The Skin Microbiome: Current Techniques, Challenges, and Future Directions
Abstract
1. Introduction
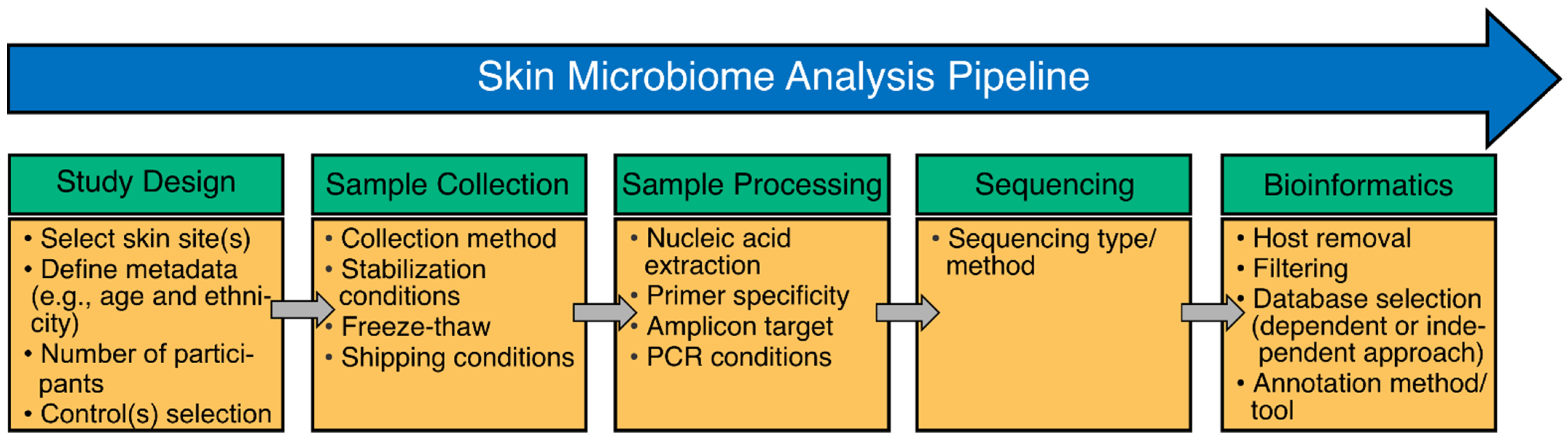
2. Methods to Study the Skin Microbiome and Associated Biases
2.1. Study Design
2.2. Sample Collection and Storage
2.3. Sample Processing: Nucleic Acid Extraction
2.4. Sample Processing: Amplification and Library Preparation
2.5. Bioinformatics: Database Selection and Annotation
3. Ongoing and Proposed Approaches to Study the Skin Microbiome
3.1. Shotgun Metagenomics
3.2. Whole 16S rRNA Gene Sequencing
3.3. Metatranscriptomics
4. Assessing Reagent and Cross-Contamination in Skin Microbiome Studies Using Controls
5. Future Directions and Applications of Skin Microbiome Research
5.1. Unraveling Multi-Kingdom Interactions
5.2. Developing Therapeutics and Diagnostics
5.3. Forensic Applications
5.4. Enabling ‘Omics Integration—Multi’omics
6. Concluding Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Sanford, J.A.; Gallo, R.L. Functions of the skin microbiota in health and disease. Semin. Immunol. 2013, 25, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S.; Zheng, Q.; Hodkinson, B.P.; SanMiguel, A.J.; Minot, S.; Bushman, F.D.; Grice, E.A. The Human Skin Double-Stranded DNA Virome: Topographical and Temporal Diversity, Genetic Enrichment, and Dynamic Associations with the Host Microbiome. MBio 2015, 6, e01578-15. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Kong, H.H.; Segre, J.A. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 550, 61–66. [Google Scholar] [CrossRef]
- Linehan, J.L.; Harrison, O.J.; Han, S.J.; Byrd, A.L.; Vujkovic-Cvijin, I.; Villarino, A.V.; Sen, S.K.; Shaik, J.; Smelkinson, M.; Tamoutounour, S.; et al. Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell 2018, 172, 784–796.e18. [Google Scholar] [CrossRef]
- Cundell, A.M. Microbial Ecology of the Human Skin. Microb. Ecol. 2018, 76, 113–120. [Google Scholar] [CrossRef]
- Saheb Kashaf, S.; Proctor, D.M.; Deming, C.; Saary, P.; Hölzer, M.; Mullikin, J.; Thomas, J.; Young, A.; Bouffard, G.; Barnabas, B.; et al. Integrating cultivation and metagenomics for a multi-kingdom view of skin microbiome diversity and functions. Nat. Microbiol. 2022, 7, 169–179. [Google Scholar] [CrossRef]
- Eisenstein, M. The skin microbiome and its relationship with the human body explained. Nature 2020, 588, S210–S211. [Google Scholar] [CrossRef] [PubMed]
- Chiller, K.; Selkin, B.A.; Murakawa, G.J. Skin microflora and bacterial infections of the skin. J. Investig. Dermatol. Symp. Proc. 2001, 6, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Findley, K.; Oh, J.; Yang, J.; Conlan, S.; Deming, C.; Meyer, J.A.; Schoenfeld, D.; Nomicos, E.; Park, M.; Becker, J.; et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013, 498, 367–370. [Google Scholar] [CrossRef]
- Galazzo, G.; van Best, N.; Bervoets, L.; Dapaah, I.O.; Savelkoul, P.H.; Hornef, M.W.; Hutton, E.K.; Morrison, K.; Holloway, A.C.; McDonald, H.; et al. Development of the Microbiota and Associations with Birth Mode, Diet, and Atopic Disorders in a Longitudinal Analysis of Stool Samples, Collected From Infancy Through Early Childhood. Gastroenterology 2020, 158, 1584–1596. [Google Scholar] [CrossRef] [PubMed]
- De Agüero, M.G.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef]
- Boxberger, M.; Cenizo, V.; Cassir, N.; La Scola, B. Challenges in exploring and manipulating the human skin microbiome. Microbiome 2021, 9, 125. [Google Scholar] [CrossRef]
- Howard, B.; Bascom, C.C.; Hu, P.; Binder, R.L.; Fadayel, G.; Huggins, T.G.; Jarrold, B.B.; Osborne, R.; Rocchetta, H.L.; Swift, D.; et al. Aging-Associated Changes in the Adult Human Skin Microbiome and the Host Factors that Affect Skin Microbiome Composition. J. Investig. Dermatol. 2022, 142, 1934–1946.e21. [Google Scholar] [CrossRef]
- Kim, H.J.; Oh, H.N.; Park, T.; Kim, H.; Lee, H.G.; An, S.; Sul, W.J. Aged related human skin microbiome and mycobiome in Korean women. Sci. Rep. 2022, 12, 2351. [Google Scholar] [CrossRef]
- Wilkinson, H.N.; Clowes, C.; Banyard, K.L.; Matteuci, P.; Mace, K.A.; Hardman, M.J. Elevated local senescence in diabetic wound healing is linked to pathological repair via CXCR2. J. Investig. Dermatol. 2019, 139, 1171–1181.e6. [Google Scholar] [CrossRef]
- Wicke, C.; Bachinger, A.; Coerper, S.; Beckert, S.; Witte, M.B.; Königsrainer, A. Aging influences wound healing in patients with chronic lower extremity wounds treated in a specialized wound care center. Wound Repair Regen. 2009, 17, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Larson, P.J.; Zhou, W.; Santiago, A.; Driscoll, S.; Fleming, E.; Voigt, A.Y.; Chun, O.K.; Grady, J.J.; Kuchel, G.A.; Robison, J.T. Associations of the skin, oral and gut microbiome with aging, frailty and infection risk reservoirs in older adults. Nat. Aging 2022, 2, 941–955. [Google Scholar] [CrossRef]
- Acosta, E.M.; Little, K.A.; Bratton, B.P.; Mao, X.; Payne, A.; Devenport, D.; Gitai, Z. Bacterial DNA on the skin surface overrepresents the viable skin microbiome. bioRxiv 2021. [Google Scholar] [CrossRef]
- Nearing, J.T.; Comeau, A.M.; Langille, M.G.I. Identifying biases and their potential solutions in human microbiome studies. Microbiome 2021, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Ogai, K.; Nagase, S.; Mukai, K.; Iuchi, T.; Mori, Y.; Matsue, M.; Sugitani, K.; Sugama, J.; Okamoto, S. A comparison of techniques for collecting skin microbiome samples: Swabbing versus tape-stripping. Front. Microbiol. 2018, 9, 2362. [Google Scholar] [CrossRef]
- Kong, H.H.; Andersson, B.; Clavel, T.; Common, J.E.; Jackson, S.A.; Olson, N.D.; Segre, J.A.; Traidl-Hoffmann, C. Performing Skin Microbiome Research: A Method to the Madness. J. Investig. Dermatol. 2017, 137, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef]
- Prast-Nielsen, S.; Tobin, A.M.; Adamzik, K.; Powles, A.; Hugerth, L.W.; Sweeney, C.; Kirby, B.; Engstrand, L.; Fry, L. Investigation of the skin microbiome: Swabs vs. biopsies. Br. J. Dermatol. 2019, 181, 572–579. [Google Scholar] [CrossRef]
- Bouevitch, A.; Macklaim, J.; Le François, B. OMNIgene®• SKIN (OMR-140): An optimized collection device for the capture and stabilization of the human skin microbiome. DNA 2020, 20, 200. [Google Scholar]
- Liang, K.; Leong, C.; Loh, J.M.; Chan, N.; Lim, L.; Lam, Y.I.; Dawson Jr, T.L.; Tey, H.L. A 3D-printed transepidermal microprojection array for human skin microbiome sampling. Proc. Natl. Acad. Sci. USA 2022, 119, e2203556119. [Google Scholar] [CrossRef]
- Young, R.R.; Jenkins, K.; Araujo-Perez, F.; Seed, P.C.; Kelly, M.S. Long-term stability of microbiome diversity and composition in fecal samples stored in eNAT medium. Microbiologyopen 2020, 9, e1046. [Google Scholar] [CrossRef]
- Manus, M.B.; Kuthyar, S.; Perroni-Marañón, A.G.; de la Mora, A.N.; Amato, K.R. Comparing different sample collection and storage methods for field-based skin microbiome research. Am. J. Hum. Biol. 2022, 34, e23584. [Google Scholar] [CrossRef] [PubMed]
- Bjerre, R.D.; Hugerth, L.W.; Boulund, F.; Seifert, M.; Johansen, J.D.; Engstrand, L. Effects of sampling strategy and DNA extraction on human skin microbiome investigations. Sci. Rep. 2019, 9, 17287. [Google Scholar] [CrossRef]
- Boulesnane, Y.; Leloup, J.; Lerch, T.Z.; Roynette, A.; Pensé-Lhéritier, A.M.; Mielcarek, C.; Changey, F. Impact of sampling and DNA extraction methods on skin microbiota assessment. J. Microbiol. Methods 2020, 171, 105880. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, J.P.; Carpenter, C.S.; Martino, C.; A Salido, R.; Minich, J.J.; Bryant, M.; Sanders, K.; Schwartz, T.; Humphrey, G.; Swafford, A.D.; et al. A comparison of six DNA extraction protocols for 16S, ITS and shotgun metagenomic sequencing of microbial communities. Biotechniques 2022, 73, 34–46. [Google Scholar] [CrossRef]
- Findley, K.; Oh, J.; Yang, J.; Conlan, S.; Deming, C.; Meyer, J.A.; Schoenfeld, D.; Nomicos, E.; Park, M.; Kong, H.H. Human skin fungal diversity. Nature 2013, 498, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Loesche, M.A.; Farahi, K.; Capone, K.; Fakharzadeh, S.; Blauvelt, A.; Duffin, K.C.; DePrimo, S.E.; Muñoz-Elías, E.J.; Brodmerkel, C.; Dasgupta, B.; et al. Longitudinal Study of the Psoriasis-Associated Skin Microbiome during Therapy with Ustekinumab in a Randomized Phase 3b Clinical Trial. J. Investig. Dermatol. 2018, 138, 1973–1981. [Google Scholar] [CrossRef]
- Khadka, V.D.; Key, F.M.; Romo-González, C.; Martínez-Gayosso, A.; Campos-Cabrera, B.L.; Gerónimo-Gallegos, A.; Lynn, T.C.; Durán-McKinster, C.; Coria-Jiménez, R.; Lieberman, T.D.; et al. The Skin Microbiome of Patients with Atopic Dermatitis Normalizes Gradually During Treatment. Front. Cell. Infect. Microbiol. 2021, 11, 720674. [Google Scholar] [CrossRef]
- De Filippis, F.; Laiola, M.; Blaiotta, G.; Ercolini, D. Different amplicon targets for sequencing-based studies of fungal diversity. Appl. Environ. Microbiol. 2017, 83, e00905-17. [Google Scholar] [CrossRef]
- Ali, N.A.B.M.; Mac Aogáin, M.; Morales, R.F.; Tiew, P.Y.; Chotirmall, S.H. Optimisation and benchmarking of targeted amplicon sequencing for mycobiome analysis of respiratory specimens. Int. J. Mol. Sci. 2019, 20, 4991. [Google Scholar] [CrossRef]
- Meisel, J.S.; Hannigan, G.D.; Tyldsley, A.S.; SanMiguel, A.J.; Hodkinson, B.P.; Zheng, Q.; Grice, E.A. Skin Microbiome Surveys Are Strongly Influenced by Experimental Design. J. Investig. Dermatol. 2016, 136, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Gohl, D.M.; Auch, B.; Certano, A.; LeFrançois, B.; Bouevitch, A.; Doukhanine, E.; Fragel, C.; Macklaim, J.; Hollister, E.; Garbe, J.; et al. Dissecting and tuning primer editing by proofreading polymerases. Nucleic Acids Res. 2021, 49, e87. [Google Scholar] [CrossRef] [PubMed]
- Multinu, F.; Harrington, S.C.; Chen, J.; Jeraldo, P.R.; Johnson, S.; Chia, N.; Walther-Antonio, M.R. Systematic Bias Introduced by Genomic DNA Template Dilution in 16S rRNA Gene-Targeted Microbiota Profiling in Human Stool Homogenates. mSphere 2018, 3, e00560-17. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.D.; Bloom, R.J.; Jiang, S.; Durand, H.K.; Dallow, E.; Mukherjee, S.; David, L.A. Measuring and mitigating PCR bias in microbiota datasets. PLoS Comput. Biol. 2021, 17, e1009113. [Google Scholar] [CrossRef]
- Sze, M.A.; Schloss, P.D. The Impact of DNA Polymerase and Number of Rounds of Amplification in PCR on 16S rRNA Gene Sequence Data. mSphere 2019, 4, e00163-19. [Google Scholar] [CrossRef]
- Mayslich, C.; Grange, P.A.; Dupin, N. Cutibacterium acnes as an Opportunistic Pathogen: An Update of Its Virulence-Associated Factors. Microorganisms 2021, 9, 303. [Google Scholar] [CrossRef]
- Dixit, K.; Davray, D.; Chaudhari, D.; Kadam, P.; Kshirsagar, R.; Shouche, Y.; Dhotre, D.; Saroj, S.D. Benchmarking of 16S rRNA gene databases using known strain sequences. Bioinformation 2021, 17, 377. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Hsu, D.K.; Fung, M.A.; Chen, H.-L. Role of skin and gut microbiota in the pathogenesis of psoriasis, an inflammatory skin disease. Med. Microecol. 2020, 4, 100016. [Google Scholar] [CrossRef]
- Wright, R.J.; Comeau, A.M.; Langille, M.G. From defaults to databases: Parameter and database choice dramatically impact the performance of metagenomic taxonomic classification tools. BioRxiv 2022, 2004–2022. [Google Scholar] [CrossRef]
- Caruso, V.; Song, X.; Asquith, M.; Karstens, L. Performance of Microbiome Sequence Inference Methods in Environments with Varying Biomass. mSystems 2019, 4, e00163-18. [Google Scholar] [CrossRef] [PubMed]
- Ahannach, S.; Delanghe, L.; Spacova, I.; Wittouck, S.; Van Beeck, W.; De Boeck, I.; Lebeer, S. Microbial enrichment and storage for metagenomics of vaginal, skin, and saliva samples. iScience 2021, 24, 103306. [Google Scholar] [CrossRef] [PubMed]
- Conwill, A.; Kuan, A.C.; Damerla, R.; Poret, A.J.; Baker, J.S.; Tripp, A.D.; Alm, E.J.; Lieberman, T.D. Anatomy promotes neutral coexistence of strains in the human skin microbiome. Cell Host Microbe 2022, 30, 171–182.e7. [Google Scholar] [CrossRef]
- Wei, Q.; Li, Z.; Gu, Z.; Liu, X.; Krutmann, J.; Wang, J.; Xia, J. Shotgun metagenomic sequencing reveals skin microbial variability from different facial sites. Front. Microbiol. 2022, 13, 2779. [Google Scholar] [CrossRef]
- Barnard, E.; Johnson, T.; Ngo, T.; Arora, U.; Leuterio, G.; McDowell, A.; Li, H. Porphyrin Production and Regulation in Cutaneous Propionibacteria. mSphere 2020, 5, e00793-19. [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.; Shi, B.; Kang, D.; Craft, N.; Li, H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 2016, 6, 39491. [Google Scholar] [CrossRef] [PubMed]
- Kilian, M.; Scholz, C.F.P.; Lomholt, H.B. Multilocus Sequence Typing and Phylogenetic Analysis of Propionibacterium acnes. J. Clin. Microbiol. 2012, 50, 1158–1165. [Google Scholar] [CrossRef]
- Zolfo, M.; Tett, A.; Jousson, O.; Donati, C.; Segata, N. MetaMLST: Multi-locus strain-level bacterial typing from metagenomic samples. Nucleic Acids Res. 2016, 45, e7. [Google Scholar] [CrossRef]
- Jin, H.; You, L.; Zhao, F.; Li, S.; Ma, T.; Kwok, L.-Y.; Xu, H.; Sun, Z. Hybrid, ultra-deep metagenomic sequencing enables genomic and functional characterization of low-abundance species in the human gut microbiome. Gut Microbes 2022, 14, 2021790. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Conlan, S.; Deming, C.; Segre, J.A.; Kong, H.H.; Korlach, J.; Oh, J.; Program, N.C.S. Resolving the Complexity of Human Skin Metagenomes Using Single-Molecule Sequencing. mBio 2016, 7, e01948-15. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. 16S rRNA Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Tse, H.; Yuen, K.-Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934. [Google Scholar] [CrossRef] [PubMed]
- Rosselli, R.; Romoli, O.; Vitulo, N.; Vezzi, A.; Campanaro, S.; de Pascale, F.; Schiavon, R.; Tiarca, M.; Poletto, F.; Concheri, G.; et al. Direct 16S rRNA-seq from bacterial communities: A PCR-independent approach to simultaneously assess microbial diversity and functional activity potential of each taxon. Sci. Rep. 2016, 6, 32165. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Fida, M.; Khalil, S.; Abu Saleh, O.; Challener, D.W.; Sohail, M.R.; Yang, J.N.; Pritt, B.S.; Schuetz, A.N.; Patel, R. Diagnostic Value of 16S Ribosomal RNA Gene Polymerase Chain Reaction/Sanger Sequencing in Clinical Practice. Clin. Infect. Dis. 2021, 73, 961–968. [Google Scholar] [CrossRef]
- Godlewska, U.; Brzoza, P.; Kwiecień, K.; Kwitniewski, M.; Cichy, J. Metagenomic Studies in Inflammatory Skin Diseases. Curr. Microbiol. 2020, 77, 3201–3212. [Google Scholar] [CrossRef]
- Callahan, B.J.; Grinevich, D.; Thakur, S.; Balamotis, M.A.; Ben Yehezkel, T. Ultra-accurate microbial amplicon sequencing with synthetic long reads. Microbiome 2021, 9, 130. [Google Scholar] [CrossRef]
- Rozas, M.; Brillet, F.; Callewaert, C.; Paetzold, B. MinION™ Nanopore Sequencing of Skin Microbiome 16S and 16S-23S rRNA Gene Amplicons. Front. Cell. Infect. Microbiol. 2022, 11, 1317. [Google Scholar] [CrossRef]
- Dabney, J.; Meyer, M. Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. Biotechniques 2012, 52, 87–94. [Google Scholar] [CrossRef]
- Boye, K.; Høgdall, E.; Borre, M. Identification of bacteria using two degenerate 16S rDNA sequencing primers. Microbiol. Res. 1999, 154, 23–26. [Google Scholar] [CrossRef]
- Aguiar-Pulido, V.; Huang, W.; Suarez-Ulloa, V.; Cickovski, T.; Mathee, K.; Narasimhan, G. Metagenomics, Metatranscriptomics, and Metabolomics Approaches for Microbiome Analysis. Evol. Bioinform. 2016, 12, EBO-S36436. [Google Scholar] [CrossRef] [PubMed]
- Wahl, A.; Huptas, C.; Neuhaus, K. Comparison of rRNA depletion methods for efficient bacterial mRNA sequencing. Sci. Rep. 2022, 12, 5765. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, S.S.; Pourang, A.; Sivamani, R.K. A review of next generation sequencing technologies used in the evaluation of the skin microbiome: What a time to be alive. Dermatol. Online J. 2019, 25, 13030/qt3hv5z3q3. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 29–34. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef]
- Prescott, S.L.; Larcombe, D.-L.; Logan, A.C.; West, C.; Burks, W.; Caraballo, L.; Levin, M.; Van Etten, E.; Horwitz, P.; Kozyrskyj, A.; et al. The skin microbiome: Impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ. J. 2017, 10, 29. [Google Scholar] [CrossRef]
- Radzieta, M.; Peters, T.J.; Dickson, H.G.; Cowin, A.J.; Lavery, L.A.; Schwarzer, S.; Roberts, T.; Jensen, S.O.; Malone, M. A metatranscriptomic approach to explore longitudinal tissue specimens from non-healing diabetes related foot ulcers. Apmis 2022, 130, 383–396. [Google Scholar] [CrossRef]
- Ojala, T.; Lindford, A.; Savijoki, K.; Lagus, H.; Tommila, J.; Medlar, A.; Kuusela, P.; Varmanen, P.; Holm, L.; Vuola, J.; et al. Metatranscriptomic assessment of burn wound infection clearance. Clin. Microbiol. Infect. 2020, 27, 144–146. [Google Scholar] [CrossRef]
- Kang, D.; Shi, B.; Erfe, M.C.; Craft, N.; Li, H. Vitamin B 12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Sci. Transl. Med. 2015, 7, 293ra103. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.C.; Hoff, J.; Olm, M.R.; West-Roberts, J.; Diamond, S.; Dired, B.A.; Morowitz, M.J.; Banfield, J.F. Using strain-resolved analysis to identify contamination in metagenomics data. Microbiome 2023, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.T.; Kelley, S.T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef]
- McGhee, J.J.; Rawson, N.; Bailey, B.A.; Fernandez-Guerra, A.; Sisk-Hackworth, L.; Kelley, S.T. Meta-SourceTracker: Application of Bayesian source tracking to shotgun metagenomics. PeerJ 2020, 8, e8783. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- McKnight, D.T.; Huerlimann, R.; Bower, D.S.; Schwarzkopf, L.; Alford, R.A.; Zenger, K.R. microDecon: A highly accurate read-subtraction tool for the post-sequencing removal of contamination in metabarcoding studies. Environ. DNA 2019, 1, 14–25. [Google Scholar] [CrossRef]
- Martí, J.M. Recentrifuge: Robust comparative analysis and contamination removal for metagenomics. PLoS Comput. Biol. 2019, 15, e1006967. [Google Scholar] [CrossRef]
- Liu, Y.; Elworth, R.A.L.; Jochum, M.D.; Aagaard, K.M.; Treangen, T.J. Squeegee: De-novo identification of reagent and laboratory induced microbial contaminants in low biomass microbiomes. bioRxiv 2021. [Google Scholar] [CrossRef]
- Caldwell, R.; Zhou, W.; Oh, J. Strains to go: Interactions of the skin microbiome beyond its species. Curr. Opin. Microbiol. 2022, 70, 102222. [Google Scholar] [CrossRef]
- Chng, K.R.; Tay, A.S.L.; Li, C.; Ng, A.H.Q.; Wang, J.; Suri, B.K.; Matta, S.A.; McGovern, N.; Janela, B.; Wong, X.F.C.C.; et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat. Microbiol. 2016, 1, 16106. [Google Scholar] [CrossRef]
- Cau, L.; Williams, M.R.; Butcher, A.M.; Nakatsuji, T.; Kavanaugh, J.S.; Cheng, J.Y.; Shafiq, F.; Higbee, K.; Hata, T.R.; Horswill, A.R.; et al. Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J. Allergy Clin. Immunol. 2020, 147, 955–966.e16. [Google Scholar] [CrossRef] [PubMed]
- Lima-Junior, D.S.; Krishnamurthy, S.R.; Bouladoux, N.; Collins, N.; Han, S.-J.; Chen, E.Y.; Constantinides, M.G.; Link, V.M.; Lim, A.I.; Enamorado, M.; et al. Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota. Cell 2021, 184, 3794–3811.e19. [Google Scholar] [CrossRef] [PubMed]
- Luganini, A.; Gribaudo, G. Retroviruses of the Human Virobiota: The Recycling of Viral Genes and the Resulting Advantages for Human Hosts During Evolution. Front. Microbiol. 2020, 11, 1140. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Amagai, M. Controlling skin microbiome as a new bacteriotherapy for inflammatory skin diseases. Inflamm. Regen. 2022, 42, 26. [Google Scholar] [CrossRef]
- Castillo-González, R.; Fernández-Delgado, I.; Comberiati, P. Bacteriotherapy with human skin commensals in atopic dermatitis. Allergy 2022, 77, 1331–1333. [Google Scholar] [CrossRef]
- Golembo, M.; Puttagunta, S.; Rappo, U.; Weinstock, E.; Engelstein, R.; Gahali-Sass, I.; Moses, A.; Kario, E.; Cohen, E.B.; Nicenboim, J.; et al. Development of a topical bacteriophage gel targeting Cutibacterium acnes for acne prone skin and results of a phase 1 cosmetic randomized clinical trial. Ski. Health Dis. 2022, 2, e93. [Google Scholar] [CrossRef] [PubMed]
- Farfán, J.; Gonzalez, J.M.; Vives, M. The immunomodulatory potential of phage therapy to treat acne: A review on bacterial lysis and immunomodulation. PeerJ 2022, 10, e13553. [Google Scholar] [CrossRef]
- Pires, D.P.; Costa, A.R.; Pinto, G.; Meneses, L.; Azeredo, J. Current challenges and future opportunities of phage therapy. FEMS Microbiol. Rev. 2020, 44, 684–700. [Google Scholar] [CrossRef]
- Grice, E.A. The skin microbiome: Potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin. Cutan. Med. Surg. 2014, 33, 98–103. [Google Scholar] [CrossRef]
- Onywera, H.; Williamson, A.-L.; Ponomarenko, J.; Meiring, T.L. The Penile Microbiota in Uncircumcised and Circumcised Men: Relationships with HIV and Human Papillomavirus Infections and Cervicovaginal Microbiota. Front. Med. 2020, 7, 383. [Google Scholar] [CrossRef]
- Price, L.B.; Liu, C.M.; Johnson, K.E.; Aziz, M.; Lau, M.K.; Bowers, J.; Ravel, J.; Keim, P.S.; Serwadda, D.; Wawer, M.J.; et al. The Effects of Circumcision on the Penis Microbiome. PLoS ONE 2010, 5, e8422. [Google Scholar] [CrossRef] [PubMed]
- Onywera, H.; Williamson, A.-L.; Cozzuto, L.; Bonnin, S.; Mbulawa, Z.Z.A.; Coetzee, D.; Ponomarenko, J.; Meiring, T.L. The penile microbiota of Black South African men: Relationship with human papillomavirus and HIV infection. BMC Microbiol. 2020, 20, 78. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.M.; A Hungate, B.; Tobian, A.A.R.; Serwadda, D.; Ravel, J.; Lester, R.; Kigozi, G.; Aziz, M.; Galiwango, R.M.; Nalugoda, F.; et al. Male Circumcision Significantly Reduces Prevalence and Load of Genital Anaerobic Bacteria. mBio 2013, 4, e00076-13. [Google Scholar] [CrossRef]
- Schneider, J.A.; Vadivelu, S.; Liao, C.; Kandukuri, S.R.; Trikamji, B.V.; Chang, E.; Antonopoulos, D.; Prasad, S.; Lakshmi, V. Increased likelihood of bacterial pathogens in the coronal sulcus and urethra of uncircumcised men in a diverse group of HIV infected and uninfected patients in India. J. Glob. Infect. Dis. 2012, 4, 6–9. [Google Scholar] [CrossRef]
- Auvert, B.; Taljaard, D.; Lagarde, E.; Sobngwi-Tambekou, J.; Sitta, R.; Puren, A. Randomized, Controlled Intervention Trial of Male Circumcision for Reduction of HIV Infection Risk: The ANRS 1265 Trial. PLoS Med. 2005, 2, e298. [Google Scholar] [CrossRef]
- Gray, R.H.; Kigozi, G.; Serwadda, D.; Makumbi, F.; Watya, S.; Nalugoda, F.; Kiwanuka, N.; Moulton, L.H.; A Chaudhary, M.; Chen, M.Z.; et al. Male circumcision for HIV prevention in men in Rakai, Uganda: A randomised trial. Lancet 2007, 369, 657–666. [Google Scholar] [CrossRef]
- Gray, R.H.; Serwadda, D.; Kong, X.; Makumbi, F.; Kigozi, G.; Gravitt, P.E.; Watya, S.; Nalugoda, F.; Ssempijja, V.; Tobian, A.A.R.; et al. Male Circumcision Decreases Acquisition and Increases Clearance of High-Risk Human Papillomavirus in HIV-Negative Men: A Randomized Trial in Rakai, Uganda. J. Infect. Dis. 2010, 201, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef]
- Fierer, N.; Hamady, M.; Lauber, C.L.; Knight, R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 17994–17999. [Google Scholar] [CrossRef]
- Gibbons, S.M.; Schwartz, T.; Fouquier, J.; Mitchell, M.; Sangwan, N.; Gilbert, J.A.; Kelley, S.T. Ecological Succession and Viability of Human-Associated Microbiota on Restroom Surfaces. Appl. Environ. Microbiol. 2015, 81, 765–773. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Zhou, N.; McDonald, D.; Costello, E.K.; Knight, R. Forensic identification using skin bacterial communities. Proc. Natl. Acad. Sci. USA 2010, 107, 6477–6481. [Google Scholar] [CrossRef]
- Schmedes, S.E.; Woerner, A.E.; Budowle, B. Forensic Human Identification Using Skin Microbiomes. Appl. Environ. Microbiol. 2017, 83, e01672-17. [Google Scholar] [CrossRef]
- Cho, H.W.; Eom, Y. Bin Forensic Analysis of Human Microbiome in Skin and Body Fluids Based on Geographic Location. Front. Cell. Infect. Microbiol. 2021, 11, 695191. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L. Estimating the postmortem interval using microbes: Knowledge gaps and a path to technology adoption. Forensic Sci. Int. Genet. 2018, 38, 211–218. [Google Scholar] [CrossRef]
- Carter, D.O.; Metcalf, J.L.; Bibat, A.; Knight, R. Seasonal variation of postmortem microbial communities. Forensic Sci. Med. Pathol. 2015, 11, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L.; Xu, Z.Z.; Weiss, S.; Lax, S.; Van Treuren, W.; Hyde, E.R.; Song, S.J.; Amir, A.; Larsen, P.; Sangwan, N.; et al. Microbial community assembly and metabolic function during mammalian corpse decomposition. Science 2016, 351, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, W.; Simayijiang, H.; Hu, P.; Yan, J. Application of Microbiome in Forensics. Genom. Proteom. Bioinform. 2022. [Google Scholar] [CrossRef]
- Carmona-Cruz, S.; Orozco-Covarrubias, L.; Sáez-De-Ocariz, M. The Human Skin Microbiome in Selected Cutaneous Diseases. Front. Cell. Infect. Microbiol. 2022, 12, 145. [Google Scholar] [CrossRef]
- Chen, Y.E.; Tsao, H. The skin microbiome: Current perspectives and future challenges. J. Am. Acad. Dermatol. 2013, 69, 143–155.e3. [Google Scholar] [CrossRef]
- Neckovic, A.; van Oorschot, R.A.; Szkuta, B.; Durdle, A. Investigation of direct and indirect transfer of microbiomes between individuals. Forensic Sci. Int. Genet. 2019, 45, 102212. [Google Scholar] [CrossRef]
- Kuehne, A.; Hildebrand, J.; Soehle, J.; Wenck, H.; Terstegen, L.; Gallinat, S.; Knott, A.; Winnefeld, M.; Zamboni, N. An integrative metabolomics and transcriptomics study to identify metabolic alterations in aged skin of humans in vivo. BMC Genom. 2017, 18, 169. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Hollister, E.B. Multi ‘omic data integration: A review of concepts, considerations, and approaches. Semin. Perinatol. 2021, 45, 151456. [Google Scholar] [CrossRef]

| Step | Key Considerations |
|---|---|
| Study design | Skin site and condition of interest (when applicable) |
| Study power (i.e., number of participants and/or samples collected; relative abundance of signal(s) of interest) | |
| Participant metadata (e.g., ethnicity, age, biological sex, health status, use of medications, hygiene products, and/or cosmetics) | |
| Robust sampling procedure: area size vs. bioload, impact of hygiene, bioburden, etc. | |
| End-to-end review of the methods for compatibility and optimal sample performance | |
| Downstream analysis strategy compatibility | |
| Additional control(s): environmental/non-collected control | |
| Sample collection/ storage | Means of sample collection (e.g., swab, scraping, biopsy, and tape-stripping) Validated and standardized for skin |
| Low bioburden within device and contamination during collection | |
| Need for immediate freezing vs. inclusion of stabilization solution Storage length and conditions | |
| Sample processing: nucleic acid extraction | Validated and standardized Optimized nucleic acid recovery |
| Recovery of Gram-positive and Gram-negative bacteria and fungal species | |
| Effective clean-up of nucleic acids and removal of enzymatic inhibitors | |
| Low bioburden | |
| Extraction negative control | |
| Sample processing: amplification and library preparation | Optimized for taxa (e.g., bacterial vs. fungal) of interest and biomass/host content Accurate capture of microbiome composition Optimal DNA input (for shotgun metagenomic and amplicon sequencing) and amplification conditions (for amplicon sequencing) Efficient removal of host and microbial rRNA and sufficient RNA input for metatranscriptomic sequencing |
| Library preparation negative control | |
| Bioinformatics: database selection | Updated and curated content source and data quality, removal or consolidation of redundant sequences, and comprehensiveness |
| Level of taxonomic or functional resolution supported (e.g., genus, species, strain, and functional hierarchies) | |
| Suitability towards analysis strategy | |
| Bioinformatics: annotation | Sensitivity and specificity of tool/approach Low false positive/false negative rate |
| Database-dependent and database-independent approaches |
| Control Type | Main Step(s) to Be Applied | Description | Purpose(s) | Expected Outcome(s) |
|---|---|---|---|---|
| Extraction blank | Nucleic acid extraction/ library preparation | Type of negative control containing no sample material that is processed in parallel with sample(s) of interest during the extraction process. | Assess the kitome and the introduction of environmental contaminants or cross-contamination; analytic assessment of sample similarity to environment/reagents. | Negligible nucleic acid concentrations; no/little amplification during library preparation; low read counts; significantly differentiated from sample in analysis. |
| Negative control (amplification) | Library preparation | Type of negative control included during sample preparation that is expected to produce no library. | Assess contamination introduced during library preparation. | No/little amplification during library preparation. |
| Positive control (amplification) | Library preparation | Type of positive control included during sample preparation. | Ensures that library preparation was successful. | Library of the expected size and yield. |
| Mock community | Type of control composed of a defined mixture and composition of cells/viruses, nucleic acids, or in silico genomes. | Assess process efficiency, accuracy, and sensitivity from nucleic acid extraction to data analysis. | The identification of expected organisms, the measurement of their proportions, and a comparison to the expected/ground-truth, measurements of sensitivity and specificity (false positive/false negative rates). | |
| Cells/Viruses | Nucleic acid extraction/ library preparation/ database selection/ annotation | Single or multiple collection of cells (e.g., bacterial and fungal) or viruses relevant for study objectives. | Assess nucleic acid extraction, library preparation, sequencing, and analysis efficiency. | End-to-end assay compatibility with taxonomic target(s) (e.g., for the development of a diagnostic measurement). |
| Nucleic acids | Library preparation/ database selection/ annotation | Type of mock community composed of nucleic acids (DNA). | Assess library preparation, sequencing, and analysis efficiency | |
| In silico genomes | Database selection/ annotation | Type of mock community composed of in silico reference genomes/known sequences. | Assess analysis efficiency and accuracy. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiago-Rodriguez, T.M.; Le François, B.; Macklaim, J.M.; Doukhanine, E.; Hollister, E.B. The Skin Microbiome: Current Techniques, Challenges, and Future Directions. Microorganisms 2023, 11, 1222. https://doi.org/10.3390/microorganisms11051222
Santiago-Rodriguez TM, Le François B, Macklaim JM, Doukhanine E, Hollister EB. The Skin Microbiome: Current Techniques, Challenges, and Future Directions. Microorganisms. 2023; 11(5):1222. https://doi.org/10.3390/microorganisms11051222
Chicago/Turabian StyleSantiago-Rodriguez, Tasha M., Brice Le François, Jean M. Macklaim, Evgueni Doukhanine, and Emily B. Hollister. 2023. "The Skin Microbiome: Current Techniques, Challenges, and Future Directions" Microorganisms 11, no. 5: 1222. https://doi.org/10.3390/microorganisms11051222
APA StyleSantiago-Rodriguez, T. M., Le François, B., Macklaim, J. M., Doukhanine, E., & Hollister, E. B. (2023). The Skin Microbiome: Current Techniques, Challenges, and Future Directions. Microorganisms, 11(5), 1222. https://doi.org/10.3390/microorganisms11051222






