.J.; Tudehope, D.I.; Mohay, H.A.; Burns, Y.R.; Gray, P.H.; Rogers, Y.M. Prevalence and aetiology of neurological impairment in extremely low birthweight infants. J. Paediatr. Child. Health 1996, 32, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.C.; Chang, J.H.; Kao, H.A.; Hsu, C.H.; Hung, H.Y.; Peng, C.C. Necrotizing enterocolitis in infants: Clinical outcome and influence on growth and neurodevelopment. J. Formos. Med. Assoc. 2004, 103, 761–766. [Google Scholar] [PubMed]
- Schulzke, S.M.; Deshpande, G.C.; Patole, S.K. Neurodevelopmental outcomes of very low-birth-weight infants with necrotizing enterocolitis: A systematic review of observational studies. Arch. Pediatr. Adolesc. Med. 2007, 161, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.A.; Nighot, P.; Reyes, C.; Rawat, M.; McKee, J.; Lemon, D.; Hanson, J.; Ma, T.Y. Intestinal barrier dysfunction in human necrotizing enterocolitis. J. Pediatr. Surg. 2016, 51, 1907–1913. [Google Scholar] [CrossRef]
- Hackam, D.; Caplan, M. Necrotizing enterocolitis: Pathophysiology from a historical context. Semin. Pediatr. Surg. 2018, 27, 11–18. [Google Scholar] [CrossRef]
- Halpern, M.D.; Denning, P.W. The role of intestinal epithelial barrier function in the development of NEC. Tissue Barriers 2015, 3, e1000707. [Google Scholar] [CrossRef]
- Shiou, S.R.; Yu, Y.; Chen, S.; Ciancio, M.J.; Petrof, E.O.; Sun, J.; Claud, E.C. Erythropoietin protects intestinal epithelial barrier function and lowers the incidence of experimental neonatal necrotizing enterocolitis. J. Biol. Chem. 2011, 286, 12123–12132. [Google Scholar] [CrossRef]
- Jilling, T.; Simon, D.; Lu, J.; Meng, F.J.; Li, D.; Schy, R.; Thomson, R.B.; Soliman, A.; Arditi, M.; Caplan, M.S. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. J. Immunol. 2006, 177, 3273–3282. [Google Scholar] [CrossRef]
- Caplan, M.S.; Jilling, T. New concepts in necrotizing enterocolitis. Curr. Opin. Pediatr. 2001, 13, 111–115. [Google Scholar] [CrossRef]
- Claud, E.C.; Walker, W.A. Bacterial colonization, probiotics, and necrotizing enterocolitis. J. Clin. Gastroenterol. 2008, 42 (Suppl. S2), S46–S52. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; Sohn, K. The Microbiota of the Extremely Preterm Infant. Clin. Perinatol. 2017, 44, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Claud, E.C.; Keegan, K.P.; Brulc, J.M.; Lu, L.; Bartels, D.; Glass, E.; Chang, E.B.; Meyer, F.; Antonopoulos, D.A. Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome 2013, 1, 20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hoenig, J.D.; Malin, K.J.; Qamar, S.; Petrof, E.O.; Sun, J.; Antonopoulos, D.A.; Chang, E.B.; Claud, E.C. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009, 3, 944–954. [Google Scholar] [CrossRef]
- Pammi, M.; Cope, J.; Tarr, P.I.; Warner, B.B.; Morrow, A.L.; Mai, V.; Gregory, K.E.; Kroll, J.S.; McMurtry, V.; Ferris, M.J.; et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: A systematic review and meta-analysis. Microbiome 2017, 5, 31. [Google Scholar] [CrossRef]
- Warner, B.B.; Deych, E.; Zhou, Y.; Hall-Moore, C.; Weinstock, G.M.; Sodergren, E.; Shaikh, N.; Hoffmann, J.A.; Linneman, L.A.; Hamvas, A.; et al. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: A prospective case-control study. Lancet 2016, 387, 1928–1936. [Google Scholar] [CrossRef]
- Sommer, F.; Backhed, F. The gut microbiota--masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Hooper, L.V. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004, 12, 129–134. [Google Scholar] [CrossRef]
- Stappenbeck, T.S.; Hooper, L.V.; Gordon, J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 2002, 99, 15451–15455. [Google Scholar] [CrossRef]
- Diaz Heijtz, R.; Wang, S.; Anuar, F.; Qian, Y.; Bjorkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef]
- Borre, Y.E.; O’Keeffe, G.W.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Microbiota and neurodevelopmental windows: Implications for brain disorders. Trends Mol. Med. 2014, 20, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Borre, Y.E.; Moloney, R.D.; Clarke, G.; Dinan, T.G.; Cryan, J.F. The impact of microbiota on brain and behavior: Mechanisms & therapeutic potential. Adv. Exp. Med. Biol. 2014, 817, 373–403. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Synowiec, S.; Lu, L.; Yu, Y.; Bretherick, T.; Takada, S.; Yarnykh, V.; Caplan, J.; Caplan, M.; Claud, E.C.; et al. Microbiota influence the development of the brain and behaviors in C57BL/6J mice. PLoS ONE 2018, 13, e0201829. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. The Microbiome-Gut-Brain Axis in Health and Disease. Gastroenterol. Clin. N. Am. 2017, 46, 77–89. [Google Scholar] [CrossRef]
- Luna, R.A.; Foster, J.A. Gut brain axis: Diet microbiota interactions and implications for modulation of anxiety and depression. Curr. Opin. Biotechnol. 2015, 32, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.M.; Surette, M.; Bercik, P. The interplay between the intestinal microbiota and the brain. Nat. Rev. Microbiol. 2012, 10, 735–742. [Google Scholar] [CrossRef]
- Vuong, H.E.; Yano, J.M.; Fung, T.C.; Hsiao, E.Y. The Microbiome and Host Behavior. Annu. Rev. Neurosci. 2017, 40, 21–49. [Google Scholar] [CrossRef]
- Diaz Heijtz, R. Fetal, neonatal, and infant microbiome: Perturbations and subsequent effects on brain development and behavior. Semin. Fetal Neonatal Med. 2016, 21, 410–417. [Google Scholar] [CrossRef]
- Yu, Y.; Lu, L.; Sun, J.; Petrof, E.O.; Claud, E.C. Preterm infant gut microbiota affects intestinal epithelial development in a humanized microbiome gnotobiotic mouse model. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, G521–G532. [Google Scholar] [CrossRef]
- Lu, J.; Lu, L.; Yu, Y.; Cluette-Brown, J.; Martin, C.R.; Claud, E.C. Effects of Intestinal Microbiota on Brain Development in Humanized Gnotobiotic Mice. Sci. Rep. 2018, 8, 5443. [Google Scholar] [CrossRef]
- Lu, L.; Yu, Y.; Guo, Y.; Wang, Y.; Chang, E.B.; Claud, E.C. Transcriptional modulation of intestinal innate defense/inflammation genes by preterm infant microbiota in a humanized gnotobiotic mouse model. PLoS ONE 2015, 10, e0124504. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Lu, L.; Yu, Y.; Oliphant, K.; Drobyshevsky, A.; Claud, E.C. Early preterm infant microbiome impacts adult learning. Sci. Rep. 2022, 12, 3310. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fan, X.; Lu, L.; Yu, Y.; Markiewicz, E.; Little, J.C.; Sidebottom, A.M.; Claud, E.C. Limosilactobacillus reuteri normalizes blood-brain barrier dysfunction and neurodevelopment deficits associated with prenatal exposure to lipopolysaccharide. Gut Microbes 2023, 15, 2178800. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. Msystems 2016, 1, e00009-15. [Google Scholar] [CrossRef]
- Yarnykh, V.L. Time-efficient, high-resolution, whole brain three-dimensional macromolecular proton fraction mapping. Magn. Reson. Med. 2016, 75, 2100–2106. [Google Scholar] [CrossRef]
- Yarnykh, V.L. Actual flip-angle imaging in the pulsed steady state: A method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn. Reson. Med. 2007, 57, 192–200. [Google Scholar] [CrossRef]
- Basser, P.J.; Jones, D.K. Diffusion-tensor MRI: Theory, experimental design and data analysis—A technical review. NMR Biomed. 2002, 15, 456–467. [Google Scholar] [CrossRef]
- Basser, P.J.; Mattiello, J.; LeBihan, D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. B 1994, 103, 247–254. [Google Scholar] [CrossRef]
- Ma, D.; Cardoso, M.J.; Modat, M.; Powell, N.; Wells, J.; Holmes, H.; Wiseman, F.; Tybulewicz, V.; Fisher, E.; Lythgoe, M.F.; et al. Automatic structural parcellation of mouse brain MRI using multi-atlas label fusion. PLoS ONE 2014, 9, e86576. [Google Scholar] [CrossRef]
- Smith, S.M.; Jenkinson, M.; Johansen-Berg, H.; Rueckert, D.; Nichols, T.E.; Mackay, C.E.; Watkins, K.E.; Ciccarelli, O.; Cader, M.Z.; Matthews, P.M.; et al. Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. Neuroimage 2006, 31, 1487–1505. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Johansen-Berg, H.; Jenkinson, M.; Rueckert, D.; Nichols, T.E.; Miller, K.L.; Robson, M.D.; Jones, D.K.; Klein, J.C.; Bartsch, A.J.; et al. Acquisition and voxelwise analysis of multi-subject diffusion data with tract-based spatial statistics. Nat. Protoc. 2007, 2, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Duhrkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Lin, P.W.; Stoll, B.J. Necrotising enterocolitis. Lancet 2006, 368, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.M.; Ferguson, J.; McElroy, S.J.; Khashu, M.; Caplan, M.S. Defining necrotizing enterocolitis: Current difficulties and future opportunities. Pediatr. Res. 2020, 88 (Suppl. S1), 10–15. [Google Scholar] [CrossRef]
- Benkoe, T.; Baumann, S.; Weninger, M.; Pones, M.; Reck, C.; Rebhandl, W.; Oehler, R. Comprehensive evaluation of 11 cytokines in premature infants with surgical necrotizing enterocolitis. PLoS ONE 2013, 8, e58720. [Google Scholar] [CrossRef]
- van der Knaap, M.S.; Valk, J.; Bakker, C.J.; Schooneveld, M.; Faber, J.A.; Willemse, J.; Gooskens, R.H. Myelination as an expression of the functional maturity of the brain. Dev. Med. Child. Neurol. 1991, 33, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, e450. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.C. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: Exploring a common ground hypothesis. J. Biomed. Sci. 2018, 25, 79. [Google Scholar] [CrossRef] [PubMed]
- Kubota, A.; Yamanaka, H.; Okuyama, H.; Shiraishi, J.; Kawahara, H.; Hasegawa, T.; Ueno, T.; Kitajima, H.; Kuwae, Y.; Nakayama, M. Focal intestinal perforation in extremely-low-birth-weight neonates: Etiological consideration from histological findings. Pediatr. Surg. Int. 2007, 23, 997–1000. [Google Scholar] [CrossRef] [PubMed]
- Hackam, D.J.; Upperman, J.S.; Grishin, A.; Ford, H.R. Disordered enterocyte signaling and intestinal barrier dysfunction in the pathogenesis of necrotizing enterocolitis. Semin. Pediatr. Surg. 2005, 14, 49–57. [Google Scholar] [CrossRef]
- Bein, A.; Eventov-Friedman, S.; Arbell, D.; Schwartz, B. Intestinal tight junctions are severely altered in NEC preterm neonates. Pediatr. Neonatol. 2018, 59, 464–473. [Google Scholar] [CrossRef]
- Gunzel, D.; Yu, A.S. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [CrossRef]
- Lu, L.; Lu, J.; Yu, Y.; Claud, E. Necrotizing enterocolitis intestinal barrier function protection by antenatal dexamethasone and surfactant-D in a rat model. Pediatr. Res. 2021, 90, 768–775. [Google Scholar] [CrossRef]
- Vereecke, L.; Beyaert, R.; van Loo, G. Enterocyte death and intestinal barrier maintenance in homeostasis and disease. Trends Mol. Med. 2011, 17, 584–593. [Google Scholar] [CrossRef]
- McElroy, S.J.; Underwood, M.A.; Sherman, M.P. Paneth cells and necrotizing enterocolitis: A novel hypothesis for disease pathogenesis. Neonatology 2013, 103, 10–20. [Google Scholar] [CrossRef]
- Lu, J.; Martin, C.R.; Claud, E.C. Neurodevelopmental outcome of infants who develop necrotizing enterocolitis: The gut-brain axis. Semin. Perinatol. 2022, 47, 151694. [Google Scholar] [CrossRef]
- Nino, D.F.; Zhou, Q.; Yamaguchi, Y.; Martin, L.Y.; Wang, S.; Fulton, W.B.; Jia, H.; Lu, P.; Prindle, T., Jr.; Zhang, F.; et al. Cognitive impairments induced by necrotizing enterocolitis can be prevented by inhibiting microglial activation in mouse brain. Sci. Transl. Med. 2018, 10, eaan0237. [Google Scholar] [CrossRef] [PubMed]
- Welker, K.M.; Patton, A. Assessment of normal myelination with magnetic resonance imaging. Semin. Neurol. 2012, 32, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Grydeland, H.; Walhovd, K.B.; Tamnes, C.K.; Westlye, L.T.; Fjell, A.M. Intracortical myelin links with performance variability across the human lifespan: Results from T1- and T2-weighted MRI myelin mapping and diffusion tensor imaging. J. Neurosci. 2013, 33, 18618–18630. [Google Scholar] [CrossRef]
- Dubois, J.; Dehaene-Lambertz, G.; Kulikova, S.; Poupon, C.; Huppi, P.S.; Hertz-Pannier, L. The early development of brain white matter: A review of imaging studies in fetuses, newborns and infants. Neuroscience 2014, 276, 48–71. [Google Scholar] [CrossRef]
- Drobyshevsky, A.; Synowiec, S.; Goussakov, I.; Lu, J.; Gascoigne, D.; Aksenov, D.; Yarnykh, V. Temporal trajectories of normal myelination and axonal development assessed by quantitative macromolecular and diffusion MRI: Ultrastructural and immunochemical validation in a rabbit model. Neuroimage 2023, 270, 119974. [Google Scholar] [CrossRef] [PubMed]
- Frankland, P.W.; Bontempi, B. The organization of recent and remote memories. Nat. Rev. Neurosci. 2005, 6, 119–130. [Google Scholar] [CrossRef]
- Kitamura, T.; Ogawa, S.K.; Roy, D.S.; Okuyama, T.; Morrissey, M.D.; Smith, L.M.; Redondo, R.L.; Tonegawa, S. Engrams and circuits crucial for systems consolidation of a memory. Science 2017, 356, 73–78. [Google Scholar] [CrossRef]
- Raymond, J.L.; Lisberger, S.G.; Mauk, M.D. The cerebellum: A neuronal learning machine? Science 1996, 272, 1126–1131. [Google Scholar] [CrossRef]
- Collin, L.; Usiello, A.; Erbs, E.; Mathis, C.; Borrelli, E. Motor training compensates for cerebellar dysfunctions caused by oligodendrocyte ablation. Proc. Natl. Acad. Sci. USA 2004, 101, 325–330. [Google Scholar] [CrossRef]
- Wang, S.; Fan, G.; Xu, K.; Wang, C. Potential of diffusion tensor MR imaging in the assessment of cognitive impairments in children with periventricular leukomalacia born preterm. Eur. J. Radiol. 2013, 82, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Yung, A.; Poon, G.; Qiu, D.Q.; Chu, J.; Lam, B.; Leung, C.; Goh, W.; Khong, P.L. White matter volume and anisotropy in preterm children: A pilot study of neurocognitive correlates. Pediatr. Res. 2007, 61, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Caldu, X.; Narberhaus, A.; Junque, C.; Gimenez, M.; Vendrell, P.; Bargallo, N.; Segarra, D.; Botet, F. Corpus callosum size and neuropsychologic impairment in adolescents who were born preterm. J. Child. Neurol. 2006, 21, 406–410. [Google Scholar] [CrossRef] [PubMed]
- De Bruine, F.T.; Van Wezel-Meijler, G.; Leijser, L.M.; Steggerda, S.J.; Van Den Berg-Huysmans, A.A.; Rijken, M.; Van Buchem, M.A.; Van Der Grond, J. Tractography of white-matter tracts in very preterm infants: A 2-year follow-up study. Dev. Med. Child. Neurol. 2013, 55, 427–433. [Google Scholar] [CrossRef]
- Vangberg, T.R.; Skranes, J.; Dale, A.M.; Martinussen, M.; Brubakk, A.M.; Haraldseth, O. Changes in white matter diffusion anisotropy in adolescents born prematurely. Neuroimage 2006, 32, 1538–1548. [Google Scholar] [CrossRef]
- van Kooij, B.J.; de Vries, L.S.; Ball, G.; van Haastert, I.C.; Benders, M.J.; Groenendaal, F.; Counsell, S.J. Neonatal tract-based spatial statistics findings and outcome in preterm infants. AJNR Am. J. Neuroradiol. 2012, 33, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Wu, X.; Jin, F. Gut-Brain Psychology: Rethinking Psychology From the Microbiota-Gut-Brain Axis. Front. Integr. Neurosci. 2018, 12, 33. [Google Scholar] [CrossRef]
- Jiang, N.M.; Cowan, M.; Moonah, S.N.; Petri, W.A., Jr. The Impact of Systemic Inflammation on Neurodevelopment. Trends Mol. Med. 2018, 24, 794–804. [Google Scholar] [CrossRef]
- Martin, C.R.; Bellomy, M.; Allred, E.N.; Fichorova, R.N.; Leviton, A. Systemic inflammation associated with severe intestinal injury in extremely low gestational age newborns. Fetal Pediatr. Pathol. 2013, 32, 222–234. [Google Scholar] [CrossRef]
- Lapidaire, W.; Lucas, A.; Clayden, J.D.; Clark, C.; Fewtrell, M.S. Human milk feeding and cognitive outcome in preterm infants: The role of infection and NEC reduction. Pediatr. Res. 2022, 91, 1207–1214. [Google Scholar] [CrossRef]
- Abrams, S.A.; Schanler, R.J.; Lee, M.L.; Rechtman, D.J. Greater mortality and morbidity in extremely preterm infants fed a diet containing cow milk protein products. Breastfeed. Med. 2014, 9, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Embleton, N.; Cleminson, J. Randomized trial of exclusive human milk versus preterm formula diets in extremely premature infants. Acta Paediatr. 2017, 106, 1538. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Liu, X.; Wang, J. Small molecules in the big picture of gut microbiome-host cross-talk. EBioMedicine 2022, 81, 104085. [Google Scholar] [CrossRef]
- Cui, Z.; Houweling, M. Phosphatidylcholine and cell death. Biochim. Biophys. Acta 2002, 1585, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Vos, J.P.; de Haas, C.G.; van Golde, L.M.; Lopes-Cardozo, M. Relationships between phosphatidylcholine, phosphatidylethanolamine, and sphingomyelin metabolism in cultured oligodendrocytes. J. Neurochem. 1997, 68, 1252–1260. [Google Scholar] [CrossRef]
- Marcucci, H.; Paoletti, L.; Jackowski, S.; Banchio, C. Phosphatidylcholine biosynthesis during neuronal differentiation and its role in cell fate determination. J. Biol. Chem. 2010, 285, 25382–25393. [Google Scholar] [CrossRef]
- Lee, H.C.; Fellenz-Maloney, M.P.; Liscovitch, M.; Blusztajn, J.K. Phospholipase D-catalyzed hydrolysis of phosphatidylcholine provides the choline precursor for acetylcholine synthesis in a human neuronal cell line. Proc. Natl. Acad. Sci. USA 1993, 90, 10086–10090. [Google Scholar] [CrossRef]
- Inazu, M. Functional Expression of Choline Transporters in the Blood-Brain Barrier. Nutrients 2019, 11, 2265. [Google Scholar] [CrossRef]
- Meck, W.H.; Williams, C.L. Metabolic imprinting of choline by its availability during gestation: Implications for memory and attentional processing across the lifespan. Neurosci. Biobehav. Rev. 2003, 27, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Ando, S.; Tadenuma, T.; Tanaka, Y.; Fukui, F.; Kobayashi, S.; Ohashi, Y.; Kawabata, T. Enhancement of learning capacity and cholinergic synaptic function by carnitine in aging rats. J. Neurosci. Res. 2001, 66, 266–271. [Google Scholar] [CrossRef]
- Liu, J.; Head, E.; Gharib, A.M.; Yuan, W.; Ingersoll, R.T.; Hagen, T.M.; Cotman, C.W.; Ames, B.N. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: Partial reversal by feeding acetyl-L-carnitine and/or R-alpha -lipoic acid. Proc. Natl. Acad. Sci. USA 2002, 99, 2356–2361. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Yang, M.; Zhou, M.; Xiao, J.; Guo, J.; He, L. L-carnitine for cognitive enhancement in people without cognitive impairment. Cochrane Database Syst. Rev. 2017, 3, CD009374. [Google Scholar] [CrossRef]
- Shortland, G.J.; Walter, J.H.; Stroud, C.; Fleming, P.J.; Speidel, B.D.; Marlow, N. Randomised controlled trial of L-carnitine as a nutritional supplement in preterm infants. Arch. Dis. Child. Fetal Neonatal Ed. 1998, 78, F185–F188. [Google Scholar] [CrossRef]
- Younge, N.E.; Newgard, C.B.; Cotten, C.M.; Goldberg, R.N.; Muehlbauer, M.J.; Bain, J.R.; Stevens, R.D.; O’Connell, T.M.; Rawls, J.F.; Seed, P.C.; et al. Disrupted Maturation of the Microbiota and Metabolome among Extremely Preterm Infants with Postnatal Growth Failure. Sci. Rep. 2019, 9, 8167. [Google Scholar] [CrossRef]
- Bartz, S.; Mody, A.; Hornik, C.; Bain, J.; Muehlbauer, M.; Kiyimba, T.; Kiboneka, E.; Stevens, R.; Bartlett, J.; St Peter, J.V.; et al. Severe acute malnutrition in childhood: Hormonal and metabolic status at presentation, response to treatment, and predictors of mortality. J. Clin. Endocrinol. Metab. 2014, 99, 2128–2137. [Google Scholar] [CrossRef] [PubMed]
- Blanton, L.V.; Charbonneau, M.R.; Salih, T.; Barratt, M.J.; Venkatesh, S.; Ilkaveya, O.; Subramanian, S.; Manary, M.J.; Trehan, I.; Jorgensen, J.M.; et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 2016, 351, aad3311. [Google Scholar] [CrossRef] [PubMed]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Schonfeld, P.; Reiser, G. Why does brain metabolism not favor burning of fatty acids to provide energy? Reflections on disadvantages of the use of free fatty acids as fuel for brain. J. Cereb. Blood Flow. Metab. 2013, 33, 1493–1499. [Google Scholar] [CrossRef]
- Pearson, T.S.; Akman, C.; Hinton, V.J.; Engelstad, K.; De Vivo, D.C. Phenotypic spectrum of glucose transporter type 1 deficiency syndrome (Glut1 DS). Curr. Neurol. Neurosci. Rep. 2013, 13, 342. [Google Scholar] [CrossRef]
- Mulak, A. Bile Acids as Key Modulators of the Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J. Alzheimers Dis. 2021, 84, 461–477. [Google Scholar] [CrossRef]
- Huang, C.; Wang, J.; Hu, W.; Wang, C.; Lu, X.; Tong, L.; Wu, F.; Zhang, W. Identification of functional farnesoid X receptors in brain neurons. FEBS Lett. 2016, 590, 3233–3242. [Google Scholar] [CrossRef] [PubMed]
- Yanguas-Casas, N.; Barreda-Manso, M.A.; Nieto-Sampedro, M.; Romero-Ramirez, L. TUDCA: An Agonist of the Bile Acid Receptor GPBAR1/TGR5 With Anti-Inflammatory Effects in Microglial Cells. J. Cell. Physiol. 2017, 232, 2231–2245. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.J.; Jiang, C.; Patterson, A.D. An Intestinal Microbiota-Farnesoid X Receptor Axis Modulates Metabolic Disease. Gastroenterology 2016, 151, 845–859. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Drobyshevsky, A.; Lu, L.; Yu, Y.; Caplan, M.S.; Claud, E.C. Microbiota from Preterm Infants Who Develop Necrotizing Enterocolitis Drives the Neurodevelopment Impairment in a Humanized Mouse Model. Microorganisms 2023, 11, 1131. https://doi.org/10.3390/microorganisms11051131
Lu J, Drobyshevsky A, Lu L, Yu Y, Caplan MS, Claud EC. Microbiota from Preterm Infants Who Develop Necrotizing Enterocolitis Drives the Neurodevelopment Impairment in a Humanized Mouse Model. Microorganisms. 2023; 11(5):1131. https://doi.org/10.3390/microorganisms11051131
Chicago/Turabian StyleLu, Jing, Alexander Drobyshevsky, Lei Lu, Yueyue Yu, Michael S. Caplan, and Erika C. Claud. 2023. "Microbiota from Preterm Infants Who Develop Necrotizing Enterocolitis Drives the Neurodevelopment Impairment in a Humanized Mouse Model" Microorganisms 11, no. 5: 1131. https://doi.org/10.3390/microorganisms11051131
APA StyleLu, J., Drobyshevsky, A., Lu, L., Yu, Y., Caplan, M. S., & Claud, E. C. (2023). Microbiota from Preterm Infants Who Develop Necrotizing Enterocolitis Drives the Neurodevelopment Impairment in a Humanized Mouse Model. Microorganisms, 11(5), 1131. https://doi.org/10.3390/microorganisms11051131





