Molecular Characterization of Dengue Virus Strains from the 2019–2020 Epidemic in Hanoi, Vietnam
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients, Sample Processing, and Laboratory Testing
2.2. Virus Isolation
2.3. Envelope Nested RT-PCR and Whole-Genome RT-PCR
2.4. Envelope and Whole-Genome Sequencing
2.5. Phylogenetic and Genetic Diversity Analyses
2.6. Statistical Tests
3. Results
3.1. DENVs in Hanoi, Vietnam, in 2019–2020
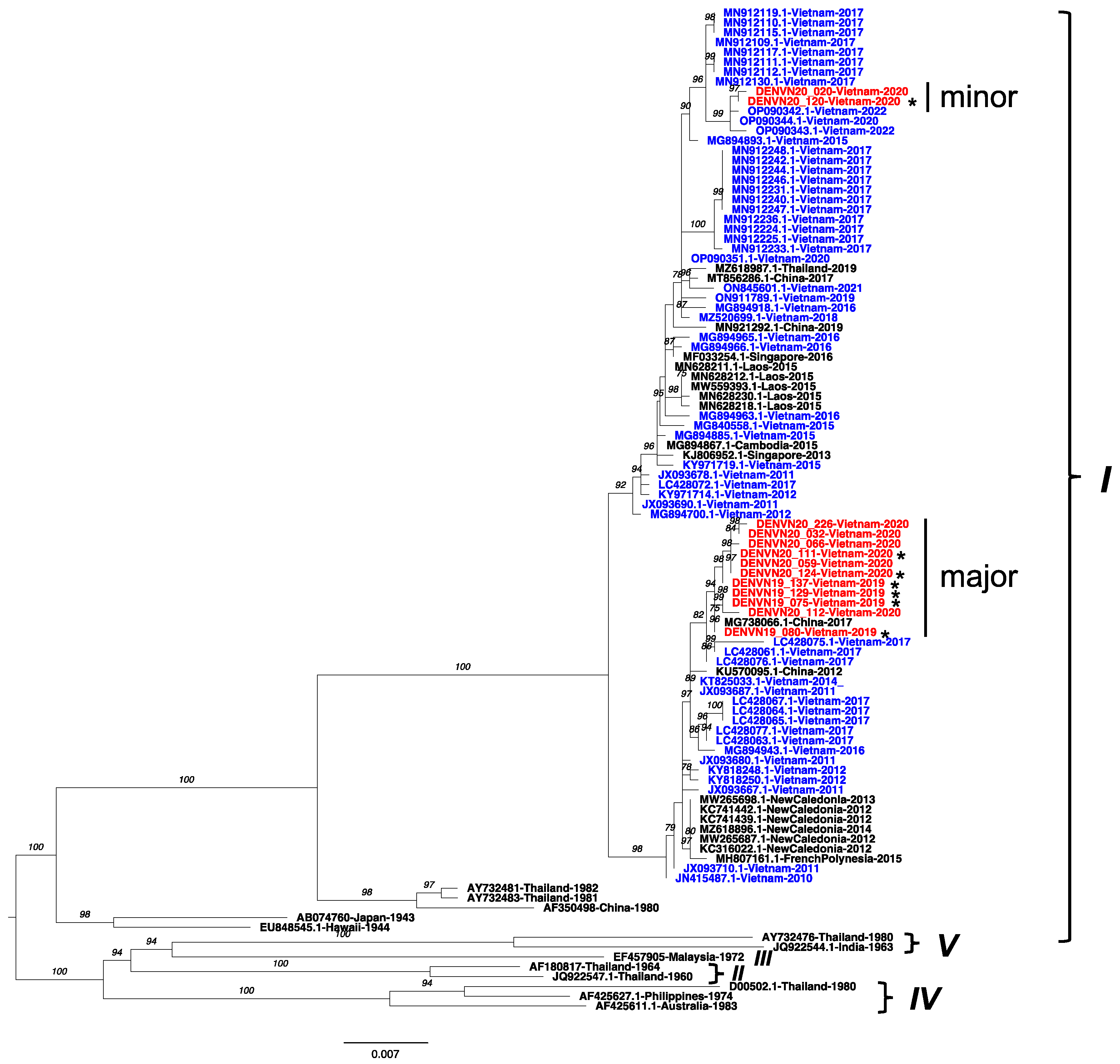
3.2. Persistence of DENV-1 Genotype I in Vietnam
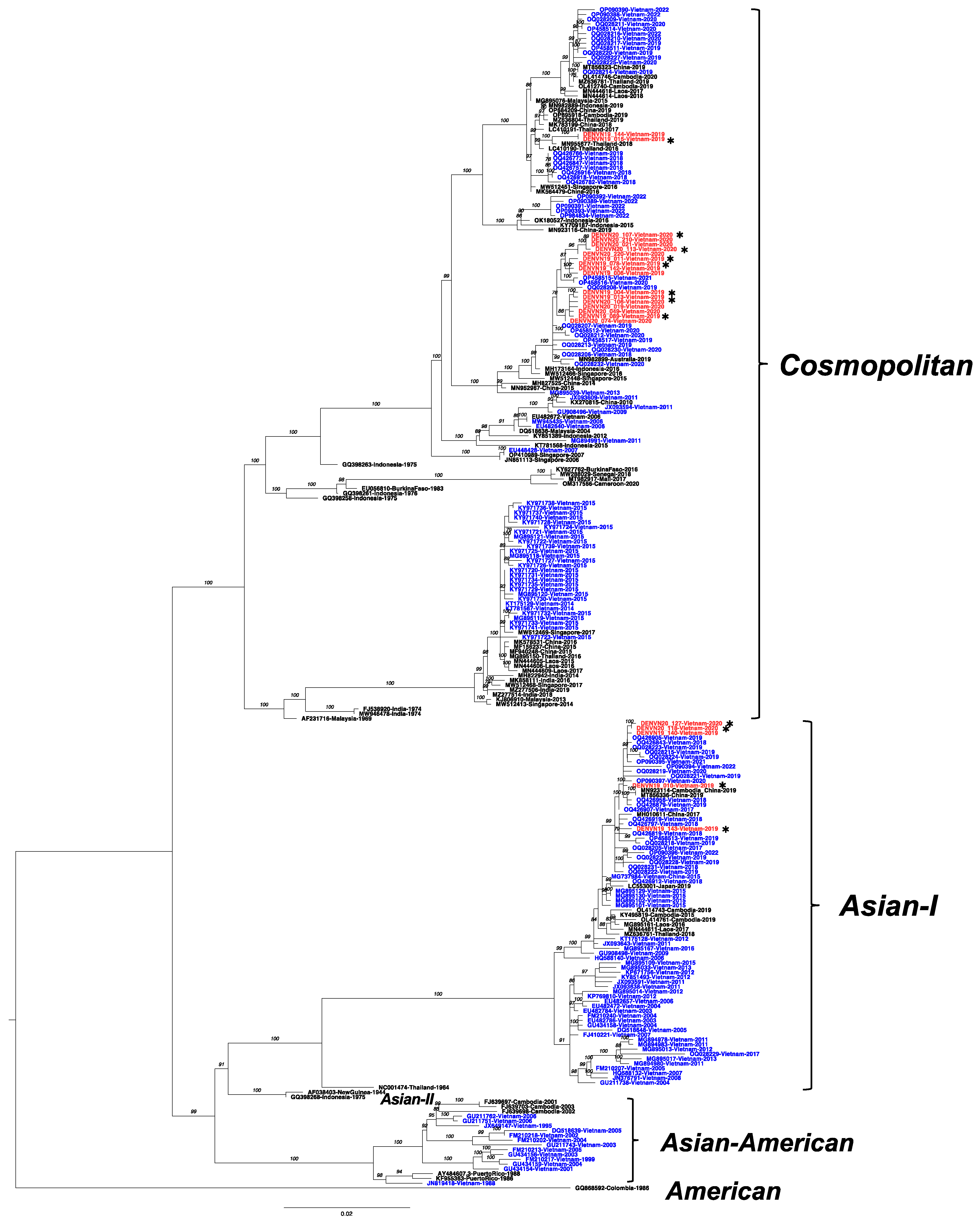
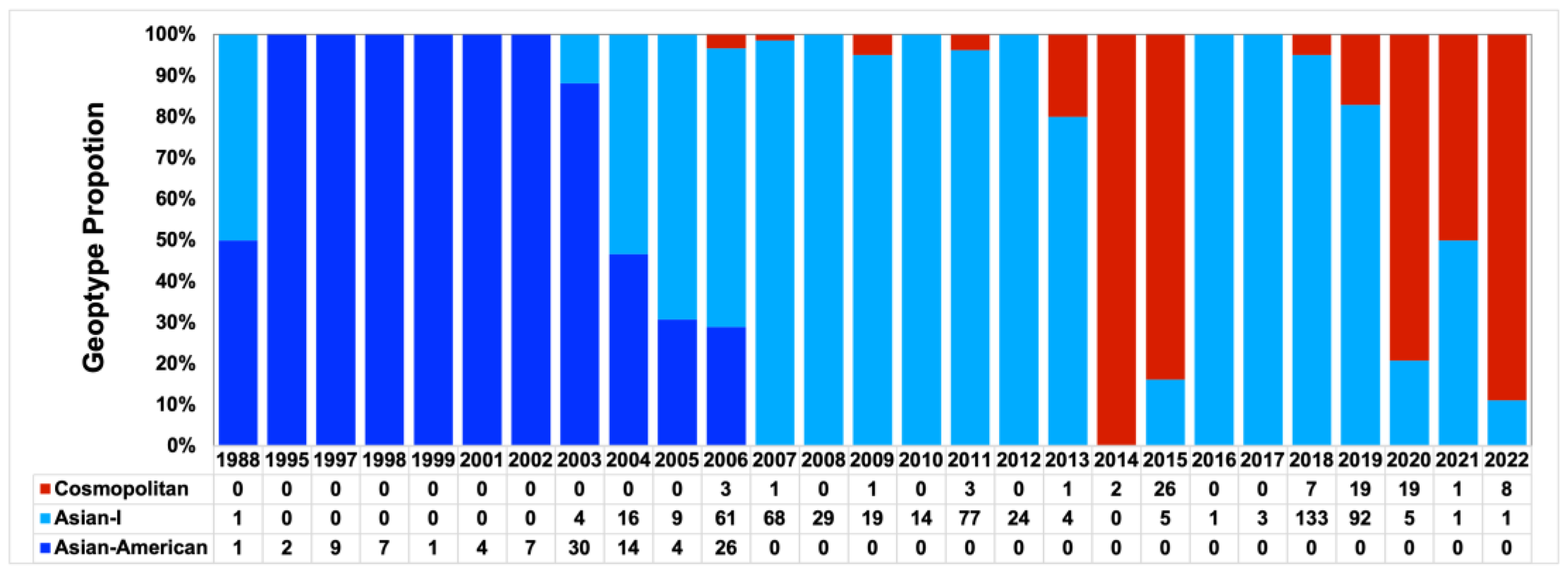
3.3. Co-Circulation of the Asian-I and Cosmopolitan Genotype of DENV-2 in Vietnam
3.4. Emergence of DENV-2 Cosmopolitan Lineage C (Asian-Pacific) in Vietnam
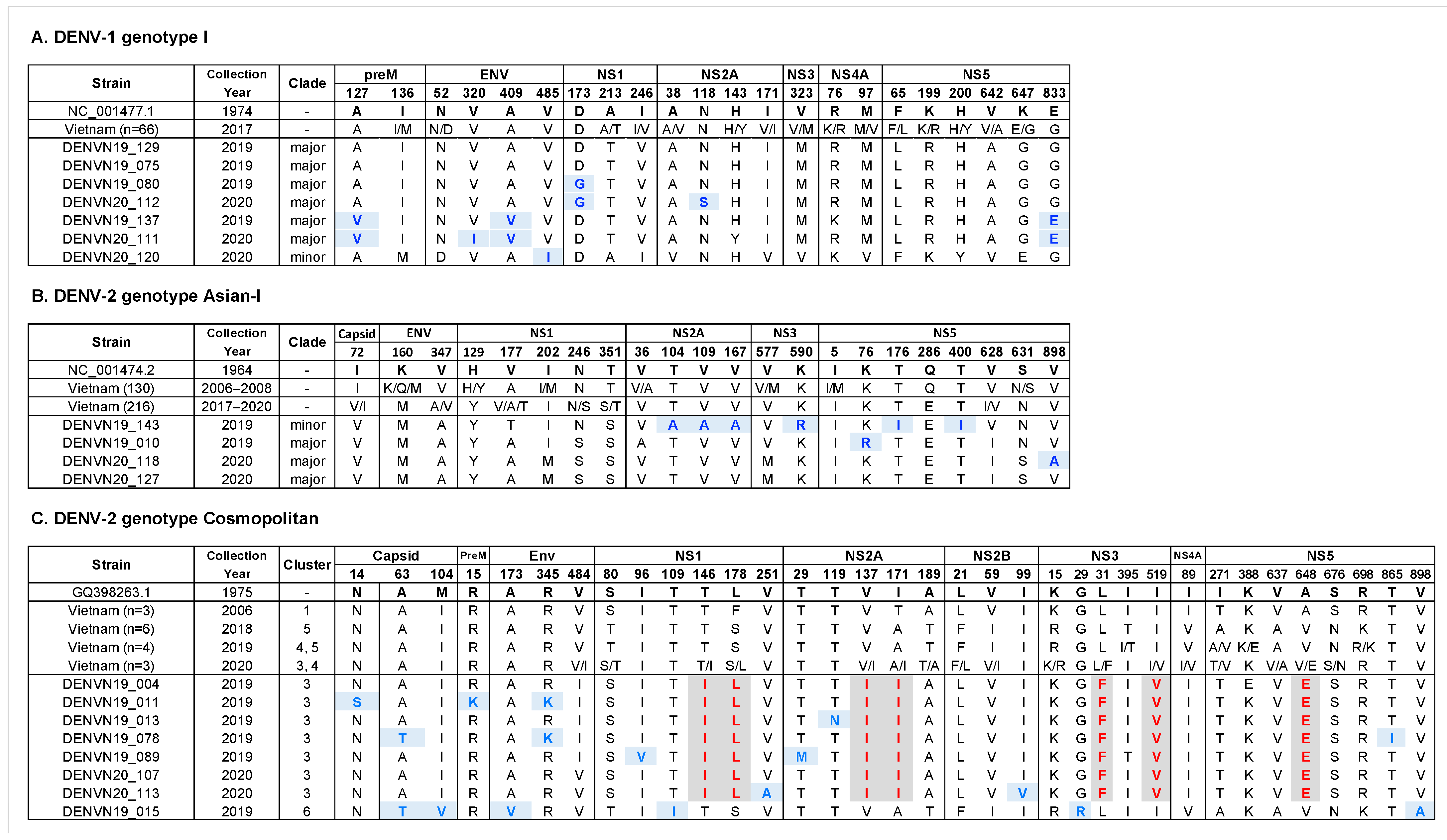
3.5. Amino Acid Polymorphisms within Genotypes of Hanoi DENV Strains
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Dengue Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition; World Health Organization: Geneva, Switzerland, 2009; p. 147. [Google Scholar]
- World Health Organization. Dengue and Severe Dengue. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 3 April 2023).
- Guzman, M.G.; Alvarez, M.; Halstead, S.B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: An historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 2013, 158, 1445–1459. [Google Scholar] [CrossRef]
- Chen, R.; Vasilakis, N. Dengue—Quo tu et quo vadis? Viruses 2011, 3, 1562–1608. [Google Scholar] [CrossRef]
- Vu, T.T.; Holmes, E.C.; Duong, V.; Nguyen, T.Q.; Tran, T.H.; Quail, M.; Churcher, C.; Parkhill, J.; Cardosa, J.; Farrar, J.; et al. Emergence of the Asian 1 genotype of dengue virus serotype 2 in viet nam: In vivo fitness advantage and lineage replacement in South-East Asia. PLoS Negl. Trop. Dis. 2010, 4, e757. [Google Scholar] [CrossRef]
- Aguas, R.; Dorigatti, I.; Coudeville, L.; Luxemburger, C.; Ferguson, N.M. Cross-serotype interactions and disease outcome prediction of dengue infections in Vietnam. Sci. Rep. 2019, 9, 9395. [Google Scholar] [CrossRef] [PubMed]
- Takemura, T.; Nguyen, C.T.; Pham, H.C.; Nguyen, T.T.; Hoang, V.M.P.; Nguyen, L.K.H.; Nabeshima, T.; Nguyen, T.T.T.; Le, T.Q.M.; Moi, M.L.; et al. The 2017 Dengue virus 1 outbreak in northern Vietnam was caused by a locally circulating virus group. Trop. Med. Health 2022, 50, 3. [Google Scholar] [CrossRef] [PubMed]
- Phadungsombat, J.; Lin, M.Y.; Srimark, N.; Yamanaka, A.; Nakayama, E.E.; Moolasart, V.; Suttha, P.; Shioda, T.; Uttayamakul, S. Emergence of genotype Cosmopolitan of dengue virus type 2 and genotype III of dengue virus type 3 in Thailand. PLoS ONE 2018, 13, e0207220. [Google Scholar] [CrossRef]
- Shu, P.Y.; Chang, S.F.; Kuo, Y.C.; Yueh, Y.Y.; Chien, L.J.; Sue, C.L.; Lin, T.H.; Huang, J.H. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J. Clin. Microbiol. 2003, 41, 2408–2416. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef]
- Baele, G.; Lemey, P.; Suchard, M.A. Genealogical Working Distributions for Bayesian Model Testing with Phylogenetic Uncertainty. Syst. Biol. 2016, 65, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Gill, M.S.; Lemey, P.; Faria, N.R.; Rambaut, A.; Shapiro, B.; Suchard, M.A. Improving Bayesian population dynamics inference: A coalescent-based model for multiple loci. Mol. Biol. Evol. 2013, 30, 713–724. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Fourie, T.; El Bara, A.; Dubot-Peres, A.; Grard, G.; Briolant, S.; Basco, L.K.; Ouldabdallahi Moukah, M.; Leparc-Goffart, I. Emergence of dengue virus serotype 2 in Mauritania and molecular characterization of its circulation in West Africa. PLoS Negl. Trop. Dis. 2021, 15, e0009829. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Regional Office for the Western, P. Dengue Situation Updates 2020. Available online: https://iris.wpro.who.int/handle/10665.10661/14461 (accessed on 3 April 2023).
- Raghwani, J.; Rambaut, A.; Holmes, E.C.; Hang, V.T.; Hien, T.T.; Farrar, J.; Wills, B.; Lennon, N.J.; Birren, B.W.; Henn, M.R.; et al. Endemic dengue associated with the co-circulation of multiple viral lineages and localized density-dependent transmission. PLoS Pathog. 2011, 7, e1002064. [Google Scholar] [CrossRef] [PubMed]
- Lytton, S.D.; Nematollahi, G.; van Tong, H.; Xuan Anh, C.; Hung, H.V.; Hoan, N.X.; Diez, G.; Schumacher, T.; Landt, O.; Melchior, W.; et al. Predominant secondary dengue infection among Vietnamese adults mostly without warning signs and severe disease. Int. J. Infect. Dis. 2020, 100, 316–323. [Google Scholar] [CrossRef]
- Dang, T.T.; Pham, M.H.; Bui, H.V.; Van Le, D. Whole genome sequencing and genetic variations in several dengue virus type 1 strains from unusual dengue epidemic of 2017 in Vietnam. Virol. J. 2020, 17, 7. [Google Scholar] [CrossRef]
- Gibbons, R.V.; Kalanarooj, S.; Jarman, R.G.; Nisalak, A.; Vaughn, D.W.; Endy, T.P.; Mammen, M.P., Jr.; Srikiatkhachorn, A. Analysis of repeat hospital admissions for dengue to estimate the frequency of third or fourth dengue infections resulting in admissions and dengue hemorrhagic fever, and serotype sequences. Am. J. Trop. Med. Hyg. 2007, 77, 910–913. [Google Scholar] [CrossRef]
- Phan, D.Q.; Nguyen, L.D.N.; Pham, S.T.; Nguyen, T.; Pham, P.T.T.; Nguyen, S.T.H.; Pham, D.T.; Pham, H.T.; Tran, D.K.; Le, S.H.; et al. The Distribution of Dengue Virus Serotype in Quang Nam Province (Vietnam) during the Outbreak in 2018. Int. J. Environ. Res. Public Health 2022, 19, 1285. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.; Le, N.M.; Simmons, C.P.; Wolbers, M.; Wertheim, H.F.; Pham, T.K.; Tran, T.H.; Trinh, T.M.; Nguyen, T.L.; Nguyen, V.T.; et al. Immunological and viral determinants of dengue severity in hospitalized adults in Ha Noi, Viet Nam. PLoS Negl. Trop. Dis. 2011, 5, e967. [Google Scholar] [CrossRef] [PubMed]
- Cuong, H.Q.; Vu, N.T.; Cazelles, B.; Boni, M.F.; Thai, K.T.; Rabaa, M.A.; Quang, L.C.; Simmons, C.P.; Huu, T.N.; Anders, K.L. Spatiotemporal dynamics of dengue epidemics, southern Vietnam. Emerg. Infect. Dis. 2013, 19, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Rabaa, M.A.; Simmons, C.P.; Fox, A.; Le, M.Q.; Nguyen, T.T.; Le, H.Y.; Gibbons, R.V.; Nguyen, X.T.; Holmes, E.C.; Aaskov, J.G. Dengue virus in sub-tropical northern and central Viet Nam: Population immunity and climate shape patterns of viral invasion and maintenance. PLoS Negl. Trop. Dis. 2013, 7, e2581. [Google Scholar] [CrossRef]
- Cuong, H.Q.; Hien, N.T.; Duong, T.N.; Phong, T.V.; Cam, N.N.; Farrar, J.; Nam, V.S.; Thai, K.T.; Horby, P. Quantifying the emergence of dengue in Hanoi, Vietnam: 1998–2009. PLoS Negl. Trop. Dis. 2011, 5, e1322. [Google Scholar] [CrossRef]
- Thanh Toan, D.T.; Hu, W.; Quang Thai, P.; Hoat, L.N.; Wright, P.; Martens, P. Hot spot detection and spatio-temporal dispersion of dengue fever in Hanoi, Vietnam. Glob. Health Action 2013, 6, 18632. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Than, P.Q.T.; Nguyen, T.H.; Vu, G.T.; Hoang, C.L.; Tran, T.T.; Truong, N.T.; Nguyen, S.H.; Do, H.P.; Ha, G.H.; et al. Knowledge, Attitude and Practice about Dengue Fever among Patients Experiencing the 2017 Outbreak in Vietnam. Int. J. Environ. Res. Public Health 2019, 16, 976. [Google Scholar] [CrossRef]
- Quyen, D.L.; Thanh Le, N.; Van Anh, C.T.; Nguyen, N.B.; Hoang, D.V.; Montgomery, J.L.; Kutcher, S.C.; Hoang Le, N.; Hien, N.T.; Hue Kien, D.T.; et al. Epidemiological, Serological, and Virological Features of Dengue in Nha Trang City, Vietnam. Am. J. Trop. Med. Hyg. 2018, 98, 402–409. [Google Scholar] [CrossRef]
- Van Tuan, L.; Van, N.T.T.; Quan, N.H.; Duoc, P.T. Phylogeny of Dengue virus type 2 isolated in the Central Highlands, Vietnam. Rev. Biol. Trop. 2017, 65, 819–826. [Google Scholar]
- Yang, C.F.; Chang, S.F.; Hsu, T.C.; Su, C.L.; Wang, T.C.; Lin, S.H.; Yang, S.L.; Lin, C.C.; Shu, P.Y. Molecular characterization and phylogenetic analysis of dengue viruses imported into Taiwan during 2011–2016. PLoS Negl. Trop. Dis. 2018, 12, e0006773. [Google Scholar] [CrossRef]
- Brook, C.E.; Li, Y.; Yek, C.; Northrup, G.R.; Lay, S.; Chea, S.; Ahyong, V.; Parker, D.M.; Man, S.; Pacheco, A.R.; et al. The Perfect Storm of 2019: An immunological and phylodynamic analysis of Cambodia’s unprecedented dengue outbreak. medRxiv 2022. [Google Scholar] [CrossRef]
- Ngwe Tun, M.M.; Muthugala, R.; Nabeshima, T.; Rajamanthri, L.; Jayawardana, D.; Attanayake, S.; Soe, A.M.; Dumre, S.P.; Ando, T.; Hayasaka, D.; et al. Unusual, neurological and severe dengue manifestations during the outbreak in Sri Lanka, 2017. J. Clin. Virol. 2020, 125, 104304. [Google Scholar] [CrossRef] [PubMed]
- Ngwe Tun, M.M.; Pandey, K.; Nabeshima, T.; Kyaw, A.K.; Adhikari, M.; Raini, S.K.; Inoue, S.; Dumre, S.P.; Pandey, B.D.; Morita, K. An Outbreak of Dengue Virus Serotype 2 Cosmopolitan Genotype in Nepal, 2017. Viruses 2021, 13, 1444. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Phadungsombat, J.; Nakayama, E.E.; Saito, A.; Egawa, A.; Sato, T.; Rahim, R.; Hasan, A.; Lin, M.Y.; Takasaki, T.; et al. Genotype replacement of dengue virus type 3 and clade replacement of dengue virus type 2 genotype Cosmopolitan in Dhaka, Bangladesh in 2017. Infect. Genet. Evol. 2019, 75, 103977. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.P.; Padilla, C.; Figueroa, D.; Manrique, C.; Cabezas, C. Emergence of the Cosmopolitan genotype of dengue virus serotype 2 (DENV2) in Madre de Dios, Peru, 2019. Rev. Peru. Med. Exp. Salud Publica. 2022, 39, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Pereira, L.A.; Santiago, G.A.; Fonseca, V.; Mendoza, M.P.G.; de Oliveira, C.; de Moraes, L.; Xavier, J.; Tosta, S.; Fristch, H.; et al. Emergence of Dengue Virus Serotype 2 Cosmopolitan Genotype, Brazil. Emerg. Infect. Dis. 2022, 28, 1725–1727. [Google Scholar] [CrossRef]
- Pollett, S.; Melendrez, M.C.; Maljkovic Berry, I.; Duchene, S.; Salje, H.; Cummings, D.A.T.; Jarman, R.G. Understanding dengue virus evolution to support epidemic surveillance and counter-measure development. Infect. Genet. Evol. 2018, 62, 279–295. [Google Scholar] [CrossRef]
- Wang, C.; Katzelnick, L.C.; Montoya, M.; Hue, K.D.; Simmons, C.P.; Harris, E. Evolutionarily Successful Asian 1 Dengue Virus 2 Lineages Contain One Substitution in Envelope That Increases Sensitivity to Polyclonal Antibody Neutralization. J. Infect. Dis. 2016, 213, 975–984. [Google Scholar] [CrossRef]
- Lambrechts, L.; Fansiri, T.; Pongsiri, A.; Thaisomboonsuk, B.; Klungthong, C.; Richardson, J.H.; Ponlawat, A.; Jarman, R.G.; Scott, T.W. Dengue-1 virus clade replacement in Thailand associated with enhanced mosquito transmission. J. Virol. 2012, 86, 1853–1861. [Google Scholar] [CrossRef]
- O’Connor, O.; Ou, T.P.; Aubry, F.; Dabo, S.; Russet, S.; Girault, D.; In, S.; Minier, M.; Lequime, S.; Hoem, T.; et al. Potential role of vector-mediated natural selection in dengue virus genotype/lineage replacements in two epidemiologically contrasted settings. Emerg. Microbes. Infect. 2021, 10, 1346–1357. [Google Scholar] [CrossRef]
- Quiner, C.A.; Parameswaran, P.; Ciota, A.T.; Ehrbar, D.J.; Dodson, B.L.; Schlesinger, S.; Kramer, L.D.; Harris, E. Increased replicative fitness of a dengue virus 2 clade in native mosquitoes: Potential contribution to a clade replacement event in Nicaragua. J. Virol. 2014, 88, 13125–13134. [Google Scholar] [CrossRef] [PubMed]


| Total DENV (n = 133) | DENV-1 (n = 22) | DENV-2 (n = 64) | p-Value | ||||
|---|---|---|---|---|---|---|---|
| Frequency | Percentage | Frequency | Percentage | Frequency | Percentage | ||
| Sex | |||||||
| Male | 55 | 41.35 | 11 | 50.00 | 28 | 43.75 | 0.6290 b |
| Female | 51 | 38.35 | 8 | 36.36 | 22 | 34.38 | >0.9999 b |
| No data | 27 | 20.30 | 3 | 13.64 | 14 | 21.88 | 0.5413 b |
| Age | |||||||
| 18–29 | 73 | 54.89 | 14 | 63.64 | 31 | 48.44 | 0.3225 b |
| 30–49 | 41 | 30.83 | 8 | 36.36 | 23 | 35.94 | >0.9999 b |
| >50 | 13 | 9.77 | 0 | 0.00 | 8 | 12.50 | 0.1071 b |
| N/A | 6 | 4.51 | 0 | 0.00 | 2 | 3.13 | >0.9999 b |
| Symptoms a | |||||||
| Fever | 99 | 98.02 | 17 | 94.44 | 48 | 97.96 | 0.4681 b |
| Fatigue | 91 | 88.35 | 17 | 94.44 | 43 | 87.76 | 0.6645 b |
| Muscle pain | 55 | 53.40 | 9 | 50.00 | 21 | 42.86 | 0.7823 b |
| Joint pain | 26 | 25.24 | 5 | 27.78 | 11 | 22.45 | 0.7488 b |
| Skin rash | 7 | 6.80 | 2 | 11.11 | 3 | 6.12 | 0.6051 b |
| Headache | 6 | 5.83 | 2 | 11.11 | 1 | 2.04 | 0.1735 b |
| Tourniquet | 5 | 4.85 | 1 | 5.56 | 3 | 6.12 | >0.9999 b |
| Abdomen pain | 4 | 3.88 | 2 | 11.11 | 2 | 4.08 | 0.2909 b |
| Joint swelling | 3 | 2.91 | 1 | 5.56 | 1 | 2.04 | 0.4681 b |
| Vomit | 2 | 1.94 | 0 | 0.00 | 2 | 4.08 | >0.9999 b |
| Other (gingival bleeding, ascites, and reduction in urination) | 3 | 2.91 | 0 | 0.00 | 3 | 6.12 | 0.5581 b |
| Chemical a | Median | IQR | Median | IQR | Median | IQR | |
| Platelets (103/μL) | 158 | 126–185 | 131.00 | 115.50–194.75 | 162 | 129–183 | 0.2470 c |
| White blood cells (103/μL) | 4.6 | 3–5.9 | 3.8 | 2.98–5.98 | 4.9 | 3.15–6.3 | 0.3337 c |
| Hemoglobin (g/L) | |||||||
| Male | 148 | 141–155.5 | 146 | 140–155 | 147 | 141–153 | 0.9431 c |
| Female | 131 | 125–141.25 | 124 | 111–145 | 131 | 126–134.25 | 0.1245 c |
| Hematocrit (%) | |||||||
| Male | 43.3 | 41.4–45.3 | 43 | 41.6–44.9 | 42.5 | 40.8–45.3 | 0.9557 c |
| Female | 38.35 | 36.75–41.35 | 36.6 | 34–43.8 | 38.45 | 36.95–39.9 | 0.2858 c |
| Strain | Resident Area | Country | Collection Date | NS1 Ag | Anti-IgM | Anti-IgG | Serotype | Genotype | Accession Number | |
|---|---|---|---|---|---|---|---|---|---|---|
| Envelope | Full Genome | |||||||||
| DENVN19_075 | ND | Vietnam | November 2019 | POS | NEG | NEG | DENV-1 | I | OQ832560 | OQ832609 |
| DENVN19_080 | Hanoi | Vietnam | December 2019 | POS | NEG | NEG | DENV-1 | I | OQ832561 | OQ832610 |
| DENVN19_129 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-1 | I | ud | OQ832611 |
| DENVN19_137 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-1 | I | OQ832562 | OQ832612 |
| DENVN20_020 | Phu Tho | Vietnam | September 2020 | POS | POS | NEG | DENV-1 | I | OQ832563 | ud |
| DENVN20_032 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-1 | I | OQ832564 | ud |
| DENVN20_059 | Thanh Hoa | Vietnam | September 2020 | POS | NEG | NEG | DENV-1 | I | OQ832565 | ud |
| DENVN20_066 | Hanoi | Vietnam | October 2020 | POS | NEG | NEG | DENV-1 | I | OQ832566 | ud |
| DENVN20_111 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-1 | I | OQ832567 | OQ832613 |
| DENVN20_112 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-1 | I | OQ832568 | OQ832614 |
| DENVN20_120 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-1 | I | OQ832569 | OQ832615 |
| DENVN20_124 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-1 | I | OQ832570 | ud |
| DENVN20_226 | Hanoi | Vietnam | October 2020 | POS | NEG | NEG | DENV-1 | I | OQ832571 | ud |
| DENVN19_004 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832572 | OQ832616 |
| DENVN19_006 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832573 | ud |
| DENVN19_010 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Asian-I | OQ832574 | OQ832617 |
| DENVN19_011 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832575 | OQ832618 |
| DENVN19_013 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832576 | OQ832619 |
| DENVN19_015 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832577 | OQ832620 |
| DENVN19_078 | ND | Vietnam | December 2019 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832578 | OQ832621 |
| DENVN19_089 | Hanoi | Vietnam | December 2019 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832579 | OQ832622 |
| DENVN19_140 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Asian-I | OQ832580 | ud |
| DENVN19_142 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832581 | ud |
| DENVN19_143 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Asian-I | OQ832582 | OQ832623 |
| DENVN19_144 | ND | Vietnam | October 2019 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832583 | ud |
| DENVN20_019 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832584 | ud |
| DENVN20_021 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832585 | ud |
| DENVN20_049 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832586 | ud |
| DENVN20_074 | Hanoi | Vietnam | October 2020 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832587 | ud |
| DENVN20_106 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832588 | ud |
| DENVN20_107 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832589 | OQ832624 |
| DENVN20_113 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832590 | OQ832625 |
| DENVN20_118 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-2 | Asian-I | OQ832591 | OQ832626 |
| DENVN20_127 | Hanoi | Vietnam | September 2020 | POS | NEG | NEG | DENV-2 | Asian-I | OQ832592 | OQ832627 |
| DENVN20_210 | Hanoi | Vietnam | October 2020 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832593 | ud |
| DENVN20_220 | Hanoi | Vietnam | October 2020 | POS | NEG | NEG | DENV-2 | Cosmopolitan | OQ832594 | ud |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phadungsombat, J.; Vu, H.T.T.; Nguyen, Q.T.; Nguyen, H.T.V.; Nguyen, H.T.N.; Dang, B.T.; Nakayama, E.E.; Ishizaki, A.; Ichimura, H.; Shioda, T.; et al. Molecular Characterization of Dengue Virus Strains from the 2019–2020 Epidemic in Hanoi, Vietnam. Microorganisms 2023, 11, 1267. https://doi.org/10.3390/microorganisms11051267
Phadungsombat J, Vu HTT, Nguyen QT, Nguyen HTV, Nguyen HTN, Dang BT, Nakayama EE, Ishizaki A, Ichimura H, Shioda T, et al. Molecular Characterization of Dengue Virus Strains from the 2019–2020 Epidemic in Hanoi, Vietnam. Microorganisms. 2023; 11(5):1267. https://doi.org/10.3390/microorganisms11051267
Chicago/Turabian StylePhadungsombat, Juthamas, Huong Thi Thu Vu, Quynh Thi Nguyen, Ha Thi Van Nguyen, Ha Thi Nhu Nguyen, Bich Thi Dang, Emi E. Nakayama, Azumi Ishizaki, Hiroshi Ichimura, Tatsuo Shioda, and et al. 2023. "Molecular Characterization of Dengue Virus Strains from the 2019–2020 Epidemic in Hanoi, Vietnam" Microorganisms 11, no. 5: 1267. https://doi.org/10.3390/microorganisms11051267
APA StylePhadungsombat, J., Vu, H. T. T., Nguyen, Q. T., Nguyen, H. T. V., Nguyen, H. T. N., Dang, B. T., Nakayama, E. E., Ishizaki, A., Ichimura, H., Shioda, T., & Pham, T. N. (2023). Molecular Characterization of Dengue Virus Strains from the 2019–2020 Epidemic in Hanoi, Vietnam. Microorganisms, 11(5), 1267. https://doi.org/10.3390/microorganisms11051267






