Host-Associated Bacteroides 16S rDNA-Based Markers for Source Tracking of Fecal Pollution in Laguna Lake, Philippines
Abstract
1. Introduction
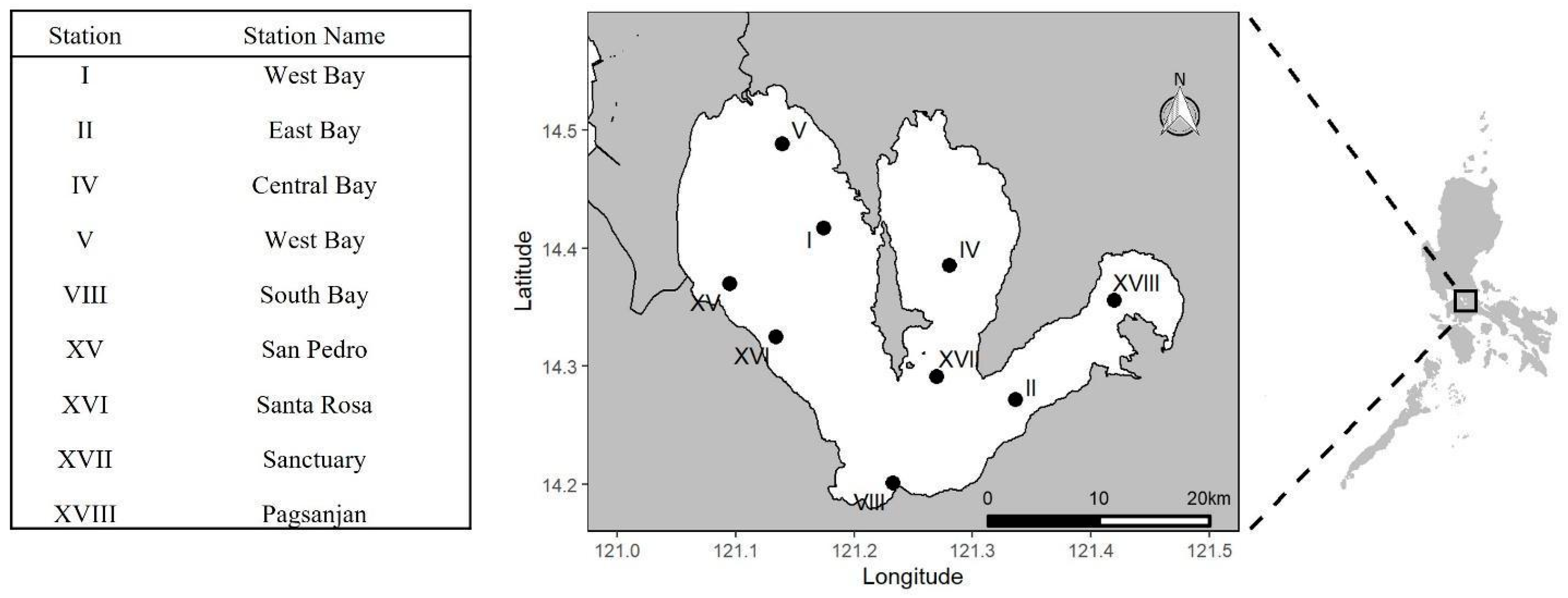
2. Materials and Methods
3. Results
3.1. Validation of Bacteroides 16S rDNA-Based Markers for MST
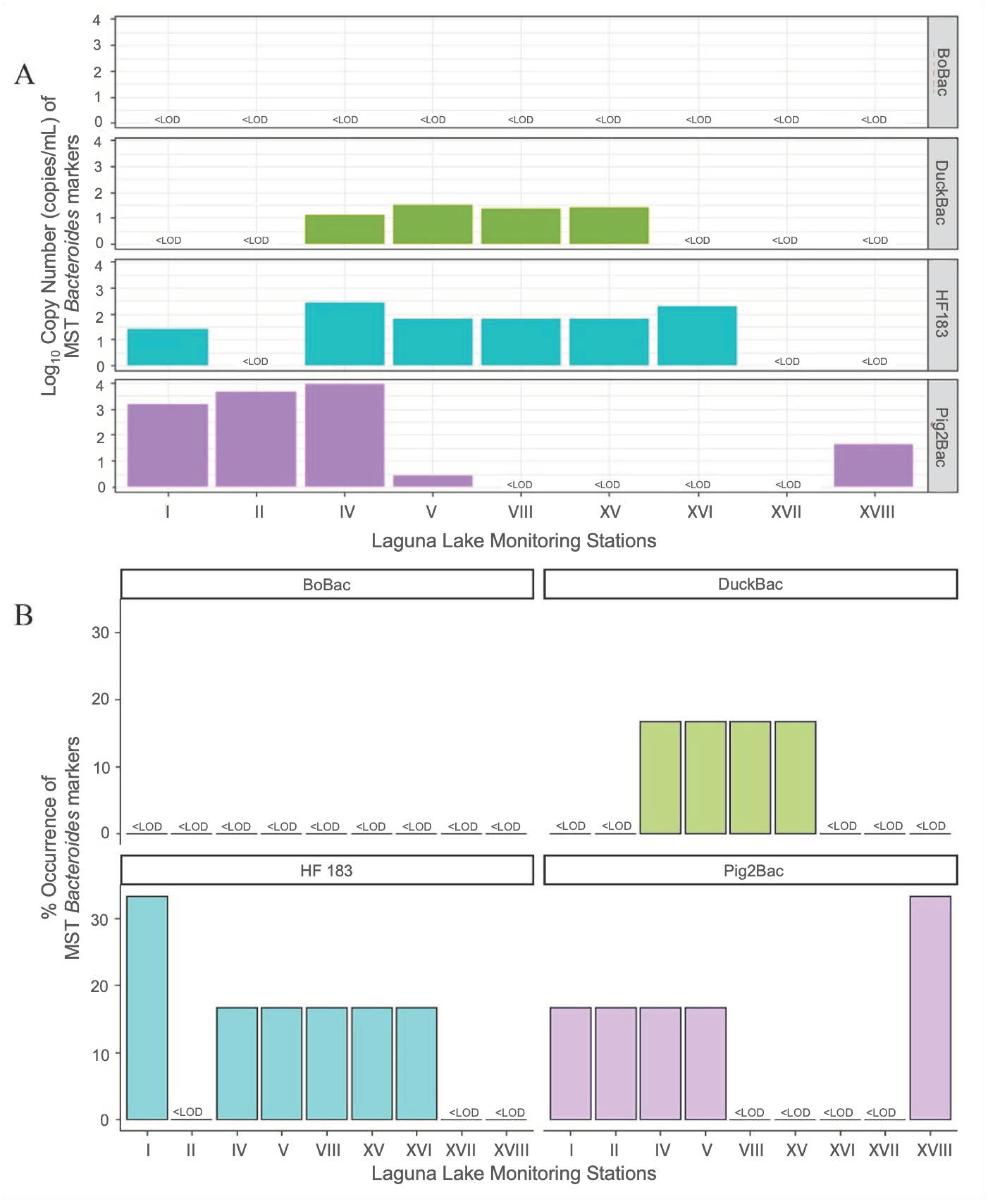
3.2. Detection and Quantification of Host-Associated Contamination in Lake Waters
3.3. Seasonal Patterns in the Abundance of Bacteroides MST Markers
3.4. Correlation between Bacteroides Markers, Fecal Coliforms, and Water Quality Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laguna Lake Development Authority. Existing Lake Uses. 2019. Available online: http://llda.gov.ph/existing-lake-uses/ (accessed on 23 April 2023).
- Laguna Lake Development Authority. Towards Climate-Resilience and Sustainable Development. 2015. Available online: http://gwhs-stg03.i.gov.ph/~s3lldagov/wp-content/dox/ldbMP2016.pdf (accessed on 23 April 2023).
- Santos-Borja, A.; Nepomuceno, D.N. Laguna de Bay: Institutional Development and Change for Lake Basin Management. Lakes Reserv. 2006, 11, 257–269. [Google Scholar] [CrossRef]
- Wealth Accounting and the Valuation of Ecosystem Services. In Pilot Ecosystem Account for Laguna de Bay Basin; The World Bank: Washington, DC, USA, 2016.
- US Environmental Protection Agency. Microbial Source Tracking Guide Document; U.S. Environmental Protection Agency: Washington, DC, USA, 2005.
- Gómez-Doñate, M.; Casanovas-Massana, A.; Muniesa, M.; Blanch, A.R. Development of New Host-Specific Bacteroides qPCRs for the Identification of Fecal Contamination Sources in Water. MicrobiologyOpen 2016, 5, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Mieszkin, S.; Furet, J.-P.; Corthier, G.; Gourmelon, M. Estimation of Pig Fecal Contamination in a River Catchment by Real-Time PCR Using Two Pig-Specific Bacteroidales 16S rRNA Genetic Markers. Appl. Environ. Microbiol. 2009, 75, 3045–3054. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, A.E.; Field, K.G. A PCR Assay to Discriminate Human and Ruminant Feces on the Basis of Host Differences in Bacteroides-Prevotella Genes Encoding 16S rRNA. Appl. Environ. Microbiol. 2000, 66, 4571–4574. [Google Scholar] [CrossRef]
- Hussein, K.R.; Waines, P.L.; Nisr, R.B.; Glegg, G.; Bradley, G. Development and Use of Bacteroides 16S rRNA Polymerase Chain Reaction Assay for Source Tracking Dog Faecal Pollution in Bathing Waters. Hydrol. Curr. Res. 2014, 5, 1–8. [Google Scholar] [CrossRef]
- Green, H.C.; Haugland, R.A.; Varma, M.; Millen, H.T.; Borchardt, M.A.; Field, K.G.; Walters, W.A.; Knight, R.; Sivaganesan, M.; Kelty, C.A.; et al. Improved HF183 Quantitative Real-Time PCR Assay for Characterization of Human Fecal Pollution in Ambient Surface Water Samples. Appl. Environ. Microbiol. 2014, 80, 3086–3094. [Google Scholar] [CrossRef]
- Layton, A.; Mckay, L.; Williams, D.; Garrett, V.; Gentry, R.; Sayler, G. Development of Bacteroides 16S rRNA Gene TaqMan-Based Real-Time PCR Assays for Estimation of Total, Human, and Bovine Fecal Pollution in Water. Appl. Environ. Microbiol. 2006, 72, 4214–4224. [Google Scholar] [CrossRef]
- Kobayashi, A.; Sano, D.; Hatori, J.; Ishii, S.; Okabe, S. Chicken- and Duck-Associated Bacteroides–Prevotella Genetic Markers for Detecting Fecal Contamination in Environmental Water. Appl. Microbiol. Biotechnol. 2013, 97, 7427–7437. [Google Scholar] [CrossRef]
- Stoeckel, D.M.; Harwood, V.J. Performance, Design, and Analysis in Microbial Source Tracking Studies. Appl. Environ. Microbiol. 2007, 73, 2405–2415. [Google Scholar] [CrossRef]
- Diston, D.; Sinreich, M.; Zimmermann, S.; Baumgartner, A.; Felleisen, R. Evaluation of Molecular- and Culture-Dependent MST Markers to Detect Fecal Contamination and Indicate Viral Presence in Good Quality Groundwater. Environ. Sci. Technol. 2015, 49, 7142–7151. [Google Scholar] [CrossRef]
- Harwood, V.J.; Stoeckel, D.M. Performance Criteria. In Microbial Source Tracking: Methods, Applications, and Case Studies; Hagedorn, C., Blanch, A.R., Harwood, V.J., Eds.; Springer Science+Business Media: Berlin/Heidelberg, Germany, 2011; pp. 7–30. [Google Scholar]
- Staley, C.; Gordon, K.V.; Schoen, M.E.; Harwood, V.J. Performance of Two Quantitative PCR Methods for Microbial Source Tracking of Human Sewage and Implications for Microbial Risk Assessment in Recreational Waters. Appl. Environ. Microbiol. 2012, 78, 7317–7326. [Google Scholar] [CrossRef] [PubMed]
- Yahya, M.; Blanch, A.R.; Meijer, W.G.; Antoniou, K.; Hmaied, F.; Ballesté, E. Comparison of the Performance of Different Microbial Source Tracking Markers Among European and North African Regions. J. Environ. Qual. 2017, 46, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Nshimyimana, J.P.; Cruz, M.C.; Thompson, R.J.; Wuertz, S. Bacteroidales Markers for Microbial Source Tracking in Southeast Asia. Water Res. 2017, 118, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Malla, B.; Ghaju Shrestha, R.; Tandukar, S.; Bhandari, D.; Inoue, D.; Sei, K.; Tanaka, Y.; Sherchand, J.B.; Haramoto, E. Validation of Host-Specific Bacteroidales Quantitative PCR Assays and Their Application to Microbial Source Tracking of Drinking Water Sources in the Kathmandu Valley, Nepal. J. Appl. Microbiol. 2018, 125, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Haramoto, E.; Osada, R. Assessment and Application of Host-Specific Bacteroidales Genetic Markers for Microbial Source Tracking of River Water in Japan. PLoS ONE 2018, 13, e0207727. [Google Scholar] [CrossRef]
- Laguna Lake Development Authority. Laguna Lake Development Authority 2016 Annual Report; Laguna Lake Development Authority: Quezon City, Philippines, 2016.
- Kildare, B.J.; Leutenegger, C.M.; McSwain, B.S.; Bambic, D.G.; Rajal, V.B.; Wuertz, S. 16S rRNA-Based Assays for Quantitative Detection of Universal, Human-, Cow-, and Dog-Specific Fecal Bacteroidales: A Bayesian Approach. Water Res. 2007, 41, 3701–3715. [Google Scholar] [CrossRef]
- Somnark, P.; Chyerochana, N.; Mongkolsuk, S.; Sirikanchana, K. Performance Evaluation of Bacteroidales Genetic Markers for Human and Animal Microbial Source Tracking in Tropical Agricultural Watersheds. Environ. Pollut. 2018, 236, 100–110. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2021; Available online: http://www.R-project.org/ (accessed on 23 April 2023).
- Helsel, D.R. Statistics for Censored Environmental Data Using Minitab and R. In Statistics in Practice, 2nd ed.; Wiley: Hoboken, NJ, USA, 2012. [Google Scholar]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Statist. 1979, 6, 65–70. [Google Scholar]
- Ahmed, W.; Hughes, B.; Harwood, V.J. Current Status of Marker Genes of Bacteroides and Related Taxa for Identifying Sewage Pollution in Environmental Waters. Water 2016, 8, 231. [Google Scholar] [CrossRef]
- Harwood, V.J.; Staley, C.; Badgley, B.D.; Borges, K.; Korajkic, A. Microbial Source Tracking Markers for Detection of Fecal Contamination in Environmental Waters: Relationships Between Pathogens and Human Health Outcomes. FEMS Microbiol. Rev. 2014, 38, 1–40. [Google Scholar] [CrossRef]
- Ahmed, W.; Masters, N.; Toze, S. Consistency in the Host Specificity and Host Sensitivity of the Bacteroides HF183 Marker for Sewage Pollution Tracking. Lett. Appl. Microbiol. 2012, 55, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Gourmelon, M.; Caprais, M.P.; Ségura, R.; Le Mennec, C.; Lozach, S.; Piriou, J.Y.; Rincé, A. Evaluation of Two Library-Independent Microbial Source Tracking Methods to Identify Sources of Fecal Contamination in French Estuaries. Appl. Environ. Microbiol. 2007, 73, 4857–4866. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liu, P.; Zheng, G.; Chen, H.; Shi, W.; Cui, Y.; Ren, H.; Zhang, X.-X. Evaluation of Five Microbial and Four Mitochondrial DNA Markers for Tracking Human and Pig Fecal Pollution in Freshwater. Sci. Rep. 2016, 6, 35311. [Google Scholar] [CrossRef] [PubMed]
- Vadde, K.K.; McCarthy, A.J.; Rong, R.; Sekar, R. Quantification of Microbial Source Tracking and Pathogenic Bacterial Markers in Water and Sediments of Tiaoxi River (Taihu Watershed). Front. Microbiol. 2019, 10, 699. [Google Scholar] [CrossRef]
- Waso, M.; Khan, S.; Khan, W. Development and Small-Scale Validation of a Novel Pigeon-Associated Mitochondrial DNA Source Tracking Marker for the Detection of Fecal Contamination in Harvested Rainwater. Sci. Total Environ. 2018, 615, 99–106. [Google Scholar] [CrossRef]
- Green, H.C.; Dick, L.K.; Gilpin, B.; Samadpour, M.; Field, K.G. Genetic Markers for Rapid PCR-Based Identification of Gull, Canada Goose, Duck, and Chicken Fecal Contamination in Water. Appl. Environ. Microbiol. 2012, 78, 503–510. [Google Scholar] [CrossRef]
- Shanks, O.C.; Atikovic, E.; Blackwood, A.D.; Lu, J.; Noble, R.T.; Domingo, J.S.; Seifring, S.; Sivaganesan, M.; Haugland, R.A. Quantitative PCR for Detection and Enumeration of Genetic Markers of Bovine Fecal Pollution. Appl. Environ. Microbiol. 2008, 74, 745–752. [Google Scholar] [CrossRef]
- Korajkic, A.; Stoeckel, D.; Griffith, J.F. Validation of Microbial Source Tracking Markers and Detection Protocols: Considerations for Effective Interpretation. In Manual of Environmental Microbiology, 4th ed.; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Ahmed, W.; Gyawali, P.; Feng, S.; McLellan, S.L. Host Specificity and Sensitivity of Established and Novel Sewage-Associated Marker Genes in Human and Nonhuman Fecal Samples. Appl. Environ. Microbiol. 2019, 85, e00641-19. [Google Scholar] [CrossRef]
- Tanganco, L.J.J.U.; Alberto, M.A.J.; Gotangco, C.K.Z. Forecast of Potential Areas of Urban Expansion in the Laguna de Bay Basin and Its Implications to Water Supply Security. Philipp. J. Sci. 2019, 148, 715–724. [Google Scholar]
- Laguna Lake Development Authority. Laguna de Bay 2013 Ecosystem Health Report Card; Laguna Lake Development Authority: Quezon City, Philippines, 2013. Available online: http://llda.gov.ph (accessed on 23 April 2023).
- Reyes, N.J.D.G.; Geronimo, F.K.F.; Redillas, M.M.; Hong, J. Water Quality Analysis and Evaluation of Management Strategies and Policies in Laguna Lake, Philippines. J. Wetl Res. 2018, 20, 43–53. [Google Scholar] [CrossRef]
- Greenpeace. The State of Water in the Philippines; Greenpeace: Quezon City, Philippines, 2007. [Google Scholar] [CrossRef]
- dela Peña, L.B.R.O.; Labrador, K.L.; Nacario, M.A.G.; Bolo, N.R.; Rivera, W.L. Microbial Source Tracking of Fecal Contamination in Laguna Lake, Philippines Using the Library-Dependent Method, Rep-PCR. J. Water Health 2021, 19, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Palanca-Tan, R. Health and Water Quality Benefits of Alternative Sewerage Systems in Metro Manila, Philippines. Environ. Urban. 2017, 29, 567–580. [Google Scholar] [CrossRef]
- Selected Statistics on Agriculture; Philippine Statistics Authority (PSA): Quezon City, Philippines, 2019.
- Angeles, M.; Catelo, O.; Dorado, M. Backyard and Commercial Piggeries in the Philippines: Environmental Consequences and Pollution Control Options; Economy and Environment Program for Southeast Asia (EEPSEA): Singapore, September 2001. [Google Scholar]
- Catelo, M.A.O.; Narrod, C.A.; Tiongco, M.M. Structural Changes in the Philippine Pig Industry and Their/Environmental Implications; The International Food Policy Research Institute (IFPRI): Washington, DC, USA, 2008. [Google Scholar]
- Resource Assessment for Livestock and Agro-Industrial Wastes—Philippines. 2009. Available online: http://www.globalmethane.org/documents/ag_philippines_res_assessment.pdf (accessed on 23 April 2023).
- Selected Statistics on Agriculture; Philippine Statistics Authority (PSA): Quezon City, Philippines, 2015.
- Atienza, M.; Rodriguez, D.P.G.M.; Babilonia, D.R.; Guimary, M.; Rebancos, C. Food Safety Study of Duck Eggs Produced Along Laguna Lake Areas, Philippines. J. Nutr. Food Sci. 2015, S3, 1. [Google Scholar] [CrossRef]
- Green, H.; Weller, D.; Johnson, S.; Michalenko, E. Microbial Source-Tracking Reveals Origins of Fecal Contamination in a Recovering Watershed. Water 2019, 11, 2162. [Google Scholar] [CrossRef]
- Abello, J.J.M.; Malajacan, G.T.; Labrador, K.L.; Nacario, M.A.G.; Galarion, L.H.; Obusan, M.C.M.; Rivera, W.L. Library-Independent Source Tracking of Fecal Contamination in Selected Stations and Tributaries of Laguna Lake, Philippines. J. Water Health 2021, 19, 846–854. [Google Scholar] [CrossRef]
- Lasco, R.D.; Espaldon, M.V.O.; Tapia, M.A. (Eds.) Ecosystem and People: The Philippine Millenium Ecosystem Assessment (MA); University of the Philippines: Los Baños, Philippines, 2005. [Google Scholar]
- Moron, V.; Lucero, A.; Hilario, F.; Lyon, B.; Robertson, A.W.; DeWitt, D. Spatio-temporal Variability and Predictability of Summer Monsoon Onset over the Philippines. Clim. Dyn. 2009, 33, 1159–1177. [Google Scholar] [CrossRef]
- Bradshaw, J.K.; Snyder, B.J.; Oladeinde, A.; Spidle, D.; Berrang, M.E.; Meinersmann, R.J.; Oakley, B.; Sidle, R.C.; Sullivan, K.; Molina, M. Characterizing Relationships Among Fecal Indicator Bacteria, Microbial Source Tracking Markers, and Associated Waterborne Pathogen Occurrence in Stream Water and Sediments in a Mixed Land Use Watershed. Water Res. 2016, 101, 498–509. [Google Scholar] [CrossRef]
- Laguna Lake Development Authority. Laguna de Bay: Characteristics of Lake Ecosystem. 2021. Available online: https://llda.gov.ph/laguna-de-bay/ (accessed on 15 August 2021).
- Herrera, E.C.; Nadaoka, K.; Blanco, A.C.; Hernandez, E.C. Hydrodynamic Investigation of a Shallow Tropical Lake Environment (Laguna Lake, Philippines) and Associated Implications for Eutrophic Vulnerability. Asean Eng. J. Part C 2015, 4, 48–62. [Google Scholar]
- Laguna Lake Development Authority. LLDA Quarterly Water Quality Monitoring Report Laguna Lake and Tributary Rivers; Laguna Lake Development Authority: Quezon City, Philippines, 2019. Available online: https://llda.gov.ph/wp-content/uploads/dox/waterqualityrpt/2019/2019q4.pdf (accessed on 23 April 2023).
- Kistemann, T.; Claβen, T.; Koch, C.; Dangendorf, F.; Fischeder, R.; Gebel, J.; Vacata, V.; Exner, M. Microbial Load of Drinking Water Reservoir Tributaries During Extreme Rainfall and Runoff. Appl. Environ. Microbiol. 2002, 68, 2188–2197. [Google Scholar] [CrossRef]
- Tornevi, A.; Bergstedt, O.; Forsberg, B. Precipitation Effects on Microbial Pollution in a River: Lag Structures and Seasonal Effect Modification. PLoS ONE 2014, 9, e98546. [Google Scholar] [CrossRef]
- Jeng, H.A.C.; Englande, A.J.; Bakeer, R.M.; Bradford, H.B. Impact of Urban Stormwater Runoff on Estuarine Environmental Quality. Estuar. Coast. Shelf Sci. 2005, 63, 513–526. [Google Scholar] [CrossRef]
- Lucas, F.S.; Therial, C.; Gonçalves, A.; Servais, P.; Rocher, V.; Mouchel, J.-M. Variation of Raw Wastewater Microbiological Quality in Dry and Wet Weather Conditions. Environ. Sci. Pollut. Res. 2014, 21, 5318–5328. [Google Scholar] [CrossRef] [PubMed]
- Santos-Borja, A.; Nepomoceno, D. Laguna de Bay: Experience and Lessons Learned Brief. World Lake Database 2006, 15, 225–258. [Google Scholar]
- Walters, S.P.; Thebo, A.L.; Boehm, A.B. Impact of Urbanization and Agriculture on the Occurrence of Bacterial Pathogens and stx Genes in Coastal Waterbodies of Central California. Water Res. 2011, 45, 1752–1762. [Google Scholar] [CrossRef]
- Ballesté, E.; Blanch, A.R. Persistence of Bacteroides Species Populations in a River as Measured by Molecular and Culture Techniques. Appl. Environ. Microbiol. 2010, 76, 7608–7616. [Google Scholar] [CrossRef]
- USGS. Streamflow and the Water Cycle. 2019. Available online: https://www.usgs.gov/special-topic/water-science-school/science/streamflow-and-water-cycle/ (accessed on 25 October 2021).
- Laguna Lake Development Authority. LLDA Quarterly Water Quality Monitoring Report Laguna Lake and Tributary Rivers; Laguna Lake Development Authority: Quezon City, Philippines, 2019. Available online: https://llda.gov.ph/wp-content/uploads/dox/waterqualityrpt/2019/2019q3.pdf (accessed on 23 April 2023).
- Laguna Lake Development Authority. LLDA Quarterly Water Quality Monitoring Report Laguna Lake and Tributary Rivers; Laguna Lake Development Authority: Quezon City, Philippines, 2020. Available online: https://llda.gov.ph/wp-content/uploads/dox/waterqualityrpt/2020/2020q1.pdf (accessed on 23 April 2023).
- PHILMINAQ. Water Quality Criteria and Standards for Freshwater and Marine Aquacultures; PHILMINAQ: Quezon City, Philippines, 2014. [Google Scholar]
- Bali, M.; Gueddari, M. Removal of Phosphorus from Secondary Effluents Using Infiltration-Percolation Process. Appl. Water Sci. 2019, 9, 54. [Google Scholar] [CrossRef]
- Bendicho, C.; Lavilla, I. Water Analysis|Sewage. In Encyclopedia of Analytical Science; Elsevier: Amsterdam, The Netherlands, 2019; pp. 371–381. [Google Scholar] [CrossRef]
- Ritter, W.; Shirmohammadi, S. Agricultural Nonpoint Source Pollution: Watershed Management and Hydrology; CRC Press: Boca Raton, FL, USA, 2000. [Google Scholar]
- Bunce, J.T.; Ndam, E.; Ofiteru, I.D.; Moore, A.; Graham, D.W.; Graham, D.W. A Review of Phosphorus Removal Technologies and Their Applicability to Small-Scale Domestic Wastewater Treatment Systems. Front. Environ. Sci. 2018, 6, 8. [Google Scholar] [CrossRef]
- van Puijenbroek, P.J.T.M.; Beusen, A.H.W.; Bouwman, A.F. Global Nitrogen and Phosphorus in Urban Waste Water Based on the Shared Socio-economic Pathways. J. Environ. Manag. 2019, 231, 446–456. [Google Scholar] [CrossRef]


| Assay | Primer Name | Sequences (5′-3′) | Amplicon Size (bp) | Annealing Temp (°C) | Reference |
|---|---|---|---|---|---|
| Cow | BoBac367 F | GAAG(G/A)CTGAACCAGCCAAGTA | 100 | 55 | [11] |
| BoBac367 R | GCTTATTCATACGGTACATACAAG | ||||
| Pig | Pig-2-Bac41F | GCATGAATTTAGCTTGCTAAATTTGAT | 116 | 55 | [6] |
| Pig-2-Bac163R | ACCTCATACGGTATTAATCCGC | ||||
| Human | HF183F | ATCATGAGTTCACATGTCCG | 126 | 58 | [10] |
| BacR287 | CTTCCTCTCAGAACCCCTATCC | ||||
| Duck | Bac366F | TTGGTCAATGGGCGGAAG | 108 | 60 | [12] |
| Duck474R | GCACATTCCCACACGTGAGA | ||||
| Chicken | C160F | AAGGGAGATTAATACCCGATGATG | 105 | 60 | [12] |
| Bac265R | CCGTTACCCCGCCTACTAC |
| Host | HF183 (Human) | BoBac (Cow) | Pig-2-Bac (Pig) | DuckBac (Duck) | ChickenBac (Chicken) |
|---|---|---|---|---|---|
| Human | 21/24 (88) | 17/20 (85) | 0/20 (0) | 0/20 (0) | 0/20 (0) |
| Cow | 0/20 (0) | 18/20 (90) | 0/20 (0) | 0/20 (0) | 15/20 (75) |
| Pig | 0/15 (0) | 12/15 (80) | 15/15 (100) | 0/15 (0) | 15/15 (100) |
| Duck | 0/18 (0) | 15/15 (100) | 0/18 (0) | 17/18 (94) | 15/15 (100) |
| Chicken | 1/20 (5) | 19/20 (95) | 0/20 (0) | 0/20 (0) | 15/23 (65) |
| Marker | Sensitivity | Specificity | Accuracy | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|---|
| Pig-2-Bac | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| DuckBac | 0.94 | 1.00 | 0.99 | 1.00 | 0.99 |
| HF183 | 0.88 | 0.99 | 0.96 | 0.95 | 0.96 |
| BoBac | 0.90 | 0.10 | 0.28 | 0.22 | 0.78 |
| ChickenBac | 0.65 | 0.31 | 0.40 | 0.25 | 0.61 |
| Environmental Parameter | Correlation (ρ) with Bacteroides Marker Concentration (log10 Copy Number mL−1) | ||
|---|---|---|---|
| DuckBac | Pig-2-Bac | HF183 | |
| Ammonia (mg/L) | −0.08 | 0.07 | 0.27 |
| BOD (mg/L) | −0.21 | −0.23 | 0.05 |
| DO (mg/L) | −0.09 | 0.04 | −0.25 |
| Fecal coliform (MPN/100 mL geomean) | 0.20 | −0.02 | 0.06 |
| Nitrate (mg/L) | 0.14 | −0.08 | −0.22 |
| pH (units) | 0.02 | 0.07 | −0.03 |
| Phosphate (mg/L) | 0.12 | 0.01 | 0.45 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malajacan, G.T.; Nacario, M.A.G.; Obusan, M.C.M.; Rivera, W.L. Host-Associated Bacteroides 16S rDNA-Based Markers for Source Tracking of Fecal Pollution in Laguna Lake, Philippines. Microorganisms 2023, 11, 1142. https://doi.org/10.3390/microorganisms11051142
Malajacan GT, Nacario MAG, Obusan MCM, Rivera WL. Host-Associated Bacteroides 16S rDNA-Based Markers for Source Tracking of Fecal Pollution in Laguna Lake, Philippines. Microorganisms. 2023; 11(5):1142. https://doi.org/10.3390/microorganisms11051142
Chicago/Turabian StyleMalajacan, Gicelle T., Mae Ashley G. Nacario, Marie Christine M. Obusan, and Windell L. Rivera. 2023. "Host-Associated Bacteroides 16S rDNA-Based Markers for Source Tracking of Fecal Pollution in Laguna Lake, Philippines" Microorganisms 11, no. 5: 1142. https://doi.org/10.3390/microorganisms11051142
APA StyleMalajacan, G. T., Nacario, M. A. G., Obusan, M. C. M., & Rivera, W. L. (2023). Host-Associated Bacteroides 16S rDNA-Based Markers for Source Tracking of Fecal Pollution in Laguna Lake, Philippines. Microorganisms, 11(5), 1142. https://doi.org/10.3390/microorganisms11051142






