Comparative Analysis Using Raman Spectroscopy of the Cellular Constituents of Lacticaseibacillus paracasei Zhang in a Normal and Viable but Nonculturable State
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Cell Culture and Induction of VBNC State
2.3. Determining VBNC State by Plate Counting, Bacterial Viability Staining, and Cell Morphology Analysis
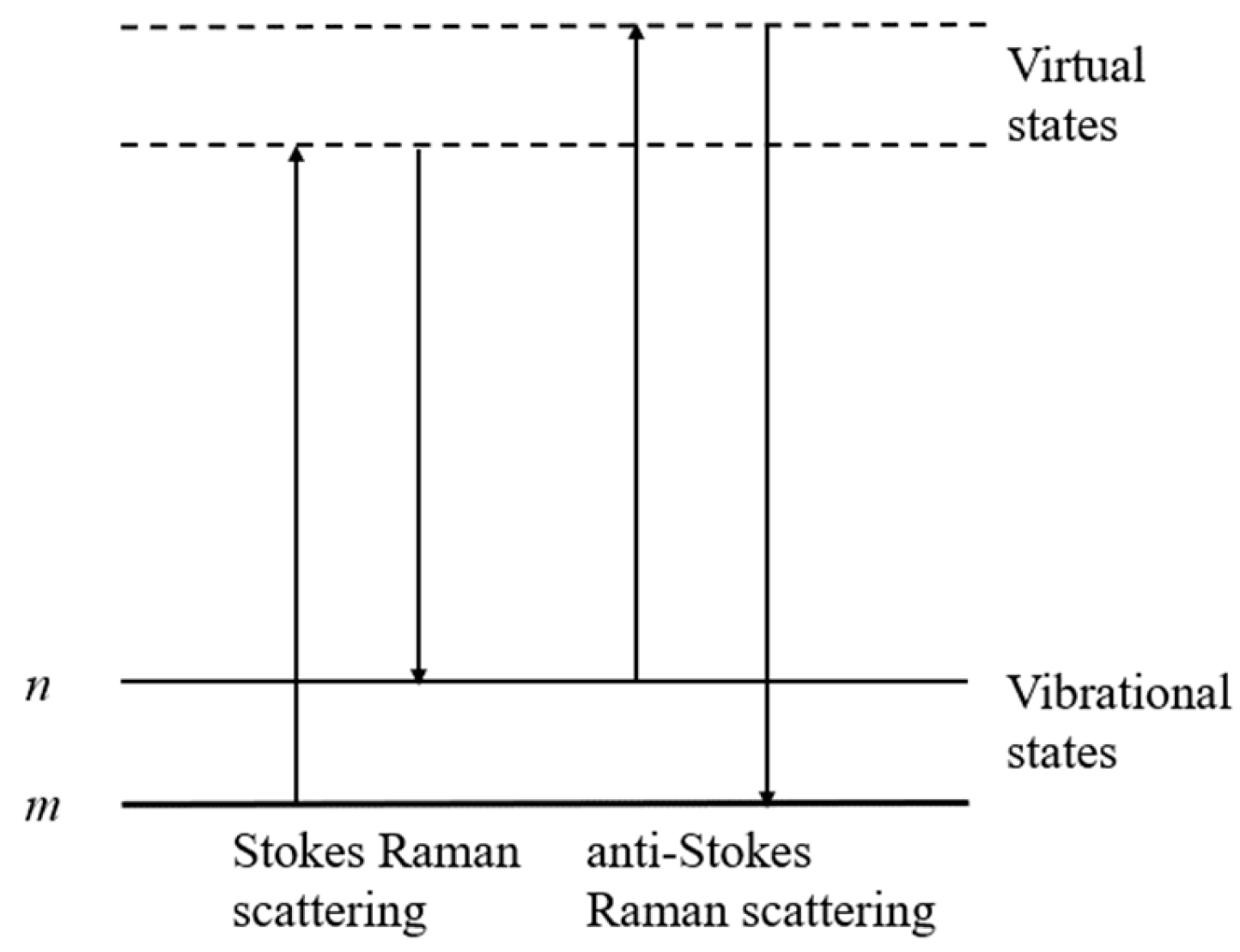
2.4. Single-Cell Raman Spectroscopy
2.5. Pre-Processing of Raman Spectra
2.6. Principal Component Analysis (PCA)
3. Results and Discussion
3.1. Cell Activity of L. paracasei Zhang in a VBNC State
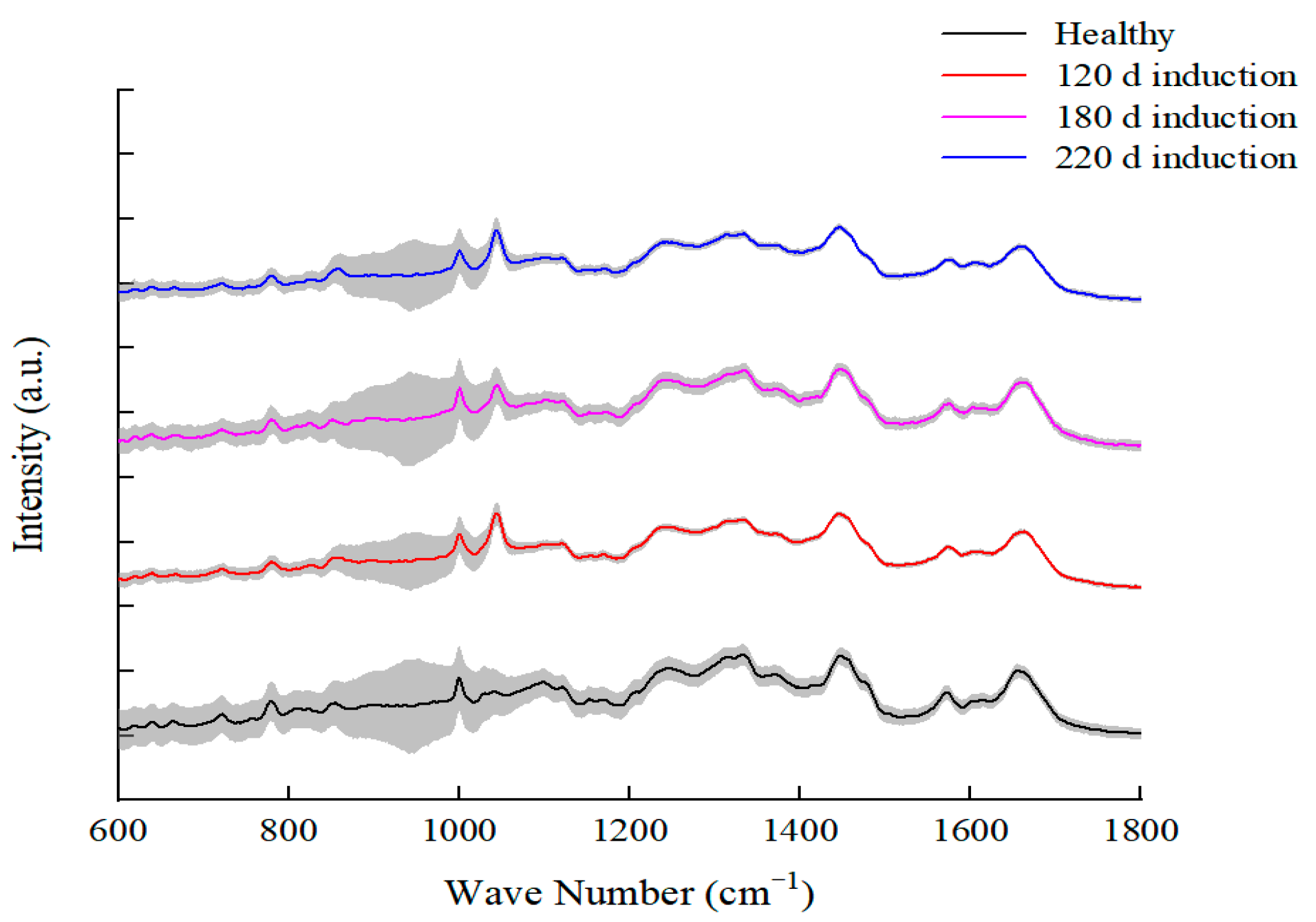
3.2. Single-Cell Raman Spectroscopy for Discrimination between Different Induction Stages of L. paracasei Zhang
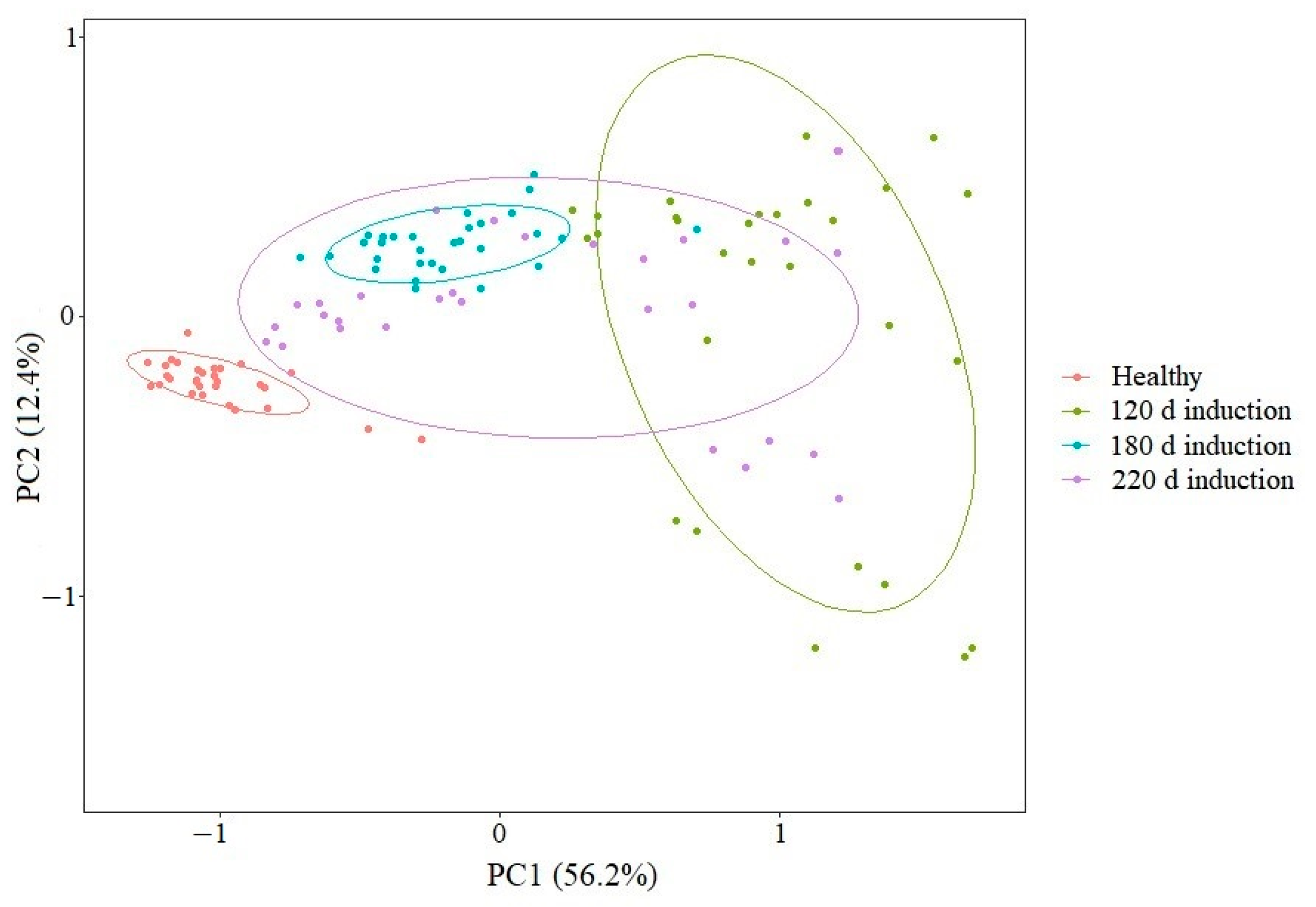
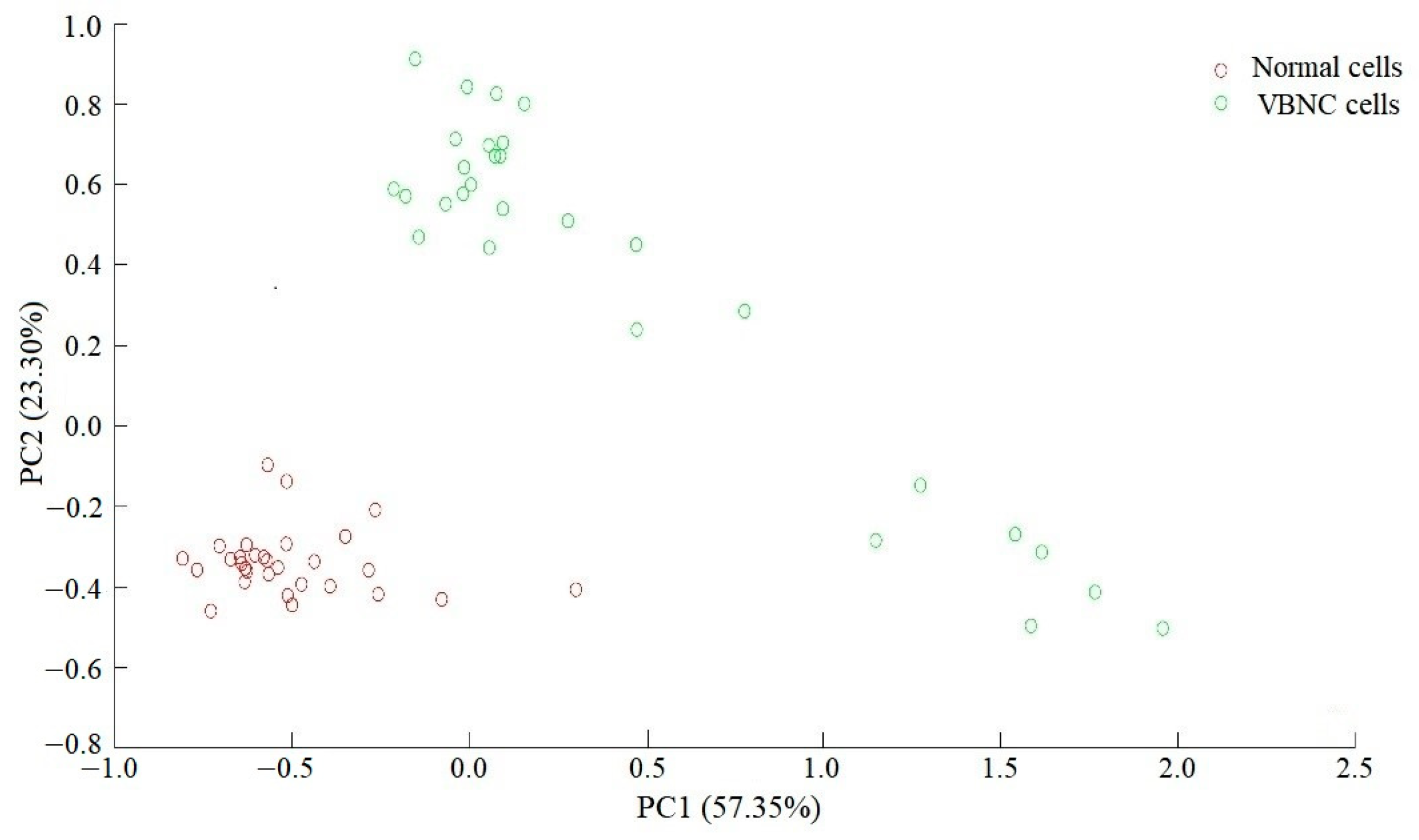
3.3. Principal Component Analysis of Raman Spectra of Normal and VBNC L. paracasei Zhang Cells
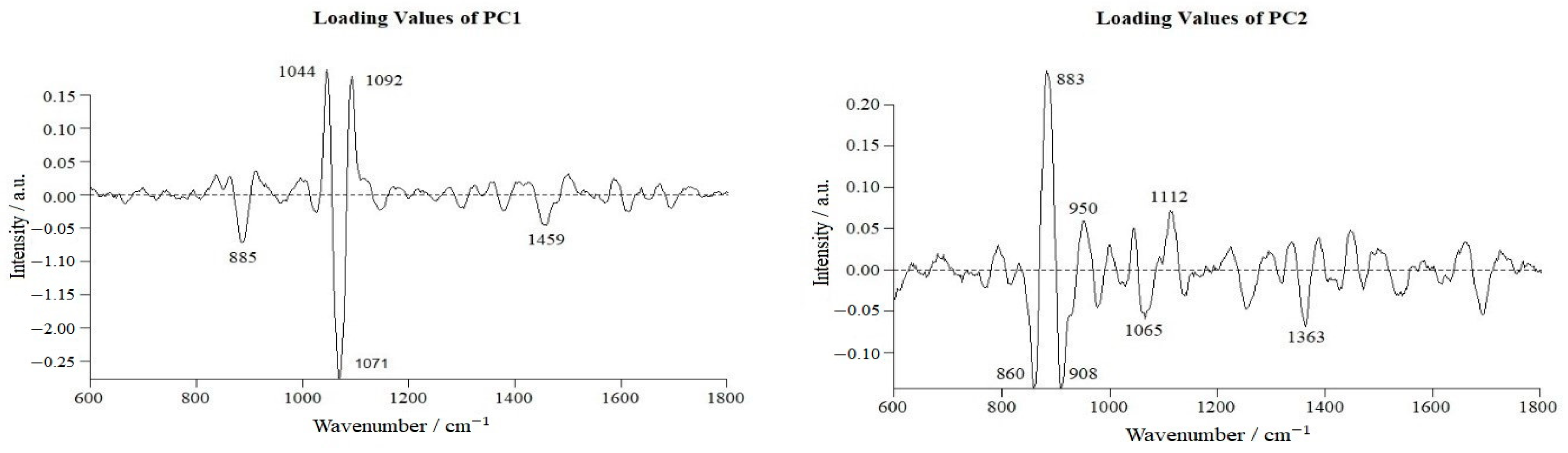
3.4. Differences in Raman Spectra between Normal and VBNC L. paracasei Zhang
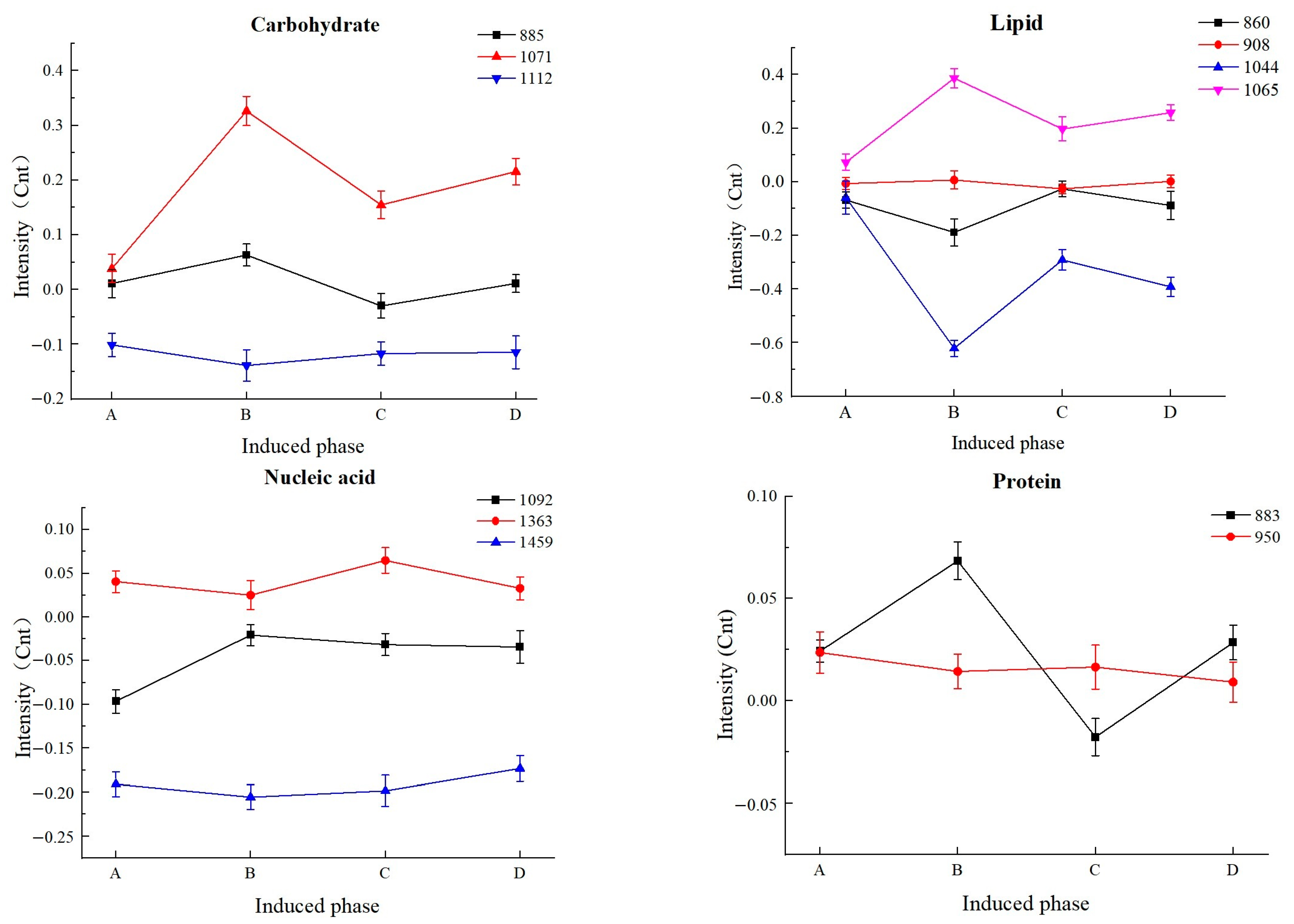
3.5. Changes in Molecular Constituents of L. paracasei Zhang after VBNC Induction
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, J.; Chen, X.; Liu, W.; Yang, M.; Zhang, H. Identification of Lactobacillus from koumiss by conventional and molecular methods. Eur. Food Res. Technol. 2008, 227, 1555–1561. [Google Scholar] [CrossRef]
- Zhu, H.; Cao, C.; Wu, Z.; Zhang, H.; Sun, Z.; Wang, M.; Xu, H.; Zhao, Z.; Wang, Y.; Pei, G.; et al. The probiotic L. casei Zhang slows the progression of acute and chronic kidney disease. Cell Metab. 2021, 33, 1926–1942.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yu, D.; Sun, Z.; Wu, R.; Chen, X.; Chen, W.; Meng, H.; Hu, S.; Zhang, H. Complete genome sequence of Lactobacillus casei Zhang, a new probiotic strain isolated from traditional homemade koumiss in Inner Mongolia, China. J. Bacteriol. 2010, 192, 5268–5269. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Huang, T.; Guo, S.; Wang, Y.; Wang, J.; Kwok, L.Y.; Dan, T.; Zhang, H.; Bilige, M. Probiotic Lactobacillus casei Zhang improved the properties of stirred yogurt. Food Biosci. 2020, 37, 100718. [Google Scholar] [CrossRef]
- Abid, S.; Farid, A.; Abid, R.; Rehman, M.U.; Alsanie, W.F.; Alhomrani, M.; Alamri, A.S.; Asdaq, S.M.B.; Hefft, D.I.; Saqib, S.; et al. Identification, Biochemical Characterization, and Safety Attributes of Locally Isolated Lactobacillus fermentum from Bubalus bubalis (buffalo) Milk as a Probiotic. Microorganisms 2022, 10, 954. [Google Scholar] [CrossRef]
- Mbye, M.; Baig, M.A.; AbuQamar, S.F.; El-Tarabily, K.A.; Obaid, R.S.; Osaili, T.M.; Al-Nabulsi, A.A.; Turner, M.S.; Shah, N.P.; Ayyash, M.M. Updates on understanding of probiotic lactic acid bacteria responses to environmental stresses and highlights on proteomic analyses. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1110–1124. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, S.; Robben, C.; Alter, T.; Rossmanith, P.; Mester, P. How to Evaluate Non-Growing Cells-Current Strategies for Determining Antimicrobial Resistance of VBNC Bacteria. Antibiotics 2021, 10, 115. [Google Scholar] [CrossRef]
- Xu, H.S.; Roberts, N.; Singleton, F.L.; Attwell, R.W.; Grimes, D.J.; Colwell, R.R. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb. Ecol. 1982, 8, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Suo, Y.; Xiang, Q.; Zhao, X.; Chen, S.; Ye, X.; Liu, D. Significance of viable but nonculturable Escherichia coli: Induction, detection, and control. J. Microbiol. Biotechnol. 2017, 27, 417–428. [Google Scholar] [CrossRef]
- Gao, R.; Liao, X.; Zhao, X.; Liu, D.; Ding, T. The diagnostic tools for viable but nonculturable pathogens in the food industry: Current status and future prospects. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2146–2175. [Google Scholar] [CrossRef]
- Yan, H.; Li, M.; Meng, L.; Zhao, F. Formation of viable but nonculturable state of Staphylococcus aureus under frozen condition and its characteristics. Int. J. Food Microbiol. 2021, 357, 109381. [Google Scholar] [CrossRef]
- Dong, K.; Pan, H.; Yang, D.; Rao, L.; Zhao, L.; Wang, Y.; Liao, X. Induction, detection, formation, and resuscitation of viable but non-culturable state microorganisms. Compr. Rev. Food Sci. Food Saf. 2020, 19, 149–183. [Google Scholar] [CrossRef]
- Lotoux, A.; Milohanic, E.; Bierne, H. The viable but non-culturable state of Listeria monocytogenes in the One-Health continuum. Front. Cell Infect. Microbiol. 2022, 12, 849915. [Google Scholar] [CrossRef]
- Serpaggi, V.; Remize, F.; Recorbet, G.; Gaudot-Dumas, E.; Sequeira-Le Grand, A.; Alexandre, H. Characterization of the “viable but nonculturable” (VBNC) state in the wine spoilage yeast Brettanomyces. Food Microbiol. 2012, 30, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Mannan, K.S.; Andrews, S. Viable but nonculturable bacteria: Food safety and public health perspective. ISRN Microbiol. 2013, 2013, 703813. [Google Scholar] [CrossRef]
- Cai, Y.; Liu, J.; Li, G.; Wong, P.K.; An, T. Formation mechanisms of viable but nonculturable bacteria through induction by light-based disinfection and their antibiotic resistance gene transfer risk: A review. Crit. Rev. Environ. Sci. Technol. 2022, 52, 3651–3688. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, K.; Zeng, Y.; Ye, Y.; Chen, L.; Huang, T. Development of a direct and rapid detection method for viable but non-culturable state of Pediococcus acidilactici. Front. Microbiol. 2021, 12, 87691. [Google Scholar] [CrossRef]
- İzgördü, Ö.K.; Darcan, C.; Kariptaş, E. Overview of VBNC, a survival strategy for microorganisms. 3 Biotech 2022, 12, 307. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Gao, L.; Wang, L.; Sun, X. Recent advances in single-cell analysis: Encapsulation materials, analysis methods and integrative platform for microfluidic technology. Talanta 2021, 234, 122671. [Google Scholar] [CrossRef]
- Zhu, Z.; Yang, C.J. Hydrogel droplet microfluidics for high-throughput single molecule/cell analysis. Acc. Chem. Res. 2017, 50, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; He, P.; Wang, Z.; Li, G.; Majed, N.; Gu, A.Z. Advances in single cell Raman spectroscopy technologies for biological and environmental applications. Curr. Opin. Biotechnol. 2020, 64, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Alraies, A.; Canetta, E.; Waddington, R.J.; Moseley, R.; Sloan, A.J. Discrimination of dental pulp stem cell regenerative heterogeneity by single-cell Raman spectroscopy. Tissue Eng. Part C Methods 2019, 25, 489–499. [Google Scholar] [CrossRef]
- Cui, D.; Kong, L.; Wang, Y.; Zhu, Y.; Zhang, C. In situ identification of environmental microorganisms with Raman spectroscopy. Environ. Sci. Ecotechnol. 2022, 11, 100187. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, P.; Gou, H.; Mou, C.; Huang, W.E.; Yang, M.; Xu, J.; Ma, B. Towards high-throughput microfluidic Raman-activated cell sorting. Analyst 2015, 140, 6163–6174. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ji, Y.; Wharfe, E.S.; Meadows, R.S.; March, P.; Goodacre, R.; Xu, J.; Huang, W.E. Raman activated cell ejection for isolation of single cells. Anal. Chem. 2013, 85, 10697–10701. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Mader, E.; Lee, T.K.; Woebken, D.; Wang, Y.; Zhu, D.; Palatinszky, M.; Schintlmeister, A.; Schmid, M.C.; Hanson, B.T.; et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. USA 2015, 112, E194–E203. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G. Raman spectroscopy in cell biology and microbiology. J. Raman Spectrosc. 2021, 52, 2348–2443. [Google Scholar] [CrossRef]
- Wang, X.; Ren, L.; Su, Y.; Ji, Y.; Liu, Y.; Li, C.; Li, X.; Zhang, Y.; Wang, W.; Hu, Q.; et al. Raman-activated droplet sorting (rads) for label-free high-throughput screening of microalgal single-cells. Anal. Chem. 2017, 89, 12569–12577. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, L.; He, T.; Cao, H.; Ren, X.; Ma, C.; Yang, J.; Huang, R.; Pan, G. Single cell Raman spectroscopy to identify different stages of proliferating human hepatocytes for cell therapy. Stem Cell Res. Ther. 2021, 12, 555. [Google Scholar] [CrossRef]
- Qi, Z.; Huang, Z.; Liu, C. Metabolism differences of biofilm and planktonic Pseudomonas aeruginosa in viable but nonculturable state induced by chlorine stress. Sci. Total Environ. 2022, 821, 153374. [Google Scholar] [CrossRef]
- Guo, L.; Ye, C.; Cui, L.; Wan, K.; Chen, S.; Zhang, S.; Yu, X. Population and single cell metabolic activity of UV-induced VBNC bacteria determined by CTC-FCM and D2O-labeled Raman spectroscopy. Environ. Int. 2019, 130, 104883. [Google Scholar] [CrossRef] [PubMed]
- Berney, M.; Hammes, F.; Bosshard, F.; Weilenmann, H.U.; Egli, T. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry. Appl. Environ. Microbiol. 2007, 73, 3283–3290. [Google Scholar] [CrossRef] [PubMed]
- Zaman, W.; Ullah, F.; Parmar, G.; Saqib, S.; Ayaz, A.; Park, S. Foliar micromorphology of selected medicinal Lamiaceae taxa and their taxonomic implication using scanning electron microscopy. Microsc. Res. Tech. 2022, 85, 3217–3236. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, Y.; Tao, Y.; Muhamadali, H.; Goodacre, R.; Zhou, N.Y.; Preston, G.M.; Xu, J.; Huang, W.E. Reverse and multiple stable isotope probing to study bacterial metabolism and interactions at the single cell level. Nal. Chem. 2016, 88, 9443–9450. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Ji, Y.; Teng, L.; Zhang, H. Using Raman spectroscopy and chemometrics to identify the growth phase of Lactobacillus casei Zhang during batch culture at the single-cell level. Microb. Cell Fact. 2017, 16, 233. [Google Scholar] [CrossRef]
- Stöckel, S.; Meisel, S.; Elschner, M.; Rösch, P.; Popp, J. Identification of Bacillus anthracis via Raman spectroscopy and chemometric approaches. Anal. Chem. 2012, 84, 9873–9880. [Google Scholar] [CrossRef]
- Liu, J.; Li, L.; Peters, B.M.; Li, B.; Chen, L.; Deng, Y.; Xu, Z.; Shirtliff, M.E. The viable but nonculturable state induction and genomic analyses of Lactobacillus casei BM-LC14617, a beer-spoilage bacterium. Microbiol. Open 2017, 6, e00506. [Google Scholar] [CrossRef]
- Kong, H.G.; Bae, J.Y.; Lee, H.J.; Joo, H.J.; Jung, E.J.; Chung, E.; Lee, S.W. Induction of the viable but nonculturable state of Ralstonia solanacearum by low temperature in the soil microcosm and its resuscitation by catalase. PLoS ONE 2014, 9, e109792. [Google Scholar] [CrossRef]
- Yoon, J.H.; Lee, S.Y. Characteristics of viable-but-nonculturable Vibrio parahaemolyticus induced by nutrient-deficiency at cold temperature. Crit. Rev. Food Sci. Nutr. 2020, 60, 1302–1320. [Google Scholar] [CrossRef]
- Pinto, D.; Santos, M.A.; Chambel, L. Thirty years of viable but nonculturable state research: Unsolved molecular mechanisms. Crit. Rev. Microbiol. 2015, 41, 61–76. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2015, 50, 46–111. [Google Scholar] [CrossRef]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, W.E.; Cui, L.; Wagner, M. Single cell stable isotope probing in microbiology using Raman microspectroscopy. Curr. Opin. Biotechnol. 2016, 41, 34–42. [Google Scholar] [CrossRef]
- Xie, M.; Xu, L.; Zhang, R.; Zhou, Y.; Xiao, Y.; Su, X.; Shen, C.; Sun, F.; Hashmi, M.Z.; Lin, H.; et al. Viable but Nonculturable State of Yeast Candida sp. Strain LN1 Induced by High Phenol Concentrations. Appl. Environ. Microbiol. 2021, 87, e0111021. [Google Scholar] [CrossRef]
- Capozzi, V.; Di Toro, M.R.; Grieco, F.; Michelotti, V.; Salma, M.; Lamontanara, A.; Russo, P.; Orrù, L.; Alexandre, H.; Spano, G. Viable but not culturable (VBNC) state of Brettanomyces bruxellensis in wine: New insights on molecular basis of VBNC behaviour using a transcriptomic approach. Food Microbiol. 2016, 59, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.L.; Wu, C.; Chen, M.J. Growth behaviors, thermostable direct hemolysin secretion and fatty acid profiles of acid-adapted and non-adapted Vibrio parahaemolyticus. Int. J Nutr. Food Eng. 2014, 8, 1099–1103. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, Y.; An, H.; Hao, Y.; Hu, X.; Liao, X. New Insights into the Formation of Viable but Nonculturable Escherichia coli O157:H7 Induced by High-Pressure CO2. mBio 2016, 7, e00961-16. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Liu, C. Metabolic characteristics and markers in viable but nonculturable state of Pseudomonas aeruginosa induced by chlorine stress. Environ. Res. 2022, 214, 114111. [Google Scholar] [CrossRef] [PubMed]
- Postnikova, O.A.; Shao, J.; Mock, N.M.; Baker, C.J.; Nemchinov, L.G. Gene expression profiling in viable but nonculturable (VBNC) cells of Pseudomonas syringae pv. syringae. Front. Microbiol. 2015, 6, 1419. [Google Scholar] [CrossRef]
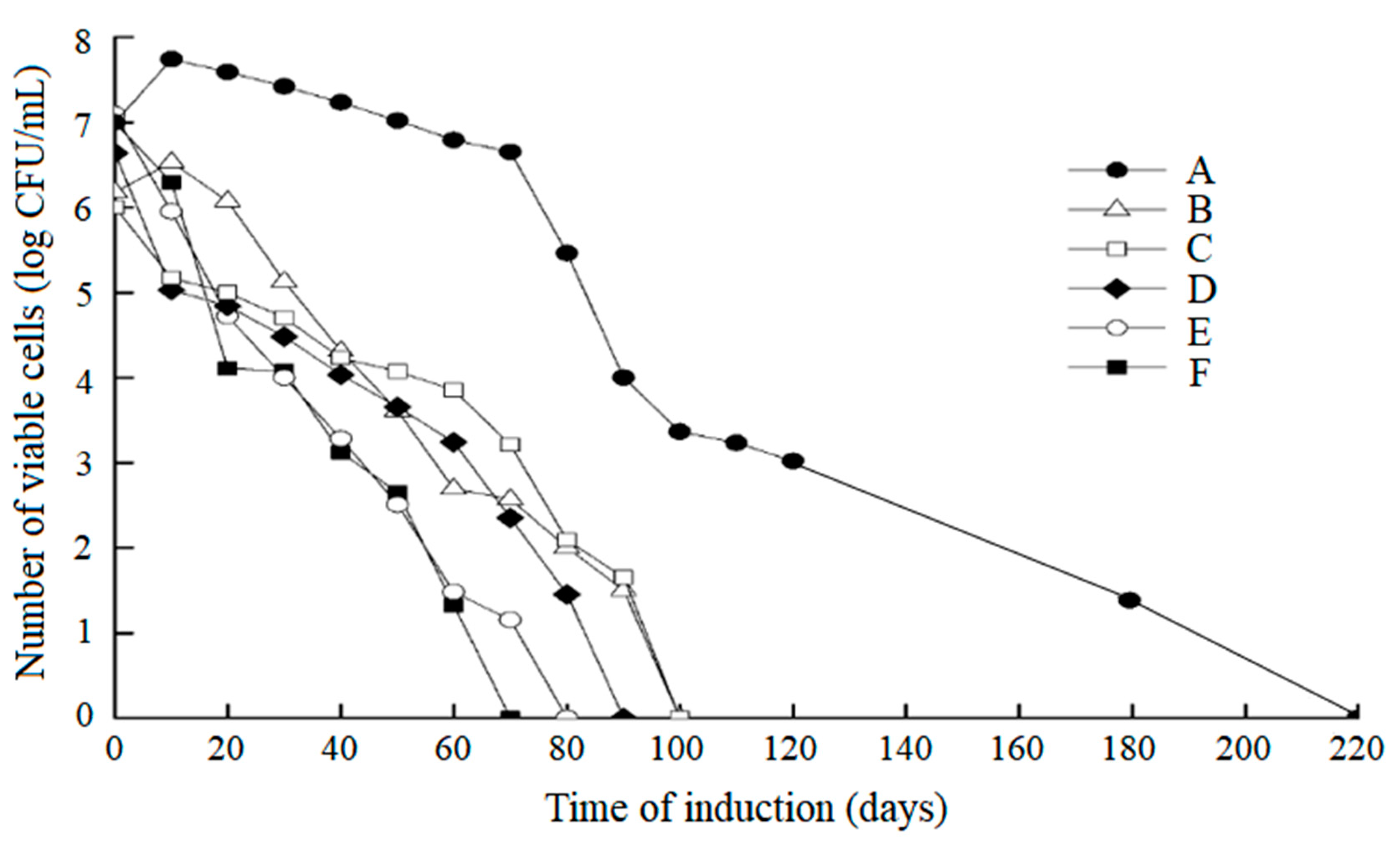
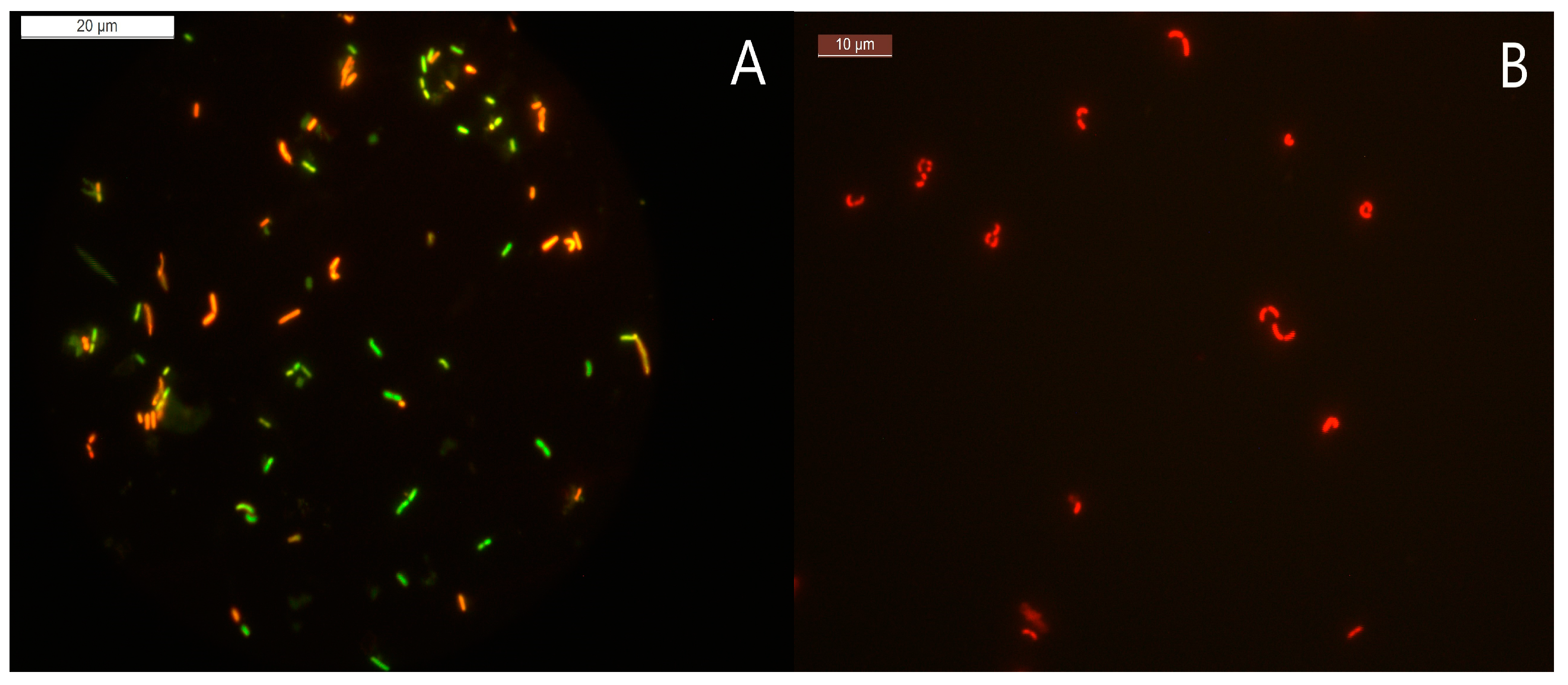
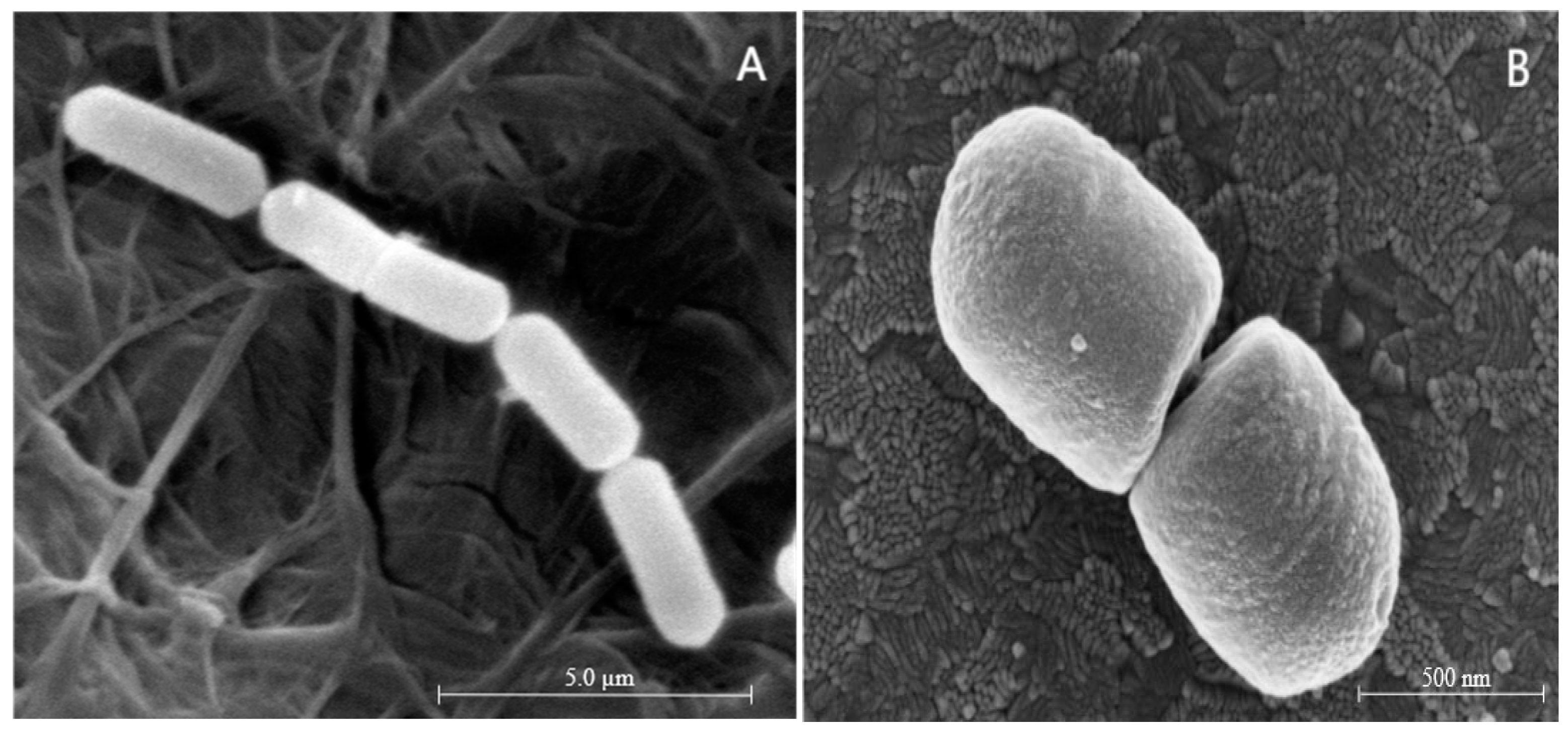

| Condition | VBNC Induction Medium | pH | Temperature |
|---|---|---|---|
| A | MRS broth | 6.8 | 4 °C |
| B | Luria–Bertani broth | 2.0 to 3.0 | 4 °C |
| C | Sterilized distilled water | 7.0 | Room temperature |
| D | Sterilized distilled water | 7.0 | 4 °C |
| E | 0.85% NaCl solution | 7.0 | 4 °C |
| F | 0.85% NaCl solution | 7.0 | −20 °C |
| Molecular Group | Raman Frequency (cm−1) | Chemical Assignment | Reference |
|---|---|---|---|
| Carbohydrate | 885 | Disaccharide (cellobiose), (C-O-C) skeletal mode | [41] |
| 1071 | D-Fructose-6 | [42] | |
| 1112 | Saccharide band (overlaps with acyl band) | [41] | |
| Lipid | 860 | Phosphate group; phosphatidic acid | [41] |
| 908 | Myristic acid | [42] | |
| 1044 | ν3PO43− (symmetric stretching vibration of ν3PO43− of HA) | [41] | |
| 1065 | Palmitic acid, fatty acid | [41] | |
| Protein | 883 | ρ (CH2) (protein assignment) | [41] |
| 950 | Most probably due to single bond stretching vibrations of the amino acids proline and valine and polysaccharides | [41] | |
| 1003 | Symmetric ring breathing mode of phenylalanine | [43] | |
| Nucleic acid | 729 | Adenine ring breathing | [43] |
| 786 | Cytosine | [35] | |
| 1092 | Phosphodioxy | [41] | |
| 1363 | Guanine (N7, B, Z-marker) | [41] | |
| 1459 | Deoxyribose; δ(CH2) | [41] | |
| Others | 540 | L-Histidine | [42] |
| 643 | Glutathione | [42] | |
| 1247 | Amide III | [43] | |
| 1328 | L-Tryptophan | [42] | |
| 1575 | Tryptophan-related bands | [27] | |
| 1660 | L-Valine | [42] | |
| 2935 | CH stretching of lipid and protein | [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, Q.; Bo, X.; Chen, L.; Ren, Y.; Wang, H.; Kwok, L.-Y.; Liu, W. Comparative Analysis Using Raman Spectroscopy of the Cellular Constituents of Lacticaseibacillus paracasei Zhang in a Normal and Viable but Nonculturable State. Microorganisms 2023, 11, 1266. https://doi.org/10.3390/microorganisms11051266
Bao Q, Bo X, Chen L, Ren Y, Wang H, Kwok L-Y, Liu W. Comparative Analysis Using Raman Spectroscopy of the Cellular Constituents of Lacticaseibacillus paracasei Zhang in a Normal and Viable but Nonculturable State. Microorganisms. 2023; 11(5):1266. https://doi.org/10.3390/microorganisms11051266
Chicago/Turabian StyleBao, Qiuhua, Xiaoyu Bo, Lu Chen, Yan Ren, Huiying Wang, Lai-Yu Kwok, and Wenjun Liu. 2023. "Comparative Analysis Using Raman Spectroscopy of the Cellular Constituents of Lacticaseibacillus paracasei Zhang in a Normal and Viable but Nonculturable State" Microorganisms 11, no. 5: 1266. https://doi.org/10.3390/microorganisms11051266
APA StyleBao, Q., Bo, X., Chen, L., Ren, Y., Wang, H., Kwok, L.-Y., & Liu, W. (2023). Comparative Analysis Using Raman Spectroscopy of the Cellular Constituents of Lacticaseibacillus paracasei Zhang in a Normal and Viable but Nonculturable State. Microorganisms, 11(5), 1266. https://doi.org/10.3390/microorganisms11051266







