Taxonomic and Phylogenetic Studies of Two Brackish Pleuronema Species (Protista, Ciliophora, Scuticociliatia) from Subtropical Coastal Waters of China, with Report of a New Species
Abstract
1. Introduction
2. Materials and Methods
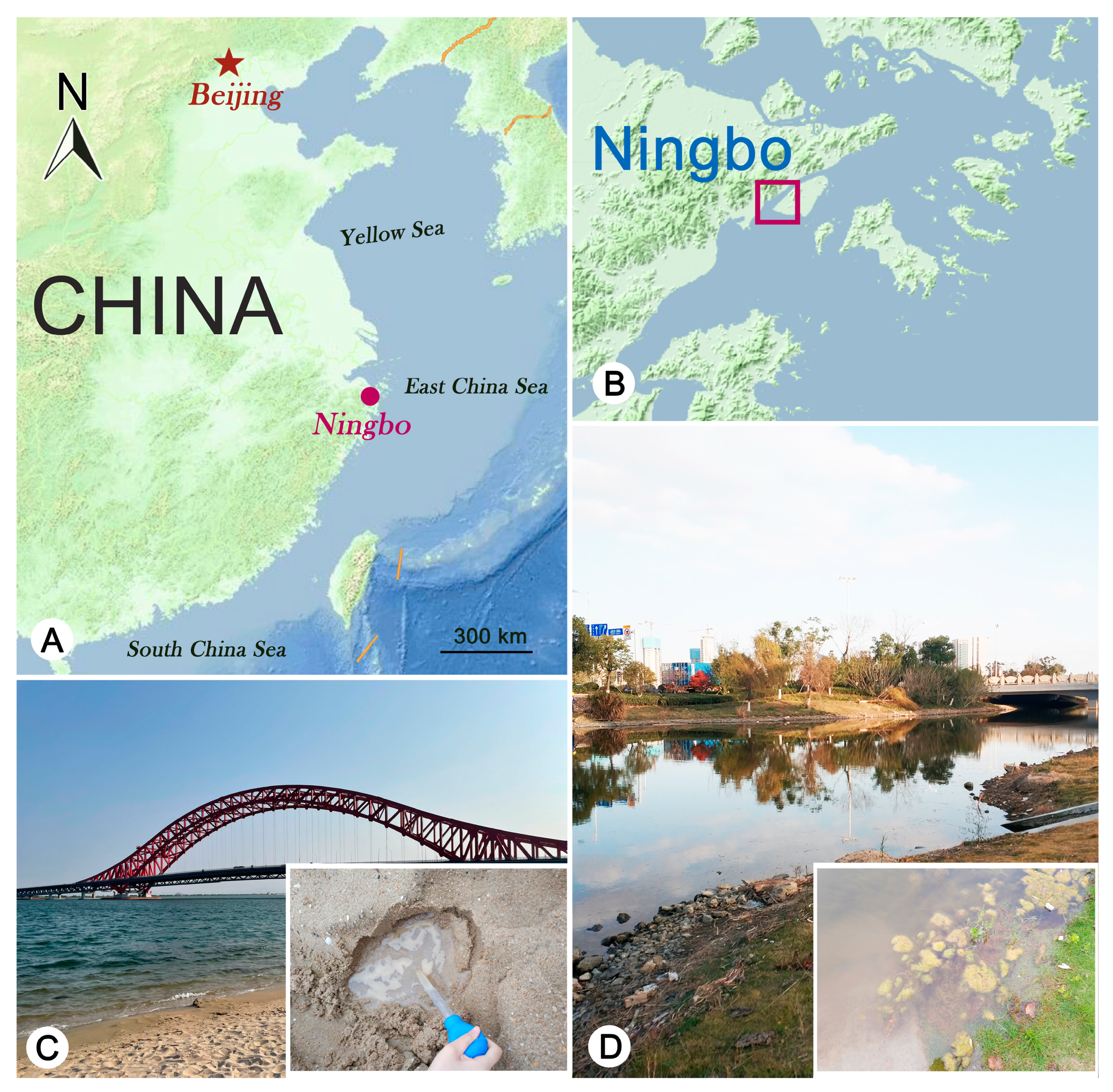
2.1. Sample Collection, Observation, and Identification
2.2. DNA Extraction, PCR Amplification, and Gene Sequencing
2.3. Phylogenetic Analyses
3. Results
3.1. Pleuronema ningboensis n. sp.
3.1.1. Diagnosis
3.1.2. Type Locality
3.1.3. Etymology
3.1.4. Type Materials
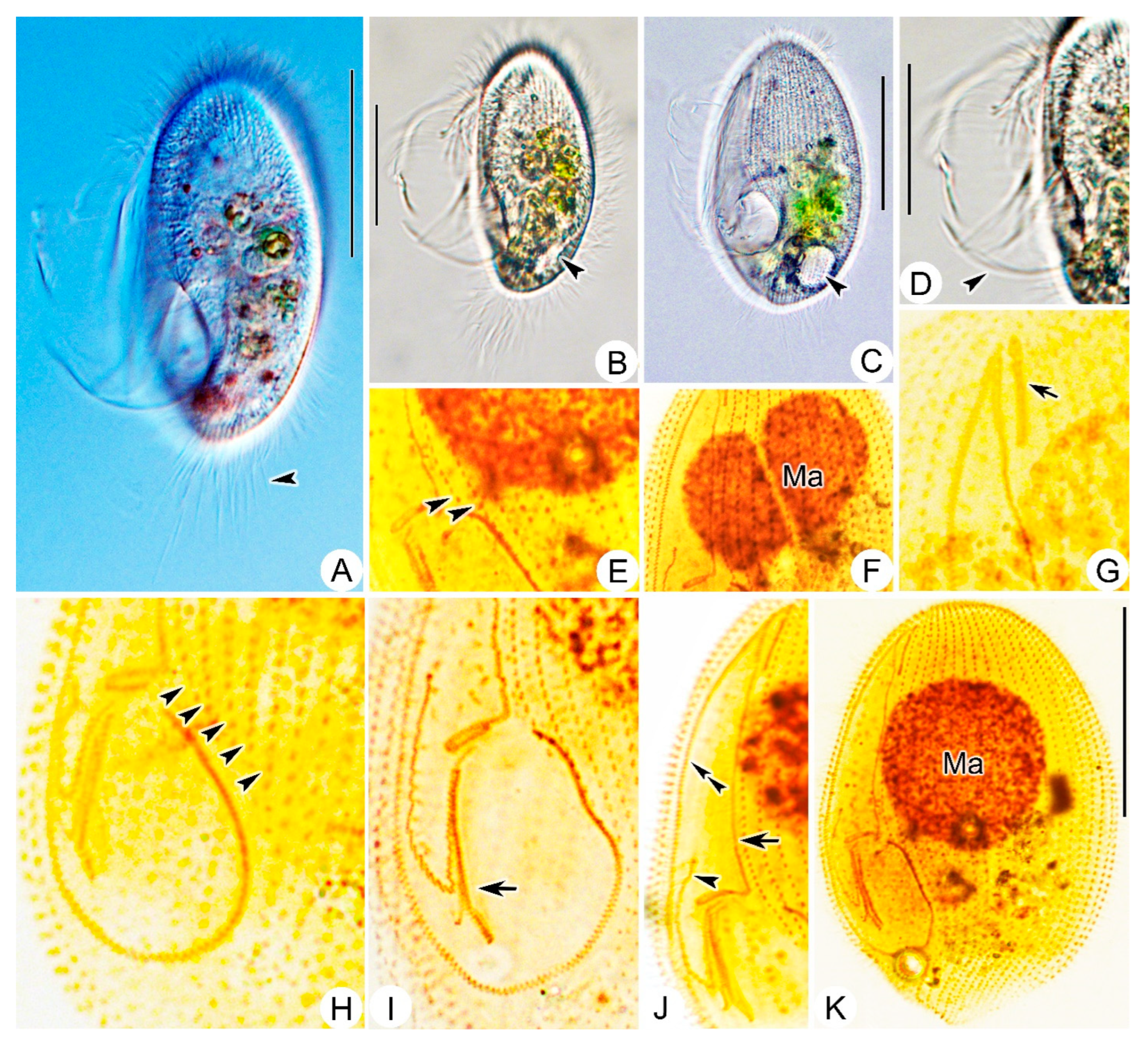
3.1.5. Morphological Description
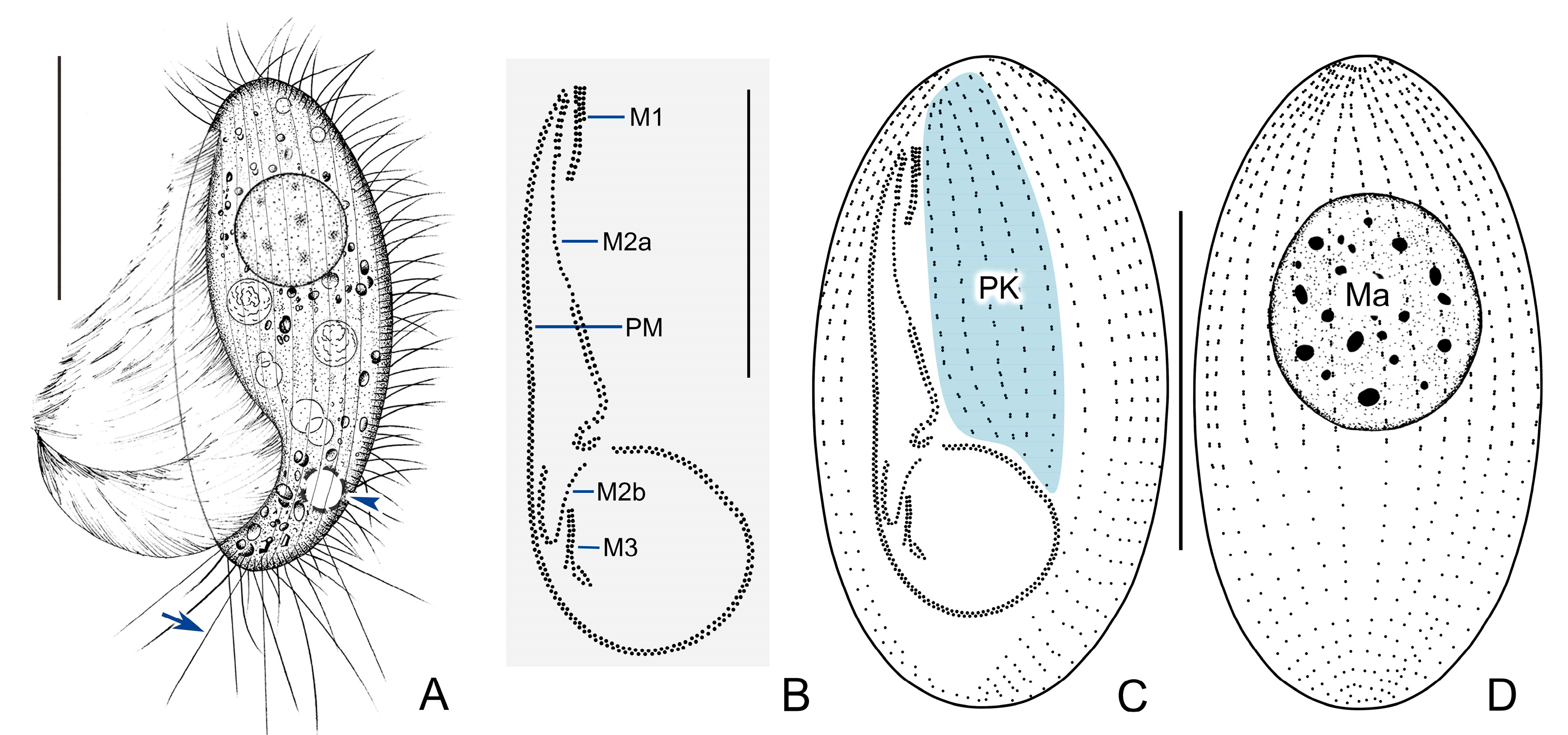

3.1.6. SSU rDNA Sequence
3.2. Pleuronema orientale Pan et al., 2015
3.2.1. Improved Diagnosis
3.2.2. Voucher Slides
3.2.3. Morphological Description of Ningbo Population
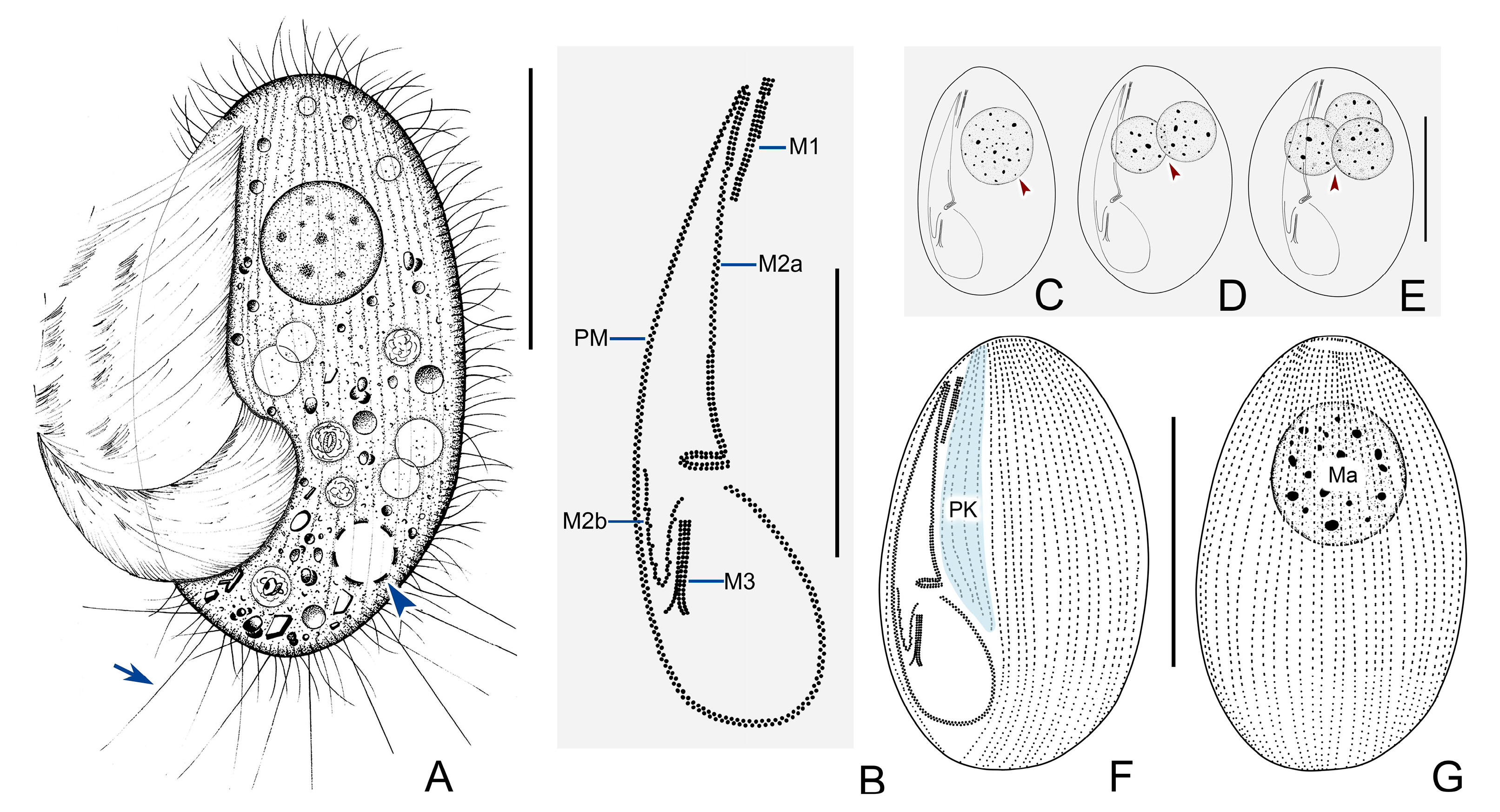
3.2.4. SSU rDNA Sequence
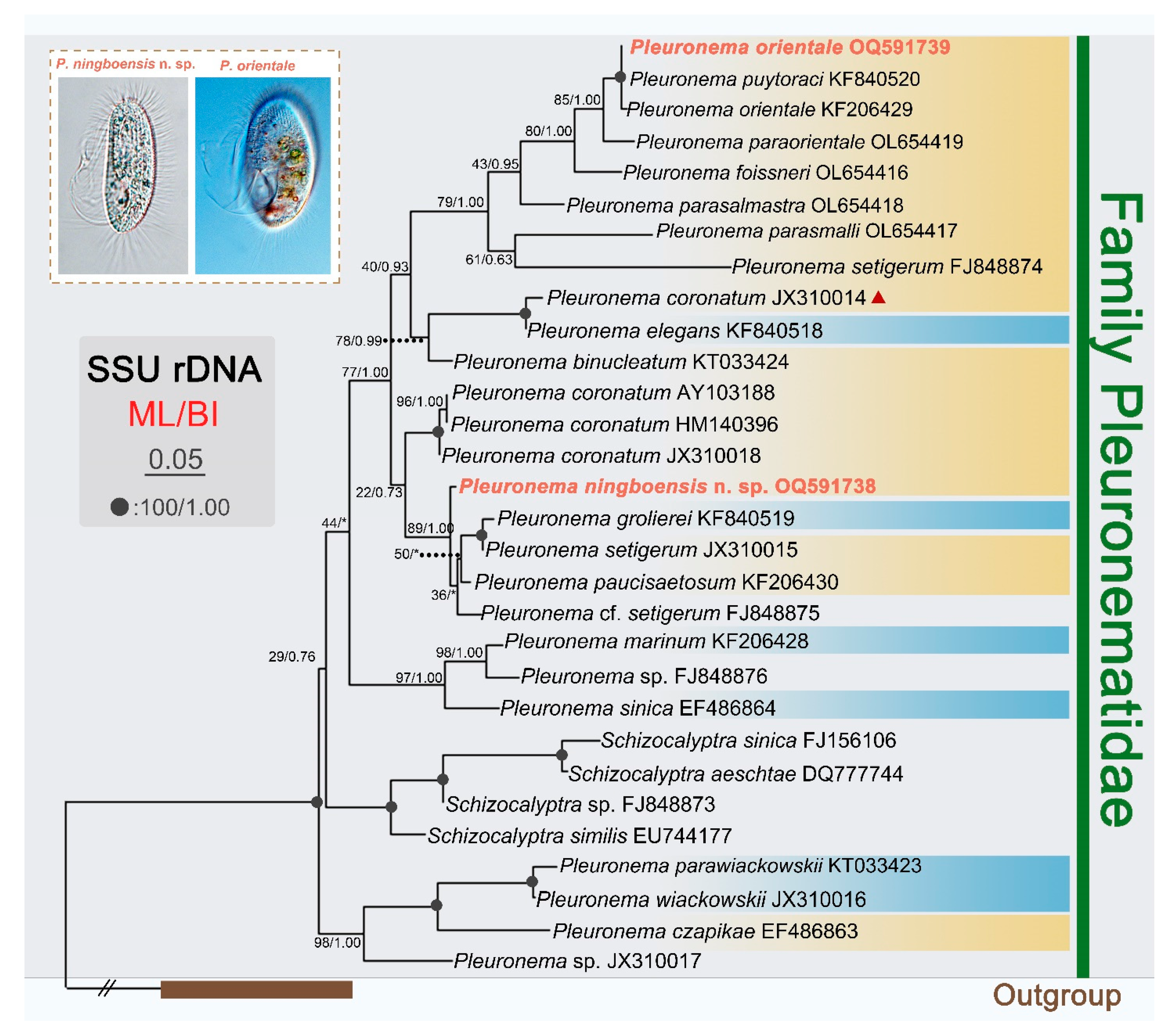
3.3. SSU rDNA Sequences and Phylogenetic Analyses
4. Discussion
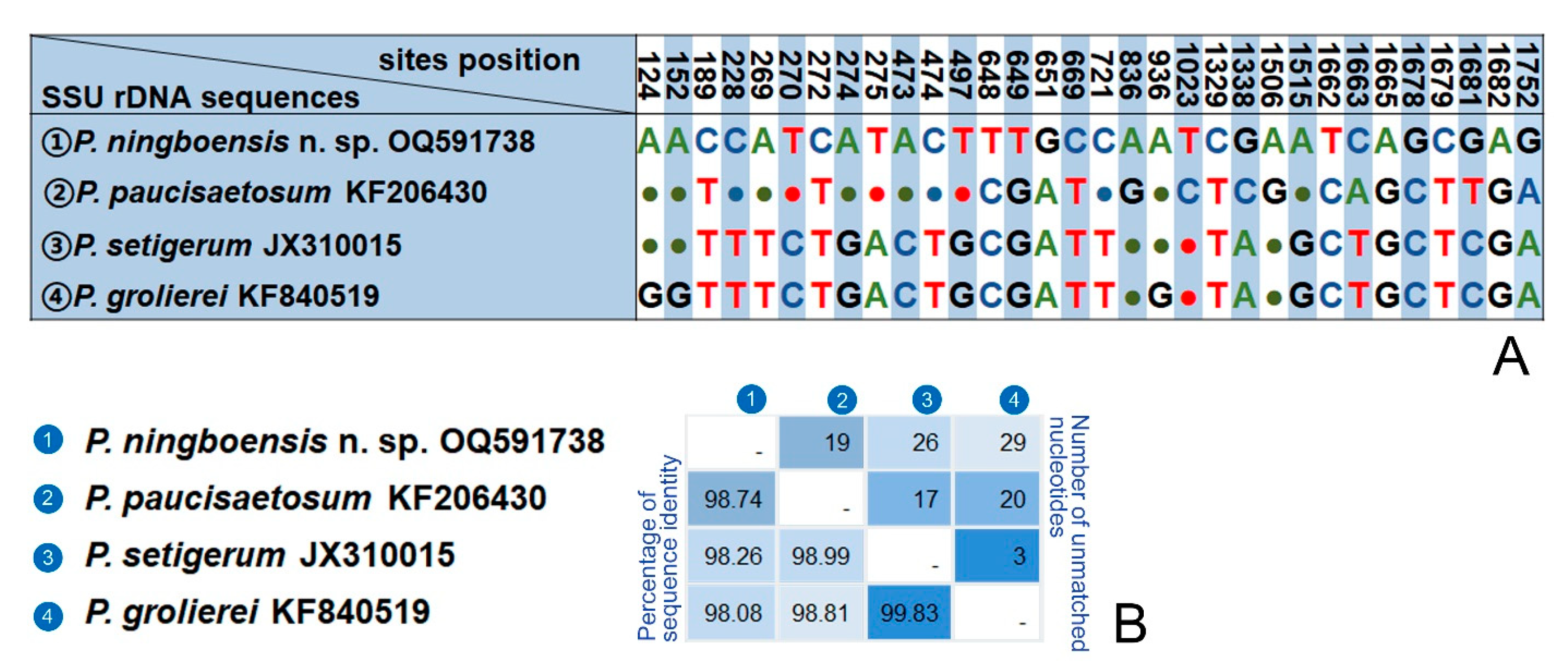
4.1. Comparison of Pleuronema ningboensis n. sp. with Related Congeners
4.2. Comparison of Two Populations of Pleuronema orientale Pan et al., 2015
4.3. Phylogenetic Relationships between Pleuronema and Schizocalyptra
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lynn, D.H. The Ciliated Protozoa: Characterization, Classification, and Guide to the Literature, 3rd ed.; Springer: Dordrecht, The Netherlands, 2008. [Google Scholar]
- Chi, Y.; Wang, Z.; Ye, T.; Wang, Y.; Zhao, J.; Song, W.; Bourland, W.A.; Chen, X. A new contribution to the taxonomy and phylogeny of the ciliate genus Spriostomum (Alveolata, Ciliophora, Heterotrichea), with comprehensive descriptions of two species from wetlands in China. Water Biol. Secur. 2022, 1, 100010. [Google Scholar] [CrossRef]
- Jin, D.; Qu, Z.; Wei, B.; Montagnes, D.; Fan, X.; Chen, X. Two parasitic ciliates (Protozoa: Ciliophora: Phyllopharyngea) isolated from respiratory-mucus of an unhealthy beluga whale: Characterization, phylogeny, and an assessment of morphological adaptations. Zool. J. Linn. Soc. 2021, 191, 941–961. [Google Scholar] [CrossRef]
- Ma, M.; Li, Y.; Yuan, Q.; Zhao, X.; Al-Rasheid, K.A.S.; Huang, J.; Ma, H.; Chen, X. New data define the molecular phylogeny and taxonomy of four freshwater suctorian ciliates with redefinition of two Families Heliophryidae and Cyclophryidae (Ciliophora, Phyllopharyngea, Suctoria). Front. Microbiol. 2021, 12, 768724. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Jiang, Y.; Chen, S.; Xu, Y.; Li, L.; Shin, M.; Chen, X. The widely reported but poorly studied ciliate family Folliculinidae (Protozoa, Ciliophora, Heterotrichea): A revision with notes on its taxonomy, morphology and phylogenetic relationships. Mar. Life Sci. Technol. 2022, 4, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Foissner, W.; Berger, H.; Blatterer, H.; Kohmann, F. Taxonomische und Ökologische Revision der Ciliaten des Saprobiensystems–Band III: Hymenostomata, Prostomatida, Nassulida; Bayer. Landesamt für Wasserwirtschaft: Munich, Germany, 1994. [Google Scholar]
- Wilbert, N.; Song, W. New contributions to the marine benthic ciliates from the Antarctic area, including description of seven new species (Protozoa, Ciliophora). J. Nat. Hist. 2005, 39, 935–973. [Google Scholar] [CrossRef]
- Corliss, J.O. The Ciliated Protozoa: Characterization, Classification and Guide to the Literature; Pergamon Press: New York, UK, USA, 1979. [Google Scholar]
- Ma, M.; Lu, B.; Fan, X.; Shi, Y.; Chen, X. Taxonomic clarification of a well-known pathogenic scuticociliate, Miamiensis avidus Thompson & Moewus, 1964 (Ciliophora, Scuticociliatia). J. Ocean Univ. China 2018, 17, 1231–1242. [Google Scholar] [CrossRef]
- Song, W.; Zhao, Y.; Xu, K.; Hu, X.; Gong, J. Pathogenic Protozoa in Mariculture; Science Press: Beijing, China, 2003. [Google Scholar]
- Xu, K.; Song, W.; Warren, A. Two new and two poorly known species of Ancistrum (Ciliophora, Scuticociliatia, Thigmotrichida) parasitizing marine molluscs from Chinese coastal waters of the Yellow Sea. Acta Protozool. 2015, 54, 195–208. [Google Scholar] [CrossRef]
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef]
- Cleven, E.J.; Königs, S.K. Growth of interstitial ciliates in association with ciliate bacterivory in a sandy hyporheic zone. Aquat. Microb. Ecol. 2007, 47, 177–189. [Google Scholar] [CrossRef]
- Fan, X.; Xu, Y.; Jiang, J.; Al-Rasheid, K.A.S.; Wang, Y.; Hu, X. Morphological descriptions of five scuticociliates including one new species of Falcicyclidium. Eur. J. Protistol. 2017, 59, 34–49. [Google Scholar] [CrossRef]
- Song, W.; Ma, H.; Al-Rasheid, K.A.S. Dexiotrichides pangi n. sp. (Protozoa Ciliophora, Scuticociliatia), a new marine ciliate from the north China Sea. J. Eukaryot. Microbiol. 2003, 50, 114–122. [Google Scholar] [CrossRef]
- Stoecker, D. Predation on protozoa: Its importance to zooplankton. J. Plankton Res. 1990, 5, 891–908. [Google Scholar] [CrossRef]
- Song, W.; Warren, A.; Hu, X. Free-Living Ciliates in the Bohai and Yellow Sea, China; Science Press: Beijing, China, 2009. [Google Scholar]
- Dujardin, F. Histoire Naturelle des Zoophytes; Librairie encyclopédique de Roret: Paris, France, 1841. [Google Scholar]
- Kahl, A. Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria), 2nd ed.; Gustav Fischer: Jena, Germany, 1931. [Google Scholar]
- Song, W. Morphological and taxonomical studies on some marine scuticociliates from China Sea, with description of two new species, Philasterides armatalis sp. n. and Cyclidium varibonneti sp. n. (Protozoa: Ciliophora: Scuticociliatida). Acta Protozool. 2000, 39, 295–322. [Google Scholar]
- Wang, Y.; Song, W.; Hu, X.; Warren, A.; Chen, X.; Al-Rasheid, K.A.S. Descriptions of two new marine species of Pleuronema, P. czapikae sp. n. and P. wiackowskii sp. n. (Ciliophora: Scuticociliatida), from the Yellow Sea, North China. Acta Protozool. 2008, 47, 35–45. [Google Scholar] [CrossRef]
- Wang, Y.; Song, W.; Warren, A.; Al-Rasheid, K.A.S.; Al-Quraishy, S.A.; Al-Farraj, S.A.; Hu, X.; Pan, H. Descriptions of two new marine scuticociliates, Pleuronema sinica n. sp. and P. wilberti n. sp. (Ciliophora: Scuticociliatida), from the Yellow Sea, China. Eur. J. Protistol. 2009, 45, 29–37. [Google Scholar] [CrossRef]
- Pan, X.; Huang, J.; Fan, X.; Ma, H.; Al-Rasheid, K.A.S.; Miao, M.; Gao, F. Morphology and phylogeny of four marine scuticociliates (Protista, Ciliophora), with descriptions of two new species: Pleuronema elegans spec. nov. and Uronema orientalis spec. nov. Acta Protozool. 2015, 54, 31–43. [Google Scholar] [CrossRef]
- Pan, H.; Hu, J.; Warren, A.; Wang, L.; Jiang, J.; Hao, R. Morphology and molecular phylogeny of Pleuronema orientale spec. nov. and P. paucisaetosum spec. nov. (Ciliophora, Scuticociliata) from Hangzhou Bay, China. Int. J. Syst. Evol. Microbiol. 2015, 65, 4800–4808. [Google Scholar] [CrossRef]
- Pan, H.; Hu, J.; Jiang, J.; Wang, L.; Hu, X. Morphology and phylogeny of three Pleuronema species (Ciliophora, Scuticociliatia) from Hangzhou Bay, China, with description of two new species, P. binucleatum n. sp. and P. parawiackowskii n. sp. J. Eukaryot. Microbiol. 2016, 63, 287–298. [Google Scholar] [CrossRef]
- Liu, M.; Liu, Y.; Zhang, T.; Lu, B.; Gao, F.; Gu, J.; Al-Farraj, S.A.; Hu, X.; Song, W. Integrative studies on the taxonomy and molecular phylogeny of four new Pleuronema species (Protozoa, Ciliophora, Scuticociliatia). Mar. Life Sci. Technol. 2022, 4, 179–200. [Google Scholar] [CrossRef]
- Wilbert, N. Eine verbesserte Technik der Protargolimprägnation für Ciliaten. Mikrokosmos 1975, 64, 171–179. [Google Scholar]
- Gao, F.; Warren, A.; Zhang, Q.; Gong, J.; Miao, M.; Sun, P.; Xu, D.; Huang, J.; Yi, Z.; Song, W. The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata). Sci. Rep. 2016, 6, 24874. [Google Scholar] [CrossRef] [PubMed]
- Medlin, L.; Hille, J.E.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Penn, O.; Privman, E.; Landan, G.; Graur, D.; Pupko, T. An alignment confidence score capturing robustness to guide tree uncertainty. Mol. Biol. Evol. 2010, 27, 1759–1767. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Res. 1999, 41, 95–98. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest V2. Program distributed by the author. Bioinformatics 2004, 24, 581–583. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Page, R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef]
- Pan, X.; Fan, X.; Al-Farraj, S.A.; Gao, S.; Chen, Y. Taxonomy and morphology of four “ophrys-related” scuticociliates (Protista, Ciliophora, Scuticociliatia), with the description of a new genus, Paramesanophrys gen. nov. Eur. J. Taxon. 2016, 191, 1–18. [Google Scholar] [CrossRef]
- Yeo, J.; Choi, J.; Omar, A.; Jung, J. New record of Pleuronema marinum Dujardin, 1841 (Protozoa, Ciliophora) from South Korea. J. Species Res. 2022, 11, 278–286. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, X.; Long, H.; Al-Rasheid, K.A.S.; Al-Farraj, S.A.; Song, W. Morphological studies indicate that Pleuronema grolierei nov. spec. and P. coronatum Kent, 1881 represent different sections of the genus Pleuronema (Ciliophora: Scuticociliatida). Eur. J. Protistol. 2008, 44, 131–140. [Google Scholar] [CrossRef]
- Agatha, S.; Spindler, M.; Wilbert, N. Ciliated protozoa (Ciliophora) from Arctic sea ice. Acta Protozool. 1993, 32, 261–268. [Google Scholar]
- Corliss, J.O.; Snyder, R.A. A preliminary description of several new ciliates from the Antarctica, including Cohnilembus grassei n. sp. and Pleuronema tardum n. sp. (Hymenostomata, Pleuronematina). Protistologica 1986, 22, 39–46. [Google Scholar]
- Dragesco, J.; Dragesco-Kernéis, A. Ciliés Libres de L’Afrique Intertropicale; Éditions de l’ORSTOM: Paris, France, 1986. [Google Scholar]
- Fan, X.; Pan, X. Ciliates of the Subclass Scuticociliatia and Peniculia in China; Science Press: Beijing, China, 2020. [Google Scholar]
- Fernandez-Leborans, G.; Novillo, A. Morphology and taxonomic position of two marine pleuronematine species: Pleuronema lynni and Schizocalyptra marina (Protozoa, Ciliophora). J. Zool. 1994, 233, 259–275. [Google Scholar] [CrossRef]
- Pan, X.; Shao, C.; Ma, H.; Fan, X.; Al-Rasheid, K.A.S.; Al-Farraj, S.A.; Hu, X. Redescriptions of two marine scuticociliates from China, with notes on stomatogenesis in Parauronema longum (Ciliophora, Scuticociliatida). Acta Protozool. 2011, 50, 301–310. [Google Scholar] [CrossRef]
- Dragesco, J. Les genres Pleuronema Dujardin, Schizocalyptra nov. gen. et Histiobalantium Stokes (ciliés holotriches hyme´nostomes). Protistologica 1968, 4, 85–106. [Google Scholar]
- Agamaliev, F.G. Materials on morphology of some psammophillic ciliates of the Caspian Sea. Acta Protozool. 1968, 6, 1–35. [Google Scholar]
- Borror, A.C. Morphology and ecology of the benthic ciliated protozoa of Alligator Harbor, Florida. Arch Protistenkd. 1963, 106, 465–534. [Google Scholar]
- Dragesco, J. Ciliés Mésopsammiques Littoraux. Systématique, Morphologie, Écologie; Imprimerie Louis-Jean: Eubagne, France, 1960. [Google Scholar]
- Kent, W.S. A Manual of the Infusoria: Including a Description of All Known Flagellate, Ciliate, and Tentaculiferous Protozoa, British and Foreign, and an Account of the Organization and Affinities of the Sponges; David Bogue: London, UK, 1881. [Google Scholar]
- Small, E.B.; Lynn, D.H. Phylum Ciliophora Doflein, 1901. In An Illustrated Guide to the Protozoa; Lee, J.J., Hunter, S.H., Bovee, E.C., Eds.; Society of Protozoologists: Lawrence, NY, USA, 1985. [Google Scholar]
- Groliere, C.; Detcheva, R. Description et stomatogenese de Pleuronema puytoraci n. sp. (Ciliata, Holotricha). Protistologica 1974, 10, 91–99. [Google Scholar]
- Yi, Z.; Song, W.; Gong, J.; Warren, A.; Al-Rasheid, K.A.S.; Al-Arifi, S.; Al-Khedhairy, A.A. Phylogeny of six oligohymenophoreans (Protozoa, Ciliophora) inferred from small subunit rRNA gene sequences. Zool. Scr. 2009, 38, 323–331. [Google Scholar] [CrossRef]
- Antipa, G.A.; Dolan, J.R.; Lynn, D.H.; Obolkina, L.A.; Strüder-Kypke, M.C. Molecular phylogeny and evolutionary relationships between the ciliate genera Peniculistoma and Mytilophilus (Peniculistomatidae, Pleuronematida). J. Eukaryot. Microbiol. 2016, 63, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Antipa, G.A.; Strüder-Kypke, M.C.; Lynn, D.H. Molecular phylogeny, taxonomic relationships and North American distribution of Conchophthirus (Conchophthiridae, Scuticociliatia). Aquat. Ecosyst. Health Manag. 2020, 23, 58–68. [Google Scholar] [CrossRef]
- Gao, F.; Strüder-Kypke, M.; Yi, Z.; Miao, M.; Al-Farraj, S.A.; Song, W. Phylogenetic analysis and taxonomic distinction of six genera of pathogenic scuticociliates (Protozoa, Ciliophora) inferred from small-subunit rRNA gene sequences. Int. J. Syst. Evol. Microbiol. 2012, 62, 246–256. [Google Scholar] [CrossRef]
- Gao, F.; Katz, L.A.; Song, W. Multigene-based analyses on evolutionary phylogeny of two controversial ciliate orders: Pleuronematida and Loxocephalida (Protista, Ciliophora, Oligohymenophorea). Mol. Phylogenet. Evol. 2013, 68, 55–63. [Google Scholar] [CrossRef]
- Gao, F.; Huang, J.; Zhao, Y.; Li, L.; Liu, W.; Miao, M.; Zhang, Q.; Li, J.; Yi, Z.; El-Serehy, H.A.; et al. Systematic studies on ciliates (Alveolata, Ciliophora) in China: Progress and achievements based on molecular information. Eur. J. Protistol. 2017, 61, 409–423. [Google Scholar] [CrossRef]
- Gao, F.; Gao, S.; Wang, P.; Katz, L.A.; Song, W. Phylogenetic analyses of cyclidiids (Protista, Ciliophora, Scuticociliatia) based on multiple genes suggest their close relationship with thigmotrichids. Mol. Phylogenet. Evol. 2014, 75, 219–226. [Google Scholar] [CrossRef]
- Zhang, T.; Fan, X.; Gao, F.; Al-Farraj, S.A.; El-Serehy, H.A.; Song, W. Further analyses on the phylogeny of the subclass Scuticociliatia (Protozoa, Ciliophora) based on both nuclear and mitochondrial data. Mol. Phylogenet. Evol. 2019, 139, 106565. [Google Scholar] [CrossRef]
| Character | Min | Max | Mean | Median | SD | CV (%) | N |
|---|---|---|---|---|---|---|---|
| Body length (μm) | 47 | 64 | 54.5 | 54 | 5.08 | 9.32 | 25 |
| 68 | 111 | 91.3 | 90 | 8.92 | 9.77 | 25 | |
| Body width (μm) | 21 | 33 | 27.0 | 27 | 3.20 | 11.85 | 25 |
| 41 | 77 | 56.2 | 58 | 7.86 | 13.99 | 25 | |
| Oral length (μm) | 37 | 47 | 40.8 | 40 | 2.85 | 6.99 | 25 |
| 53 | 84 | 68.3 | 67 | 5.69 | 8.33 | 25 | |
| Oral width (μm) | 11 | 16 | 13.6 | 13 | 1.35 | 9.93 | 25 |
| 16 | 30 | 21.9 | 22 | 2.62 | 11.96 | 25 | |
| Oral length/body length | 0.61 | 0.87 | 0.75 | 0.75 | 0.06 | 8.00 | 25 |
| 0.66 | 0.87 | 0.75 | 0.75 | 0.05 | 6.67 | 25 | |
| Number of somatic kineties | 16 | 22 | 18.7 | 19 | 1.65 | 8.82 | 25 |
| 36 | 51 | 46.6 | 47 | 3.04 | 6.52 | 25 | |
| Number of preoral kineties | 3 | 5 | 4.1 | 4 | 0.76 | 18.54 | 25 |
| 1 | 5 | 2.3 | 2 | 0.79 | 34.35 | 25 | |
| Number of kineties rows in M3 | 3 | 3 | 3.0 | 3 | 0 | 0 | 22 |
| 3 | 3 | 3.0 | 3 | 0 | 0 | 25 | |
| Number of Ma | 1 | 1 | 1.0 | 1 | 0 | 0 | 25 |
| 1 | 3 | 1.2 | 1 | 0.50 | 41.67 | 25 | |
| Diameter of macronucleus (μm) | 18 | 31 | 23.9 | 23 | 3.50 | 14.64 | 25 |
| 21 | 67 | 32.2 | 31 | 1.68 | 5.22 | 25 |
| Species | P. ningboensis | P. paucisaetosum | P. coronatum | P. smalli | P. foissneri | P. parasmalli |
|---|---|---|---|---|---|---|
| Body size in vivo (μm) | 55–65 × 25–30 | 55–85 × 25–55 | 60–125 × 30–60 | - | 60–75 × 30–40 | 55–85 × 25–35 |
| Body size after staining (μm) | 47–64 × 21–33 | 60–80 × 36–58 | 50–115 × 25–65 | 49–70 × 28–41 | 60–90 × 35–50 | 60–80 × 30–40 |
| Right ventrolateral side in vivo | Basically straight | - | Straight | - | Convex | Basically straight |
| Number of caudal cilia | 10–17 | 12–15 | 10–15 | - | About 15 | 15–18 |
| Number of somatic kineties | 16–22 | 21–23 | 35–48 | 28–36 | 32–40 | 26–32 |
| Number of preoral kineties | 3–5 | 4 or 5 | 3–7 | 2–5 | 4–8 | 4–6 |
| Number of kinety rows in M3 | 3 | 3 | 3 | 2 | 3 | 3 |
| Number of kinety rows in M1 | 3 | 3 | 2 | - | 3 | 3 |
| Mid-portion of M2a | Single-rowed | Single-rowed | Zig-zag pattern | - | Zig-zag pattern | Zig-zag pattern |
| Habitat | Brackish water | Brackish water | Freshwater and marine water | Brackish and marine water | Brackish water | Freshwater |
| Data sources | Present study | [24] | [26] | [46] | [26] | [26] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Zhao, X.; Ye, T.; Wu, Z.; Wu, F.; Chen, X.; Liu, M. Taxonomic and Phylogenetic Studies of Two Brackish Pleuronema Species (Protista, Ciliophora, Scuticociliatia) from Subtropical Coastal Waters of China, with Report of a New Species. Microorganisms 2023, 11, 1422. https://doi.org/10.3390/microorganisms11061422
Zhang H, Zhao X, Ye T, Wu Z, Wu F, Chen X, Liu M. Taxonomic and Phylogenetic Studies of Two Brackish Pleuronema Species (Protista, Ciliophora, Scuticociliatia) from Subtropical Coastal Waters of China, with Report of a New Species. Microorganisms. 2023; 11(6):1422. https://doi.org/10.3390/microorganisms11061422
Chicago/Turabian StyleZhang, Hui, Xuetong Zhao, Tingting Ye, Zehao Wu, Fan Wu, Xiangrui Chen, and Mingjian Liu. 2023. "Taxonomic and Phylogenetic Studies of Two Brackish Pleuronema Species (Protista, Ciliophora, Scuticociliatia) from Subtropical Coastal Waters of China, with Report of a New Species" Microorganisms 11, no. 6: 1422. https://doi.org/10.3390/microorganisms11061422
APA StyleZhang, H., Zhao, X., Ye, T., Wu, Z., Wu, F., Chen, X., & Liu, M. (2023). Taxonomic and Phylogenetic Studies of Two Brackish Pleuronema Species (Protista, Ciliophora, Scuticociliatia) from Subtropical Coastal Waters of China, with Report of a New Species. Microorganisms, 11(6), 1422. https://doi.org/10.3390/microorganisms11061422






