Emergence of a Hybrid IncI1-Iα Plasmid-Encoded blaCTX-M-101 Conferring Resistance to Cephalosporins in Salmonella enterica Serovar Enteritidis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Antimicrobial Susceptibility Testing
2.3. Whole-Genome Sequencing, Assembling, and Annotation
2.4. Phylogenetic Analysis
2.5. Gene Cloning
2.6. Plasmid Conjugation and Transformation Experiment
2.7. S1-PFGE Experiment
2.8. Nucleotide Sequence Accession Numbers
3. Results and Discussion
3.1. Emergence of blaCTX-M-101 in S. Enteritidis Isolate
3.2. blaCTX-M-101 Mediated the Resistance to Cephalosporin
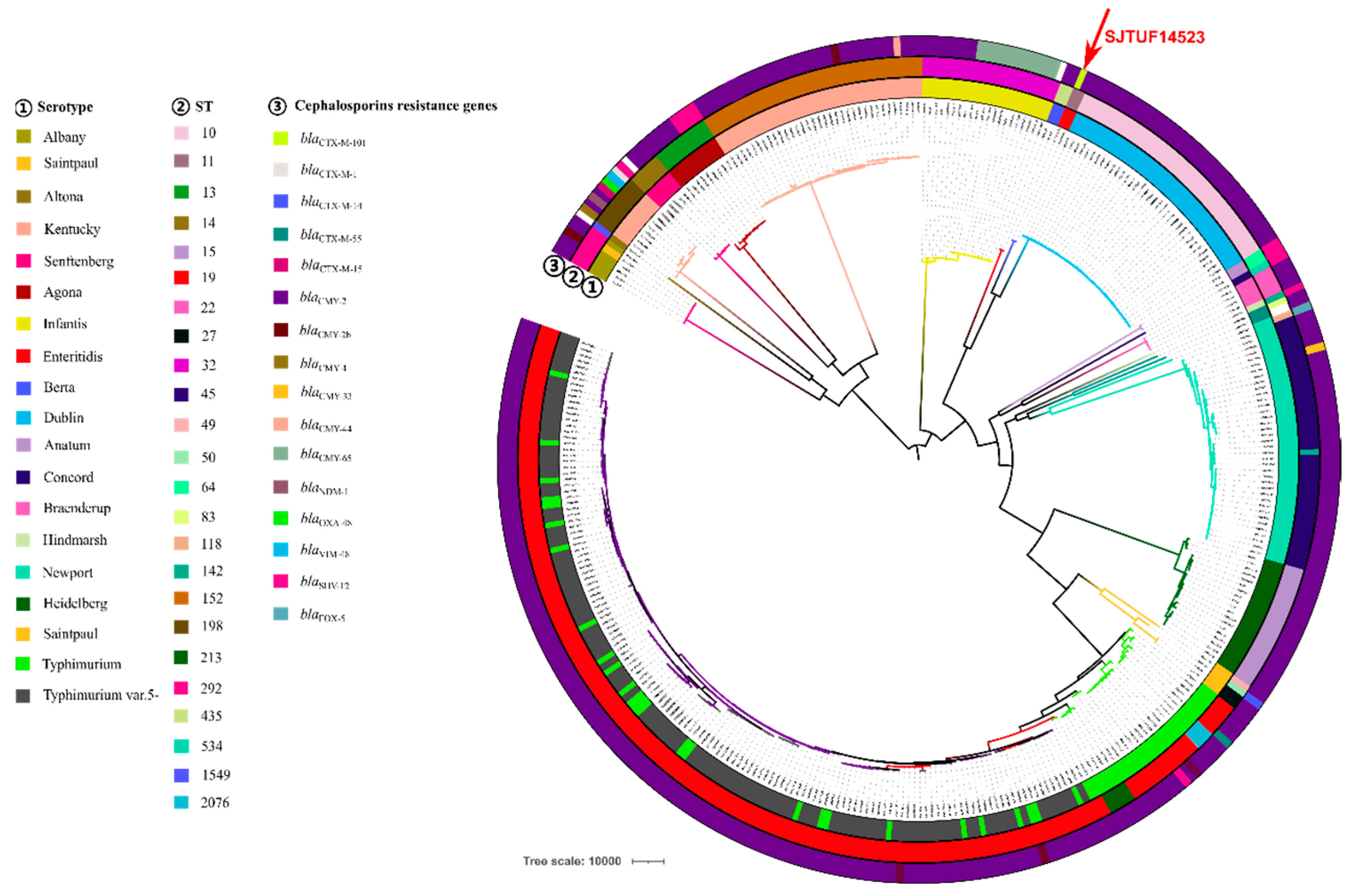
3.3. Phylogenetic Analysis of blaCTX-M-101-Positive S. Enteritidis Isolate
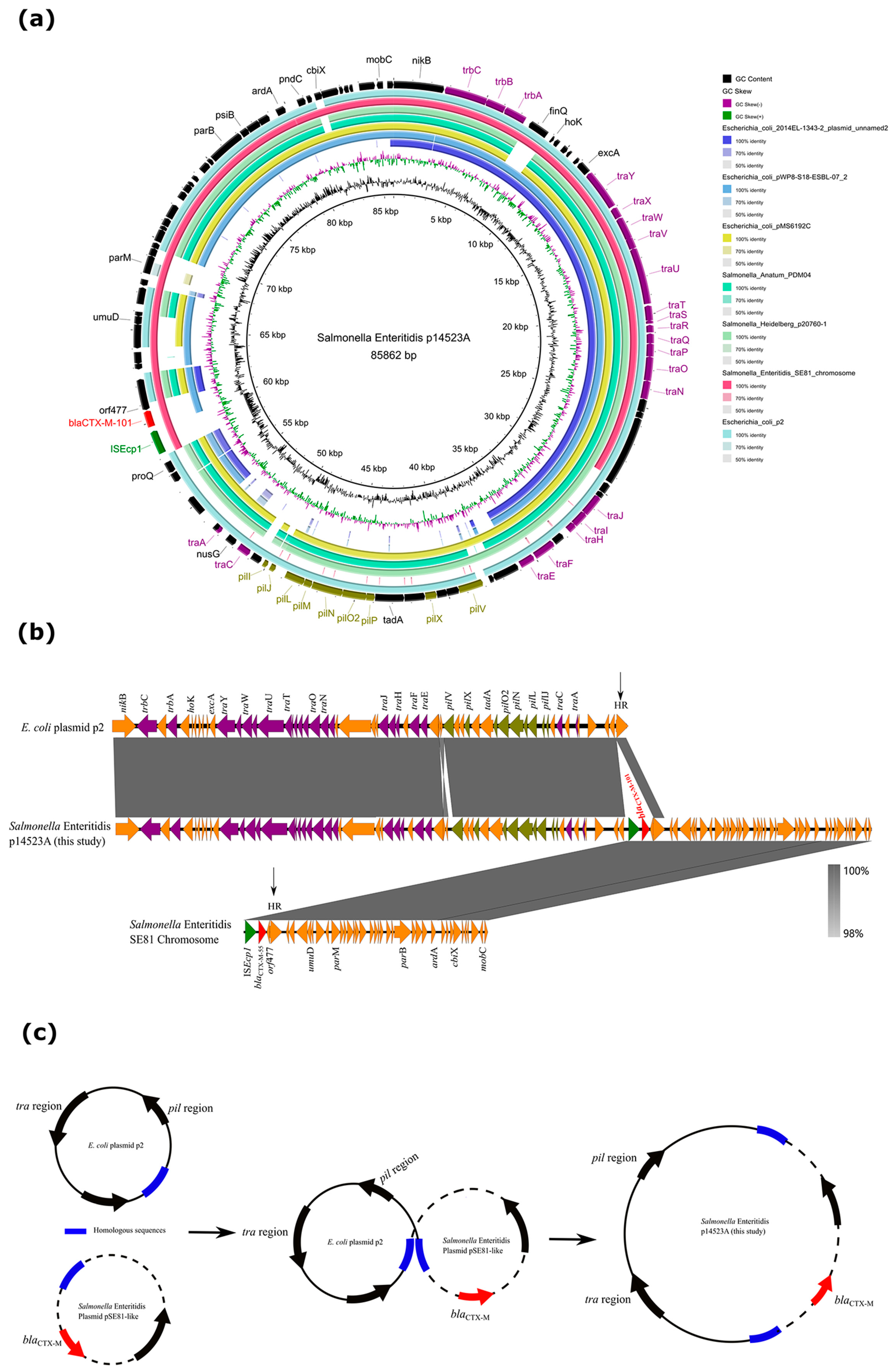
3.4. Characterization of a Novel Hybrid Plasmid Carrying blaCTX-M-101
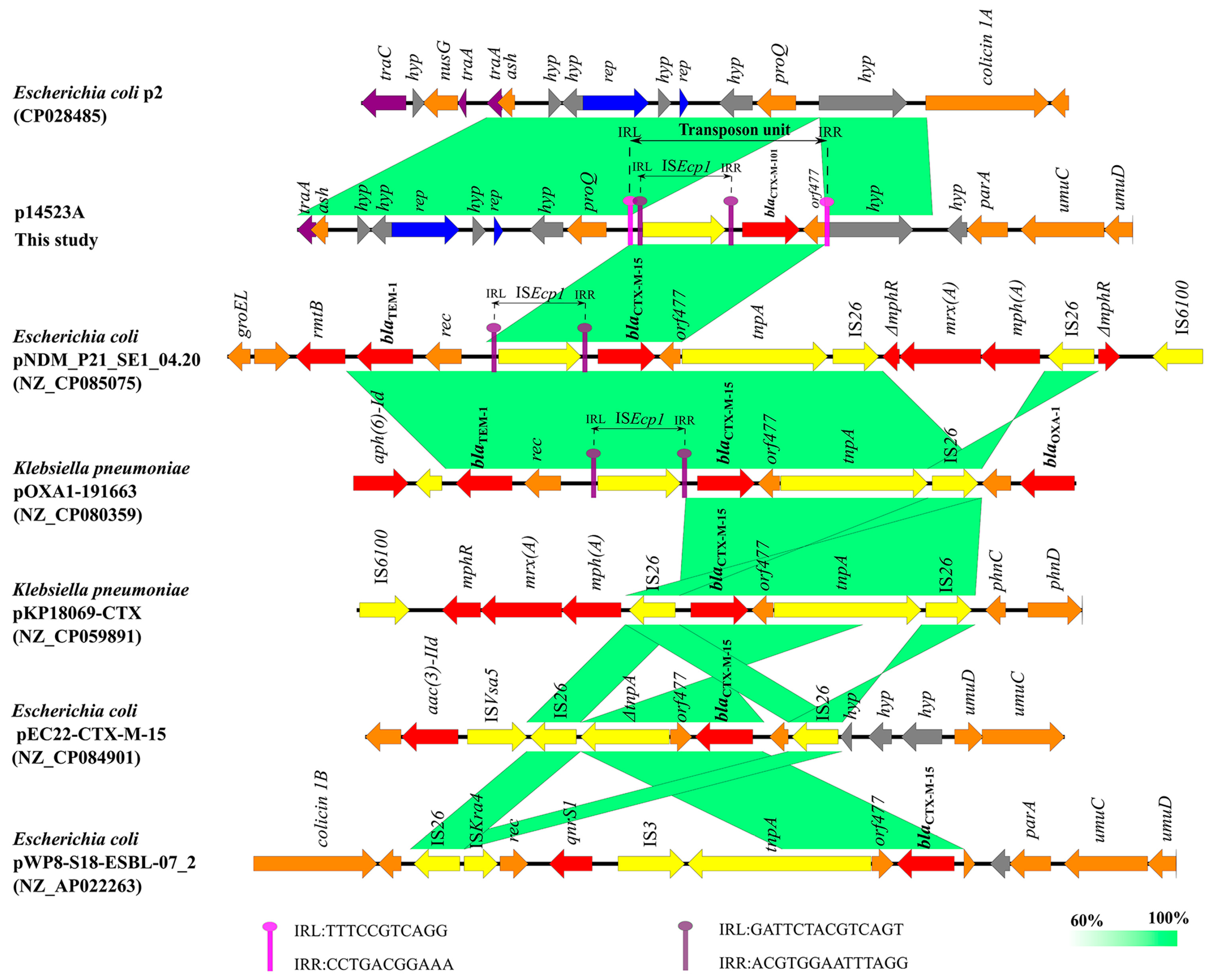
3.5. Composite-Transposon-Mediated Capture of blaCTX-M-101 by Plasmid
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. London: Review on Antimicrobial Resistance. 2014. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 17 June 2022).
- Hsueh, P.R.; Hoban, D.J.; Carmeli, Y.; Chen, S.Y.; Desikan, S.; Alejandria, M.; Ko, W.C.; Binh, T.Q. Consensus Review of the Epidemiology and Appropriate Antimicrobial Therapy of Complicated Urinary Tract Infections in Asia-Pacific Region. J. Infect. 2011, 63, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Cornaglia, G. Cephalosporins: A Microbiological Update. Clin. Microbiol. Infect. 2000, 6, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Moghnieh, R.A.; Moussa, J.A.; Aziz, M.A.; Matar, G.M. Phenotypic and Genotypic Characterisation of Cephalosporin-, Carbapenem- and Colistin-Resistant Gram-Negative Bacterial Pathogens in Lebanon, Jordan and Iraq. J. Glob. Antimicrob. Re 2021, 27, 175–199. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, E. Rise in the Prevalence of Resistance to Extended-Spectrum Cephalosporins in the USA, Nursing Homes and Antibiotic Prescribing in Outpatient and Inpatient Settings. J. Antimicrob. Chemoth 2021, 76, 2745–2747. [Google Scholar] [CrossRef] [PubMed]
- Begier, E.; Rosenthal, N.A.; Gurtman, A.; Kartashov, A.; Donald, R.G.K.; Lockhart, S.P. Epidemiology of Invasive Escherichia Coli Infection and Antibiotic Resistance Status Among Patients Treated in US Hospitals: 2009–2016. Clin. Infect. Dis. 2021, 73, 565–574. [Google Scholar] [CrossRef]
- Hu, F.P.; Guo, Y.; Zhu, D.M.; Wang, F.; Jiang, X.F.; Xu, Y.C.; Zhang, X.J.; Zhang, C.X.; Ji, P.; Xie, Y.; et al. Resistance Trends among Clinical Isolates in China Reported from CHINET Surveillance of Bacterial Resistance, 2005–2014. Clin. Microbiol. Infec 2016, 22, S9–S14. [Google Scholar] [CrossRef]
- Seif, Y.; Kavvas, E.; Lachance, J.C.; Yurkovich, J.T.; Nuccio, S.P.; Fang, X.; Catoiu, E.; Raffatellu, M.; Palsson, B.O.; Monk, J.M. Genome-scale metabolic reconstructions of multiple Salmonella strains reveal serovar-specific metabolic traits. Nat. Commun. 2018, 9, 3771. [Google Scholar] [CrossRef]
- Sholpan, A.; Lamas, A.; Cepeda, A.; Franco, C.M. Salmonella spp. quorum sensing: An overview from environmental persistence to host cell invasion. AIMS Microbiol. 2021, 7, 238–256. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Center for Disease Prevention and Control. The European Union one health 2018zoonoses report. EFSA J. 2019, 17, 5926. [Google Scholar] [CrossRef]
- Pearce, M.E.; Alikhan, N.F.; Dallman, T.J.; Zhou, Z.M.; Grant, K.; Maiden, M.C.J. Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar Enteritidis outbreak. Int. J. Food Microbiol. 2018, 274, 1–11. [Google Scholar] [CrossRef]
- Ahmed, N.; EI-Fateh, M.; Amer, M.S.; EI-Shafei, R.A.; Bilal, M.; Diarra, M.S.; Zhao, X. Antioxidative and cytoprotective efficacy of ethanolic extracted cranberry pomace against Salmonella Enteritidis infection in chicken liver cells. Antioxidants 2023, 12, 460. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, M.; Xu, X.; Fu, Y.; Xiong, Z.; Zhang, L.; Qu, X.; Zhang, H.; Wei, Y.; Zhan, Z.; et al. High-Levels of Resistance to Quinolone and Cephalosporin Antibiotics in MDR-ACSSuT Salmonella enterica Serovar Enteritidis Mainly Isolated from Patients and Foods in Shanghai, China. Int. J. Food Microbiol. 2018, 286, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chang, J.; Xu, X.; Zhou, M.; Shi, C.; Liu, Y.; Shi, X. Dissemination of IncFII Plasmids Carrying fosA3 and blaCTX-M-55 in Clinical Isolates of Salmonella Enteritidis. Zoonoses Public. Hlth 2021, 68, 760–768. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, L.; Hu, Y.; Dottorini, T.; Fanning, S.; Xu, J.; Li, F. Epidemiological Study on Prevalence, Serovar Diversity, Multidrug Resistance, and CTX-M-Type Extended-Spectrum Beta-Lactamases of Salmonella Spp. from Patients with Diarrhea, Food of Animal Origin, and Pets in Several Provinces of China. Antimicrob. Agents Chemother. 2020, 64, e00092-20. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Dai, H.; Liao, W.; Zhao, D.; Shi, Q.; Zhang, L.; Shi, K.; Akova, M.; Yu, Y. Etiology and Prevalence of ESBLs in Adult Community-Onset Urinary Tract Infections in East China: A Prospective Multicenter Study. J. Infect. 2021, 83, 175–181. [Google Scholar] [CrossRef]
- Zhang, J.; Zheng, B.; Zhao, L.; Wei, Z.; Ji, J.; Li, L.; Xiao, Y. Nationwide High Prevalence of CTX-M and an Increase of CTX-M-55 in Escherichia coli Isolated from Patients with Community-Onset Infections in Chinese County Hospitals. BMC Infect. Dis. 2014, 14, 659. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Z.; Xu, X.; He, S.; Zhao, X.; Cui, Y.; Zhou, X.; Shi, C.; Liu, Y.; Zhou, M.; et al. Molecular Characterization of Cephalosporin-Resistant Salmonella Enteritidis ST11 Isolates Carrying blaCTX-M from Children with Diarrhea. Foodborne Pathog. Dis. 2021, 18, 702–711. [Google Scholar] [CrossRef]
- Shen, Z.; Ding, B.; Bi, Y.; Wu, S.; Xu, S.; Xu, X.; Guo, Q.; Wang, M. CTX-M-190, a Novel Beta-Lactamase Resistant to Tazobactam and Sulbactam, Identified in an Escherichia coli Clinical Isolate. Antimicrob. Agents Chemother. 2017, 61, e01848-16. [Google Scholar] [CrossRef]
- Hernandez-Garcia, M.; Leon-Sampedro, R.; Perez-Viso, B.; Morosini, M.I.; Lopez-Fresnena, N.; Diaz-Agero, C.; Coque, T.M.; Ruiz-Garbajosa, P.; Canton, R. First Report of an OXA-48-and CTX-M-213-Producing Kluyvera Species Clone Recovered from Patients Admitted in a University Hospital in Madrid, Spain. Antimicrob. Agents Chemother. 2018, 62, e01238-18. [Google Scholar] [CrossRef]
- Huang, Y.T.; Yeh, T.K.; Chen, W.H.; Shih, P.W.; Mao, Y.C.; Lu, M.C.; Chen, C.M.; Liu, P.Y. Genome Analysis of Enterobacter hormaechei Identified ISEcp1 in Association with BlaCTX-M-236, a New BlaCTX-M Variant, Located Both in the Chromosome and a Plasmid. J. Glob. Antimicrob. Resist. 2021, 25, 37–39. [Google Scholar] [CrossRef]
- Manageiro, V.; Graça, R.; Ferreira, E.; Clemente, L.; Bonnet, R.; Caniça, M. Biochemical Characterization of CTX-M-166, a New CTX-M β-Lactamase Produced by a Commensal Escherichia coli Isolate. J. Antibiot. 2017, 70, 809–810. [Google Scholar] [CrossRef] [PubMed]
- Ghiglione, B.; Margarita Rodriguez, M.; Brunetti, F.; Papp-Wallace, K.M.; Yoshizumi, A.; Ishii, Y.; Bonomo, R.A.; Gutkind, G.; Klinke, S.; Power, P. Structural and Biochemical Characterization of the Novel CTX-M-151 Extended-Spectrum Beta-Lactamase and Its Inhibition by Avibactam. Antimicrob. Agents Chemother. 2021, 65, e01757-20. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, A.A.; Abdelaziz, N.A.; Amin, M.A.; Aziz, R.K. Novel BlaCTX-M Variants and Genotype-Phenotype Correlations among Clinical Isolates of Extended Spectrum Beta Lactamase-Producing Escherichia coli. Sci. Rep. 2019, 9, 4224. [Google Scholar] [CrossRef] [PubMed]
- Isgren, C.M.; Edwards, T.; Pinchbeck, G.L.; Winward, E.; Adams, E.R.; Norton, P.; Timofte, D.; Maddox, T.W.; Clegg, P.D.; Williams, N.J. Emergence of Carriage of CTX-M-15 in Faecal Escherichia coli in Horses at an Equine Hospital in the UK.; Increasing Prevalence over a Decade (2008–2017). BMC Vet. Res. 2019, 15, 268. [Google Scholar] [CrossRef]
- Elkenany, R.M.; Eladl, A.H.; El-Shafei, R.A. Genetic characterisation of class 1 integrons among multidrug-resistant Salmonella serotypes in broiler chicken farms. J. Glob. Antimicrob. Re 2018, 14, 202–208. [Google Scholar] [CrossRef]
- Glenn, L.M.; Lindsey, R.L.; Frank, J.F.; Meinersmann, R.J.; Englen, M.D.; Fedorka-Cray, P.J.; Frye, J.G. Analysis of antimicrobial resistance genes detected in multidrug-resistant Salmonella enterica serovar Typhimurium isolated from food animals. Microb. Drug. Resist. 2011, 17, 407–418. [Google Scholar] [CrossRef]
- Glenn, L.M.; Lindsey, R.L.; Folster, J.P.; Pecic, G.; Boerlin, P.; Gilmour, M.W.; Harbottle, H.; Zhao, S.; McDermott, P.F.; Fedorka-Cray, P.J.; et al. Antimicrobial resistance genes in multidrug-resistant Salmonella enterica isolated from animals, retail meats, and humans in the United States and Canada. Microb. Drug. Resist. 2013, 19, 175–184. [Google Scholar] [CrossRef]
- Wang, J.; Zeng, Z.L.; Huang, X.Y.; Ma, Z.B.; Guo, Z.W.; Lv, L.C.; Xia, Y.B.; Zeng, L.; Song, Q.H.; Liu, J.H. Evolution and Comparative Genomics of F33:A-:B- Plasmids Carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae Isolated from Animals, Food Products, and Humans in China. mSphere 2018, 3, e00137-18. [Google Scholar] [CrossRef]
- Xia, L.; Liu, Y.; Xia, S.; Kudinha, T.; Xiao, S.; Zhong, N.; Ren, G.; Zhuo, C. Prevalence of ST1193 Clone and IncI1/ST16 Plasmid in E-Coli Isolates Carrying blaCTX-M-55 Gene from Urinary Tract Infections Patients in China. Sci. Rep. 2017, 7, 44866. [Google Scholar] [CrossRef]
- Zhang, C.Z.; Ding, X.M.; Lin, X.L.; Sun, R.Y.; Lu, Y.W.; Cai, R.M.; Webber, M.A.; Ding, H.Z.; Jiang, H.X. The Emergence of Chromosomally Located blaCTX-M-55 in Salmonella from Foodborne Animals in China. Front. Microbiol. 2019, 10, 1268. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, J.; Xu, X.; Zhou, X.; Shi, C.; Zhao, X.; Liu, Y.; Shi, X. Co-Existence of mphA, oqxAB and blaCTX-M-65 on the IncHI2 Plasmid in Highly Drug-Resistant Salmonella enterica Serovar Indiana ST17 Isolated from Retail Foods and Humans in China. Food Control. 2020, 118, 107269. [Google Scholar] [CrossRef]
- Lartigue, M.-F.; Poirel, L.; Nordmann, P. Diversity of Genetic Environment of blaCTX-M Genes. FEMS Microbiol. Lett. 2004, 234, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Qu, T.T.; Yu, Y.S.; Zhou, J.Y.; Li, L.J. Insertion Sequence ISEcp1-like Element Connected with a Novel Aph(2″) Allele [Aph(2″)-Ie] Conferring High-Level Gentamicin Resistance and a Novel Streptomycin Adenylyltransferase Gene in Enterococcus. J. Med. Microbiol. 2006, 55, 1521–1525. [Google Scholar] [CrossRef] [PubMed]
- Miriagou, V.; Carattoli, A.; Fanning, S. Antimicrobial Resistance Islands: Resistance Gene Clusters in Salmonella Chromosome and Plasmids. Microbes Infect. 2006, 8, 1923–1930. [Google Scholar] [CrossRef]
- Sun, J.; Li, X.P.; Yang, R.S.; Fang, L.X.; Huo, W.; Li, S.M.; Jiang, P.; Liao, X.P.; Liu, Y.H. Complete Nucleotide Sequence of an IncI2 Plasmid Coharboring blaCTX-M-55 and mcr-1. Antimicrob. Agents Chemother. 2016, 60, 5014–5017. [Google Scholar] [CrossRef]
- Zhang, Z.F.; Chang, J.; Xu, X.B.; Hu, M.J.; He, S.K.; Qin, X.J.; Zhou, M.; Shi, C.L.; Shi, X.M. Phylogenomic analysis of Salmonella enterica Serovar Indiana ST17, an Emerging Multidrug-Resistant Clone in China. Microbiol. Spectr. 2022, 10, e0011522. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, 112963. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying Bacterial Genes and Endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A Self-Training Method for Prediction of Gene Starts in Microbial Genomes. Implications for Finding Sequence Motifs in Regulatory Regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid Phylogenetic Analysis of Large Samples of Recombinant Bacterial Whole Genome Sequences Using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhao, J.; Gan, X.; Wang, J.; Zhang, X.; Cui, S.; Xia, S.; Hu, Y.; Yan, S.; Wang, J.; et al. Emergence and Diversity of Salmonella enterica Serovar Indiana Isolates with Concurrent Resistance to Ciprofloxacin and Cefotaxime from Patients and Food-Producing Animals in China. Antimicrob. Agents Chemother. 2016, 60, 3365–3371. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, A.; Yang, Y.; Lei, C.; Jiang, W.; Liu, B.; Shi, H.; Kong, L.; Cheng, G.; Zhang, X.; et al. Emergence of Salmonella enterica Serovar Indiana and California Isolates with Concurrent Resistance to Cefotaxime, Amikacin and Ciprofloxacin from Chickens in China. Int. J. Food Microbiol. 2017, 262, 23–30. [Google Scholar] [CrossRef]
- Wong, M.H.Y.; Yan, M.; Chan, E.W.C.; Biao, K.; Chen, S. Emergence of Clinical Salmonella enterica Serovar Typhimurium Isolates with Concurrent Resistance to Ciprofloxacin, Ceftriaxone, and Azithromycin. Antimicrob. Agents Chemother. 2014, 58, 3752–3756. [Google Scholar] [CrossRef]
- Zhang, Z.; Cao, C.; Liu, B.; Xu, X.; Yan, Y.; Cui, S.; Chen, S.; Meng, J.; Yang, B. Comparative Study on Antibiotic Resistance and DNA Profiles of Salmonella enterica Serovar Typhimurium Isolated from Humans, Retail Foods, and the Environment in Shanghai, China. Foodborne Pathog. Dis. 2018, 15, 481–488. [Google Scholar] [CrossRef]
- Zhang, Q.Q.; Ying, G.G.; Pan, C.G.; Liu, Y.S.; Zhao, J.L. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Env. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H.; Kan, B.; Chan, E.W.; Yan, M.; Chen, S. IncI1 Plasmids Carrying Various bla(CTX-M) Genes Contribute to Ceftriaxone Resistance in Salmonella enterica Serovar Enteritidis in China. Antimicrob. Agents Chemother. 2016, 60, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H.; Liu, L.; Yan, M.; Chan, E.W.; Chen, S. Dissemination of IncI2 Plasmids That Harbor the bla(CTX-M) Element among Clinical Salmonella Isolates. Antimicrob. Agents Chemother. 2015, 59, 5026–5028. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ye, C.; Chang, W.; Sun, S. Serotype Distribution, Antimicrobial Resistance, and Class 1 Integrons Profiles of Salmonella from Animals in Slaughterhouses in Shandong Province, China. Front. Microbiol. 2017, 8, 1049. [Google Scholar] [CrossRef]
- Jiang, H.X.; Song, L.; Liu, J.; Zhang, X.H.; Ren, Y.N.; Zhang, W.H.; Zhang, J.Y.; Liu, Y.H.; Webber, M.A.; Ogbolu, D.O.; et al. Multiple Transmissible Genes Encoding Fluoroquinolone and Third-Generation Cephalosporin Resistance Co-Located in Non-Typhoidal Salmonella Isolated from Food-Producing Animals in China. Int. J. Antimicrob. Agents 2014, 43, 242–247. [Google Scholar] [CrossRef]
- Xie, M.; Chen, K.; Ye, L.; Yang, X.; Xu, Q.; Yang, C.; Dong, N.; Chan, E.W.C.; Sun, Q.; Shu, L.; et al. Conjugation of Virulence Plasmid in Clinical Klebsiella pneumoniae Strains through Formation of a Fusion Plasmid. Adv. Biosyst. 2020, 4, 1900239. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, Y.; Wang, C.; Liu, W.; Li, R.; Chen, F.; Luan, T.; Zhang, Y.; Schwarz, S.; Liu, S. Characterization of a Multidrug-Resistant Porcine Klebsiella pneumoniae Sequence Type 11 Strain Coharboring bla(KPC-2) and fosA3 on Two Novel Hybrid Plasmids. mSphere 2019, 4, e00590-19. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Partridge, S.R. Analysis of Antibiotic Resistance Regions in Gram-Negative Bacteria. FEMS Microbiol. Rev. 2011, 35, 820–855. [Google Scholar] [CrossRef]
- Zhao, Q.Y.; Chen, P.X.; Yang, L.; Cai, R.M.; Zhu, J.H.; Fang, L.X.; Webber, M.A.; Jiang, H.X. Transmission of Plasmid-Borne and Chromosomal blaCTX-M-64 among Escherichia coli and Salmonella Isolates from Food-Producing Animals via ISEcp1-Mediated Transposition. J. Antimicrob. Chemother. 2020, 75, 1424–1427. [Google Scholar] [CrossRef]
- Lartigue, M.F.; Poirel, L.; Aubert, D.; Nordmann, P. In Vitro Analysis of ISEcp1B-Mediated Mobilization of Naturally Occurring β-Lactamase Gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 2006, 50, 1282–1286. [Google Scholar] [CrossRef] [PubMed]
- Dhanji, H.; Doumith, M.; Hope, R.; Livermore, D.M.; Woodford, N. ISEcp1-Mediated Transposition of Linked blaCTX-M-3 and blaTEM-1b from the IncI1 Plasmid PEK204 Found in Clinical Isolates of Escherichia coli from Belfast, UK. J. Antimicrob. Chemoth 2011, 66, 2263–2265. [Google Scholar] [CrossRef] [PubMed]
| Strains | MIC | Plasmid Types | Plasmid Sizes (Kb) | |||
|---|---|---|---|---|---|---|
| FOX | CAZ | CTX | FEP | |||
| S. Enteritidis strain | ||||||
| SJTUF14253 | 8 | 64 | 256 | 16 | I1-Iα, FIIs-FIB, X1 | ~22, ~65, ~85 |
| E. coli strains | ||||||
| C600 | 2 | 0.125 | 0.06 | 0.03 | ||
| C600/p14523A (CTX-M-101) | 16 | 32 | 128 | 8 | I1-Iα | ~85 |
| DH5α | 2 | 0.25 | 0.03 | 0.03 | ||
| DH5α/p14523A (CTX-M-101) | 8 | 32 | 128 | 8 | I1-Iα | ~85 |
| DH5α/pMD19-T | 2 | 0.25 | 0.03 | 0.03 | ||
| DH5α/pMD19-T-CTX-M-101 | 4 | 32 | 64 | 4 | ||
| DH5α/pMD19-T-CTX-M-101P a | 8 | 64 | 256 | 64 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, X.; Zhang, Z. Emergence of a Hybrid IncI1-Iα Plasmid-Encoded blaCTX-M-101 Conferring Resistance to Cephalosporins in Salmonella enterica Serovar Enteritidis. Microorganisms 2023, 11, 1275. https://doi.org/10.3390/microorganisms11051275
Qin X, Zhang Z. Emergence of a Hybrid IncI1-Iα Plasmid-Encoded blaCTX-M-101 Conferring Resistance to Cephalosporins in Salmonella enterica Serovar Enteritidis. Microorganisms. 2023; 11(5):1275. https://doi.org/10.3390/microorganisms11051275
Chicago/Turabian StyleQin, Xiaojie, and Zengfeng Zhang. 2023. "Emergence of a Hybrid IncI1-Iα Plasmid-Encoded blaCTX-M-101 Conferring Resistance to Cephalosporins in Salmonella enterica Serovar Enteritidis" Microorganisms 11, no. 5: 1275. https://doi.org/10.3390/microorganisms11051275
APA StyleQin, X., & Zhang, Z. (2023). Emergence of a Hybrid IncI1-Iα Plasmid-Encoded blaCTX-M-101 Conferring Resistance to Cephalosporins in Salmonella enterica Serovar Enteritidis. Microorganisms, 11(5), 1275. https://doi.org/10.3390/microorganisms11051275






