Persister Cell Formation and Elevated lsrA and lsrC Gene Expression upon Hydrogen Peroxide Exposure in a Periodontal Pathogen Aggregatibacter actinomycetemcomitans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation of A. actinomycetemcomitans Y4 and Minimum Inhibitory Concentration of Hydrogen Peroxide
2.2. Bactericidal Effect and Genetically Antiseptic Resistant Confirmation of Hydrogen Peroxide against A. actinomycetemcomitans Y4
2.3. Persister Cells Resuscitation Time on Agarose Gel Pads
2.4. Sterilization of A. actinomycetemcomitans Y4 Persister Using Mitomycin C
2.5. Transcriptome Analysis of A. actinomycetemcomitans Y4 Using RNA Sequencing
3. Results
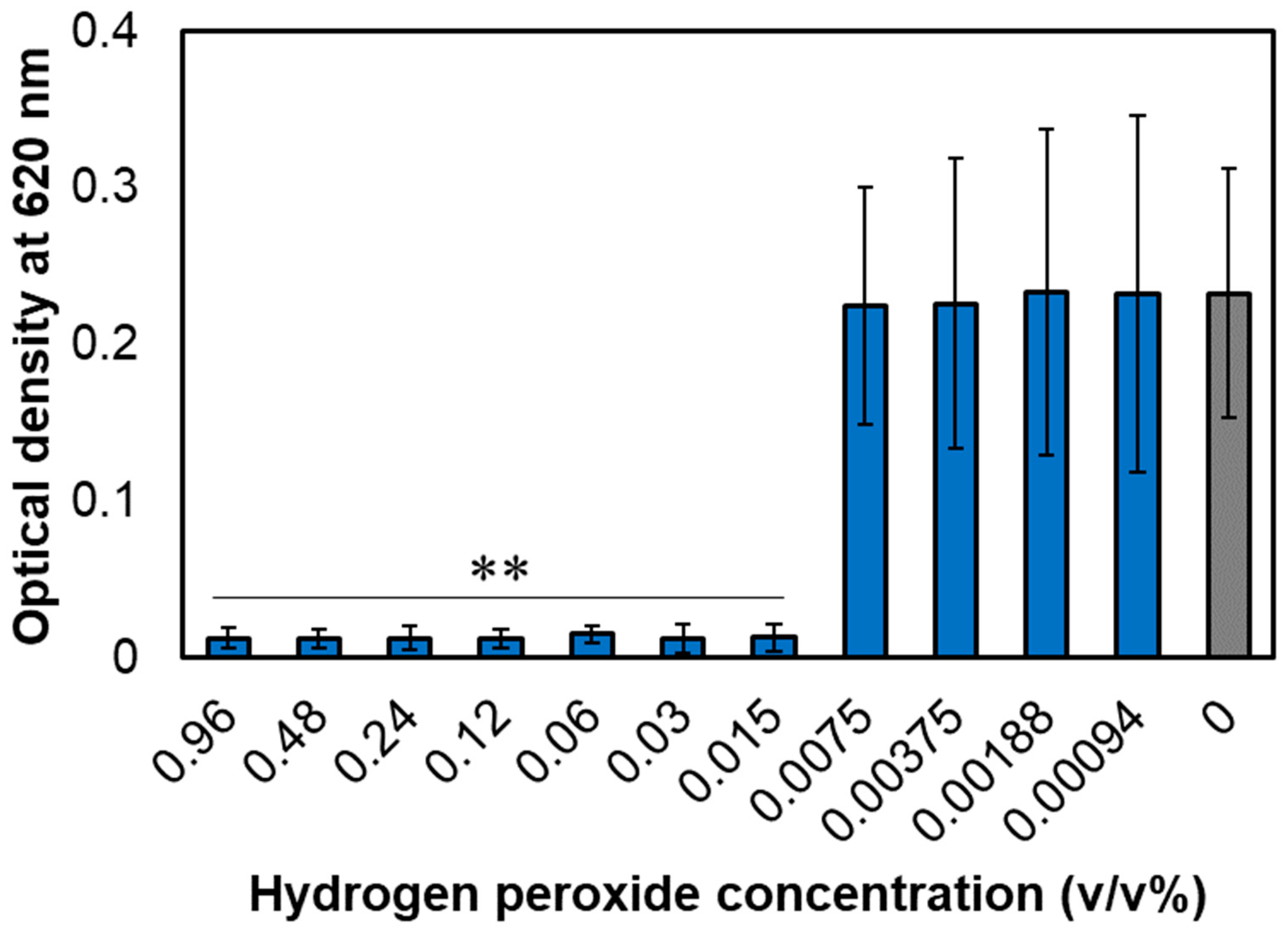
3.1. MIC of Hydrogen Peroxide for A. actinomycetemcomitans
3.2. Bactericidal Effect of Hydrogen Peroxide on A. actinomycetemcomitans and the Confirmation of Persistence
3.3. Resuscitation Time of A. actinomycetemcomitans Persister Cells
3.4. Mitomycin C Kills Persister Cells
3.5. RNA Sequencing of Gene Expression Levels after Hydrogen Peroxide Treatment
4. Discussion
4.1. The Concept of “Persister” Is Important in the Treatment of Periodontal Disease
4.2. Inference of Persister Formation Mechanism from Gene Expression Levels
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dahlen, G.; Basic, A.; Bylund, J. Importance of virulence factors for the persistence of oral bacteria in the inflamed gingival crevice and in the pathogenesis of periodontal disease. J. Clin. Med. 2019, 8, 1339. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-C.; Ho, Y.-P.; Chou, Y.-S.; Ho, K.-Y.; Wu, Y.-M.; Lin, Y.-C. Aggregatibacter (Actinobacillus) actimycetemcomitans leukotoxin and human periodontitis–A historic review with emphasis on JP2. Kaohsiung J. Med. Sci. 2018, 34, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. S1), S173–S182. [Google Scholar] [CrossRef] [PubMed]
- Tsuzukibashi, O.; Saito, M.; Kobayashi, T.; Umezawa, K.; Nagahama, F.; Hiroi, T.; Hirasawa, M.; Takada, K. A gene cluster for the synthesis of serotype g-specific polysaccharide antigen in Aggregatibacter actinomycetemcomitans. Arch. Microbiol. 2014, 196, 261–265. [Google Scholar] [CrossRef]
- Ibrahim, I.H.; Attia, A.M.; Fouad, M.; Edrees, M.A.; Hammad, H.A. Detection of JP2 and Non-JP2 Genotype strains of Aggregatibacter Actinomycetemcomitans in localized aggressive periodontitis patients among the Egyptian population. Acta Sci. Dent. Sci. 2022, 6, 149–155. [Google Scholar] [CrossRef]
- Doungudomdacha, S.; Volgina, A.; DiRienzo, J.M. Evidence that the cytolethal distending toxin locus was once part of a genomic island in the periodontal pathogen Aggregatibacter (Actinobacillus) actinomycetemcomitans strain Y4. J. Med. Microbiol. 2007, 56, 1519–1527. [Google Scholar] [CrossRef]
- Raja, M.; Ummer, F.; Dhivakar, C. Aggregatibacter actinomycetemcomitans—A tooth killer? J. Clin. Diagn. Res. 2014, 8, ZE13–ZE16. [Google Scholar] [CrossRef]
- Tang, G.; Kitten, T.; Munro, C.L.; Wellman, G.C.; Mintz, K.P. EmaA, a potential virulence determinant of Aggregatibacter actinomycetemcomitans in infective endocarditis. Infect. Immun. 2008, 76, 2316–2324. [Google Scholar] [CrossRef]
- Revest, M.; Egmann, G.; Cattoir, V.; Tattevin, P. HACEK endocarditis: State-of-the-art. Expert Rev. Anti Infect. Ther. 2016, 14, 523–530. [Google Scholar] [CrossRef]
- Liljestrand, J.M.; Paju, S.; Pietiainen, M.; Buhlin, K.; Persson, G.R.; Nieminen, M.S.; Sinisalo, J.; Mantyla, P.; Pussinen, P.J. Immunologic burden links periodontitis to acute coronary syndrome. Atherosclerosis 2018, 268, 177–184. [Google Scholar] [CrossRef]
- Al-Nafeesah, A. Aggregatibacter actinomycetemcomitans pneumonia mimicking lung cancer in a previously healthy 12-year-old child from Saudi Arabia: A case report. Pan Afr. Med. J. 2020, 36, 89. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Zúñiga, J.; Muñoz, Y.; Melgar-Rodríguez, S.; More, J.; Bruna, B.; Lobos, P.; Monasterio, G.; Vernal, R.; Paula-Lima, A. Serotype b of Aggregatibacter actinomycetemcomitans triggers pro-inflammatory responses and amyloid beta secretion in hippocampal cells: A novel link between periodontitis and Alzheimer’s disease? J. Oral Microbiol. 2019, 11, 1586423. [Google Scholar] [CrossRef] [PubMed]
- Konig, M.F.; Abusleme, L.; Reinholdt, J.; Palmer, R.J.; Teles, R.P.; Sampson, K.; Rosen, A.; Nigrovic, P.A.; Sokolove, J.; Giles, J.T.; et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016, 8, 369ra176. [Google Scholar] [CrossRef] [PubMed]
- Demmer, R.T.; Jacobs, D.R., Jr.; Singh, R.; Zuk, A.; Rosenbaum, M.; Papapanou, P.N.; Desvarieux, M. Periodontal bacteria and prediabetes prevalence in ORIGINS: The oral infections, glucose intolerance, and insulin resistance study. J. Dent. Res. 2015, 94, 201S–211S. [Google Scholar] [CrossRef]
- Kelk, P.; Abd, H.; Claesson, R.; Sandström, G.; Sjöstedt, A.; Johansson, A. Cellular and molecular response of human macrophages exposed to Aggregatibacter actinomycetemcomitans leukotoxin. Cell Death Dis. 2011, 2, e126. [Google Scholar] [CrossRef]
- Ristow, L.C.; Tran, V.; Schwartz, K.J.; Pankratz, L.; Mehle, A.; Sauer, J.-D.; Welch, R.A. The extracellular domain of the β2 integrin β subunit (CD18) is sufficient for Escherichia coli hemolysin and Aggregatibacter actinomycetemcomitans leukotoxin cytotoxic activity. mBio 2019, 10, e01459-19. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Pormohammad, A.; Eslami, H.; Shokouhi, B.; Fakhrzadeh, V.; Kafil, H.S. Oral pathogenesis of Aggregatibacter actinomycetemcomitans. Microb. Pathog. 2017, 113, 303–311. [Google Scholar] [CrossRef]
- Rahamat-Langendoen, J.C.; van Vonderen, M.G.; Engstrom, L.J.; Manson, W.L.; van Winkelhoff, A.J.; Mooi-Kokenberg, E.A. Brain abscess associated with Aggregatibacter actinomycetemcomitans: Case report and review of literature. J. Clin. Periodontol. 2011, 38, 702–706. [Google Scholar] [CrossRef]
- Oscarsson, J.; Claesson, R.; Lindholm, M.; Höglund Åberg, C.; Johansson, A. Tools of Aggregatibacter actinomycetemcomitans to evade the host response. J. Clin. Med. 2019, 8, 1079. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef]
- Lewis, K. Persister Cells: Molecular Mechanisms Related to Antibiotic Tolerance; Part of the Handbook of Experimental Pharmacology book series; Springer: Berlin/Heidelberg, Germany, 2012; Volume 211, pp. 121–133. [Google Scholar] [CrossRef]
- Wood, T.K.; Song, S.; Yamasaki, R. Ribosome dependence of persister cell formation and resuscitation. J. Microbiol. 2019, 57, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Kaldalu, N.; Hauryliuk, V.; Turnbull, K.J.; La Mensa, A.; Putrinš, M.; Tenson, T. In vitro studies of persister cells. Microbiol. Mol. Biol. Rev. 2020, 84, e00070-20. [Google Scholar] [CrossRef] [PubMed]
- Hobby, G.L.; Meyer, K.; Chaffee, E. Observations on the mechanism of action of penicillin. Proc. Soc. Exp. Biol. Med. 1942, 50, 281–285. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]
- Kim, J.S.; Chowdhury, N.; Yamasaki, R.; Wood, T.K. Viable but non-culturable and persistence describe the same bacterial stress state. Environ. Microbiol. 2018, 20, 2038–2048. [Google Scholar] [CrossRef]
- Maisonneuve, E.; Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 2014, 157, 539–548. [Google Scholar] [CrossRef]
- Yamasaki, R.; Song, S.; Benedik, M.J.; Wood, T.K. Persister cells resuscitate using membrane sensors that activate chemotaxis, lower cAMP levels, and revive ribosomes. iScience 2020, 23, 100792. [Google Scholar] [CrossRef] [PubMed]
- Aïder, M.; Martel, A.-A.; Ferracci, J.; de Halleux, D. Purification of whole brown flaxseed meal from coloring pigments by treatment in hydrogen peroxide solutions: Impact on meal color. Food Bioproc. Technol. 2012, 5, 3051–3065. [Google Scholar] [CrossRef]
- Farzaneh, H.; Loganathan, K.; Saththasivam, J.; McKay, G. Ozone and ozone/hydrogen peroxide treatment to remove gemfibrozil and ibuprofen from treated sewage effluent: Factors influencing bromate formation. Emerg. Contam. 2020, 6, 225–234. [Google Scholar] [CrossRef]
- Hage, R.; Lienke, A. Applications of transition-metal catalysts to textile and wood-pulp bleaching. Angew. Chem. Int. Ed. Engl. 2005, 45, 206–222. [Google Scholar] [CrossRef]
- Schwartz, A.; Stiegel, M.; Greeson, N.; Vogel, A.; Thomann, W.; Brown, M.; Sempowski, G.D.; Alderman, T.S.; Condreay, J.P.; Burch, J. Decontamination and reuse of N95 respirators with hydrogen peroxide vapor to address worldwide personal protective equipment shortages during the SARS-CoV-2 (COVID-19) pandemic. Appl. Biosaf. 2020, 25, 67–70. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, G. The use of hydrogen peroxide for disinfection and sterilization applications. In PATAI’S Chemistry of Functional Groups; John Wiley & Sons: Hoboken, NJ, USA, 2009; pp. 1–34. [Google Scholar] [CrossRef]
- Juven, B.J.; Pierson, M.D. Antibacterial effects of hydrogen peroxide and methods for its detection and quantitation. J. Food Prot. 1996, 59, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Kawano, A.; Yamasaki, R.; Sakakura, T.; Takatsuji, Y.; Haruyama, T.; Yoshioka, Y.; Ariyoshi, W. Reactive oxygen species penetrate persister cell membranes of Escherichia coli for effective cell killing. Front. Cell. Infect. Microbiol. 2020, 10, 496. [Google Scholar] [CrossRef] [PubMed]
- Walsh, L.J. Safety issues relating to the use of hydrogen peroxide in dentistry. Aust. Dent. J. 2000, 45, 257–269. [Google Scholar] [CrossRef]
- Kim, J.S.; Yamasaki, R.; Song, S.; Zhang, W.; Wood, T.K. Single cell observations show persister cells wake based on ribosome content. Environ. Microbiol. 2018, 20, 2085–2098. [Google Scholar] [CrossRef] [PubMed]
- Kwan, B.W.; Chowdhury, N.; Wood, T.K. Combatting bacterial infections by killing persister cells with mitomycin C. Environ. Microbiol. 2015, 17, 4406–4414. [Google Scholar] [CrossRef]
- Van der Velden, U. What exactly distinguishes aggressive from chronic periodontitis: Is it mainly a difference in the degree of bacterial invasiveness? Periodontol. 2000 2017, 75, 24–44. [Google Scholar] [CrossRef]
- Kuboniwa, M.; Lamont, R.J. Subgingival biofilm formation. Periodontol. 2000 2010, 52, 38. [Google Scholar] [CrossRef]
- Verweij, J.; Pinedo, H.M. Mitomycin C: Mechanism of action, usefulness and limitations. Anticancer Drugs 1990, 1, 5–13. [Google Scholar] [CrossRef]
- Pan, J.; Bahar, A.A.; Syed, H.; Ren, D. Reverting antibiotic tolerance of Pseudomonas aeruginosa PAO1 persister cells by (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2(5H)-one. PLoS ONE 2012, 7, e45778. [Google Scholar] [CrossRef]
- Pan, J.; Ren, D. Structural effects on persister control by brominated furanones. Bioorg. Med. Chem. Lett. 2013, 23, 6559–6562. [Google Scholar] [CrossRef] [PubMed]
- Walawalkar, Y.D.; Vaidya, Y.; Nayak, V. Response of Salmonella Typhi to bile-generated oxidative stress: Implication of quorum sensing and persister cell populations. Pathog. Dis. 2016, 74, ftw090. [Google Scholar] [CrossRef] [PubMed]
- Xavier, K.B.; Bassler, B.L. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 2005, 187, 238–248. [Google Scholar] [CrossRef]
- Grant, C.M. Regulation of translation by hydrogen peroxide. Antioxid. Redox Signal. 2011, 15, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Kim, J.S.; Yamasaki, R.; Oh, S.; Benedik, M.J.; Wood, T.K. Escherichia coli cryptic prophages sense nutrients to influence persister cell resuscitation. Environ. Microbiol. 2021, 23, 7245–7254. [Google Scholar] [CrossRef]
- Lamont, R.J.; Hajishengallis, G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 2015, 21, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Mysak, J.; Podzimek, S.; Sommerova, P.; Lyuya-Mi, Y.; Bartova, J.; Janatova, T.; Prochazkova, J.; Duskova, J. Porphyromonas gingivalis: Major periodontopathic pathogen overview. J. Immunol. Res. 2014, 2014, 476068. [Google Scholar] [CrossRef] [PubMed]




| Gene | Fold-Change | Description |
|---|---|---|
| lsrA | 2.51 | Autoinducer 2 ABC transporter ATP-binding protein |
| lsrC | 5.28 | Autoinducer 2 ABC transporter permease |
| lsrD | 1.56 | Autoinducer 2 ABC transporter permease |
| lsrB | 0.86 | Autoinducer 2 ABC transporter substrate-binding protein |
| lsrR | 0.75 | Transcriptional regulator |
| luxS | 0.95 | S-ribosylhomocysteine lyase |
| HMPREF9996_RS08555 | 2.52 | Catalase |
| crp | 0.97 | cAMP-activated global transcriptional regulator |
| hfq | 0.91 | RNA chaperone |
| ssrA | 0.31 | Transfer-messenger RNA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamura, Y.; Watanabe, K.; Yoshioka, Y.; Ariyoshi, W.; Yamasaki, R. Persister Cell Formation and Elevated lsrA and lsrC Gene Expression upon Hydrogen Peroxide Exposure in a Periodontal Pathogen Aggregatibacter actinomycetemcomitans. Microorganisms 2023, 11, 1402. https://doi.org/10.3390/microorganisms11061402
Nakamura Y, Watanabe K, Yoshioka Y, Ariyoshi W, Yamasaki R. Persister Cell Formation and Elevated lsrA and lsrC Gene Expression upon Hydrogen Peroxide Exposure in a Periodontal Pathogen Aggregatibacter actinomycetemcomitans. Microorganisms. 2023; 11(6):1402. https://doi.org/10.3390/microorganisms11061402
Chicago/Turabian StyleNakamura, Yohei, Koji Watanabe, Yoshie Yoshioka, Wataru Ariyoshi, and Ryota Yamasaki. 2023. "Persister Cell Formation and Elevated lsrA and lsrC Gene Expression upon Hydrogen Peroxide Exposure in a Periodontal Pathogen Aggregatibacter actinomycetemcomitans" Microorganisms 11, no. 6: 1402. https://doi.org/10.3390/microorganisms11061402







