Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype
Abstract
1. Introduction
2. Materials and Methods
2.1. WGS Data Retrieval
2.2. Genome Assembly and Annotations
2.3. Pangenome Construction and Analysis
2.4. Phylogenetic Analysis
2.5. Statistical Analysis
3. Results
4. Discussion
| Analyzed Genomes | Core Genome | Accessory Genome Genes | Pangenome Open/Closed | Bioinformatic Tools | Reference |
|---|---|---|---|---|---|
| 36 | 3679 | 2086 | Open | PGAP pipeline | [11] |
| 47 | 3650 | 1196 | Open | BLASTP, GET_HOMOLOGUES, Perl script | [16] |
| 96 | 2066 | 6033 | Open | BLAST2GO and Perl scripts | [12] |
| 145 | 3736 | 708 | NA 4 | Roary | [38] |
| 150 | 1251 | 3758 | Nearly closed | BPGA tool | [15] |
| 183 2 | 1166 | 5870 | Closed | BLASTPBLOSUM62 | [10] |
| 1595 3 | 3419 | 7620 | Closed | CD-hit package v4.6 | [14] |
| 500 | 2231 | 3729 | Open | Roary | This study |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Tuberculosis. Available online: https://www.who.int/news-room/fact-sheets/detail/tuberculosis (accessed on 30 January 2021).
- Shah, M.; Dorman, S.E. Latent tuberculosis infection. N. Engl. J. Med. 2021, 385, 2271–2280. [Google Scholar] [CrossRef]
- Ben Ayed, H.; Koubaa, M.; Marrakchi, C.; Rekik, K.; Hammami, F.; Smaoui, F. Extrapulmonary tuberculosis: Update on the epidemiology, risk factors and prevention strategies. Int. J. Trop. Dis. 2018, 1, 1–6. [Google Scholar] [CrossRef]
- Coscolla, M. Biological and epidemiological consequences of MTBC diversity. Adv. Exp. Med. Biol. 2017, 1019, 95–116. [Google Scholar] [CrossRef]
- Saw, S.H.; Tan, J.L.; Chan, X.Y.; Chan, K.G.; Ngeow, Y.F. Chromosomal rearrangements and protein globularity changes in Mycobacterium tuberculosis isolates from cerebrospinal fluid. PeerJ 2016, 4, e2484. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Verma, R.; Advani, J.; Chatterjee, O.; Solanki, H.S.; Sharma, A.; Varma, S.; Modi, M.; Ray, P.; Mukherjee, K.K.; et al. Whole genome sequencing of Mycobacterium tuberculosis isolates from extrapulmonary sites. Omics 2017, 21, 413–425. [Google Scholar] [CrossRef]
- Ruesen, C.; Chaidir, L.; van Laarhoven, A.; Dian, S.; Ganiem, A.R.; Nebenzahl-Guimaraes, H.; Huynen, M.A.; Alisjahbana, B.; Dutilh, B.E.; van Crevel, R. Large-scale genomic analysis shows association between homoplastic genetic variation in Mycobacterium tuberculosis genes and meningeal or pulmonary tuberculosis. BMC Genom. 2018, 19, 122. [Google Scholar] [CrossRef]
- Faksri, K.; Xia, E.; Ong, R.T.; Tan, J.H.; Nonghanphithak, D.; Makhao, N.; Thamnongdee, N.; Thanormchat, A.; Phurattanakornkul, A.; Rattanarangsee, S.; et al. Comparative whole-genome sequence analysis of Mycobacterium tuberculosis isolated from tuberculous meningitis and pulmonary tuberculosis patients. Sci. Rep. 2018, 8, 4910. [Google Scholar] [CrossRef]
- Laing, C.; Buchanan, C.; Taboada, E.N.; Zhang, Y.; Kropinski, A.; Villegas, A.; Thomas, J.E.; Gannon, V.P. Pan-genome sequence analysis using Panseq: An online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinform. 2010, 11, 461. [Google Scholar] [CrossRef] [PubMed]
- Zakham, F.; Sironen, T.; Vapalahti, O.; Kant, R. Pan and core genome analysis of 183 Mycobacterium tuberculosis strains revealed a high inter-species diversity among the human adapted strains. Antibiotics 2021, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhong, J.; Zhang, J.; Li, C.; Yu, X.; Xiao, J.; Jia, X.; Ding, N.; Ma, G.; Wang, G.; et al. Pan-genomic study of Mycobacterium tuberculosis reflecting the primary/secondary genes, generality/individuality, and the interconversion through copy number variations. Front. Microbiol. 2018, 9, 1886. [Google Scholar] [CrossRef]
- Periwal, V.; Patowary, A.; Vellarikkal, S.K.; Gupta, A.; Singh, M.; Mittal, A.; Jeyapaul, S.; Chauhan, R.K.; Singh, A.V.; Singh, P.K.; et al. Comparative whole-genome analysis of clinical isolates reveals characteristic architecture of Mycobacterium tuberculosis pangenome. PLoS ONE 2015, 10, e0122979. [Google Scholar] [CrossRef]
- Wan, X.; Koster, K.; Qian, L.; Desmond, E.; Brostrom, R.; Hou, S.; Douglas, J.T. Genomic analyses of the ancestral Manila family of Mycobacterium tuberculosis. PLoS ONE 2017, 12, e0175330. [Google Scholar] [CrossRef]
- Kavvas, E.S.; Catoiu, E.; Mih, N.; Yurkovich, J.T.; Seif, Y.; Dillon, N.; Heckmann, D.; Anand, A.; Yang, L.; Nizet, V.; et al. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat. Commun. 2018, 9, 4306. [Google Scholar] [CrossRef] [PubMed]
- Dar, H.A.; Zaheer, T.; Ullah, N.; Bakhtiar, S.M.; Zhang, T.; Yasir, M.; Azhar, E.I.; Ali, A. Pangenome analysis of Mycobacterium tuberculosis reveals core-drug targets and screening of promising lead compounds for drug discovery. Antibiotics 2020, 9, 819. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Páez, U.; Zuluaga, N.Á.; Isaza, R.E.A.; Contreras-Moreira, B.; Rouzaud, F.; Robledo, J. Pan-genome association study of Mycobacterium tuberculosis lineage-4 revealed specific genes related to the high and low prevalence of the disease in patients from the North-Eastern area of Medellín, Colombia. Front Microbiol. 2022, 13, 1076797. [Google Scholar] [CrossRef] [PubMed]
- Negrete-Paz, A.M.; Vázquez-Marrufo, G.; Vázquez-Garcidueñas, M.S. Whole-genome comparative analysis at the lineage/sublineage level discloses relationships between Mycobacterium tuberculosis genotype and clinical phenotype. PeerJ 2021, 9, e12128. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Tonkin-Hill, G.; MacAlasdair, N.; Ruis, C.; Weimann, A.; Horesh, G.; Lees, J.A.; Gladstone, R.A.; Lo, S.; Beaudoin, C.; Floto, R.A.; et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020, 21, 1–21. [Google Scholar] [CrossRef]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016, 17, 1–9. [Google Scholar] [CrossRef]
- Lees, J.A.; Galardini, M.; Bentley, S.D.; Weiser, J.N.; Corander, J. Pyseer: A comprehensive tool for microbial pangenome-wide association studies. Bioinformatics 2018, 34, 4310–4312. [Google Scholar] [CrossRef]
- Lees, J.A.; Mai, T.T.; Galardini, M.; Wheeler, N.E.; Horsfield, S.T.; Parkhill, J.; Corander, J. Improved prediction of bacterial genotype-phenotype associations using interpretable pangenome-spanning regressions. mBio 2020, 11, e01344-20. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Kohl, T.A.; Utpatel, C.; Schleusener, V.; De Filippo, M.R.; Beckert, P.; Cirillo, D.M.; Niemann, S. MTBseq: A comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 2018, 6, e5895. [Google Scholar] [CrossRef]
- Wajid, B.; Serpedin, E. Do it yourself guide to genome assembly. Brief. Funct. Genom. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Guillén-Nepita, A.L.; Negrete-Paz, A.M.; Vázquez-Marrufo, G.; Cruz-Hernández, A.; Fresia, P.; Naya, H.; Vázquez-Garcidueñas, M.S. Sequencing and annotation of the genome of Mycobacterium tuberculosis MYC004, a strain causing meningitis in Mexico. Genome Announ. 2018, 6, e00523-18. [Google Scholar] [CrossRef]
- Pepperell, C.S. Evolution of tuberculosis pathogenesis. Annu. Rev. Microbiol. 2022, 76, 661–680. [Google Scholar] [CrossRef]
- Achtman, M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 2008, 62, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Sreevatsan, S.; Pan, X.; Stockbauer, K.E.; Connell, N.D.; Kreiswirth, B.N.; Whittam, T.S.; Musser, J.M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc. Natl. Acad. Sci. USA 1997, 94, 9869–9874. [Google Scholar] [CrossRef]
- Gagneux, S. Genetic diversity in Mycobacterium tuberculosis. Curr. Top. Microbiol. Immunol. 2013, 374, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Warner, D.F.; Koch, A.; Mizrahi, V. Diversity and disease pathogenesis in Mycobacterium tuberculosis. Trends Microbiol. 2015, 23, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Galagan, J.E. Genomic insights into tuberculosis. Nat. Genet. 2014, 15, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Bottai, D.; Frigui, W.; Sayes, F.; Di Luca, M.; Spadoni, D.; Pawlik, A.; Zoppo, M.; Orgeur, M.; Khanna, V.; Hardy, D.; et al. TbD1 deletion as a driver of the evolutionary success of modern epidemic Mycobacterium tuberculosis lineages. Nat. Commun. 2020, 11, 684. [Google Scholar] [CrossRef]
- Kayani, M.R.; Zheng, Y.C.; Xie, F.C.; Kang, K.; Li, H.Y.; Zhao, H.T. Genome sequences and comparative analysis of two extended-spectrum extensively-drug resistant Mycobacterium tuberculosis strains. Front. Pharmacol. 2018, 9, 1492. [Google Scholar] [CrossRef]
- Zhang, J.; Angala, S.K.; Pramanik, P.K.; Li, K.; Crick, D.C.; Liav, A.; Jozwiak, A.; Swiezewska, E.; Jackson, M.; Chatterjee, D. Reconstitution of functional mycobacterial arabinosyltransferase AftC proteoliposome and assessment of decaprenylphosphorylarabinose analogues as arabinofuranosyl donors. ACS Chem. Biol. 2011, 6, 819–828. [Google Scholar] [CrossRef]
- Sundararajan, S.; Muniyan, R. Latent tuberculosis: Interaction of virulence factors in Mycobacterium tuberculosis. Mol. Biol. Rep. 2021, 48, 6181–6196. [Google Scholar] [CrossRef]
- Serafini, A.; Tan, L.; Horswell, S.; Howell, S.; Greenwood, D.J.; Hunt, D.M.; Phan, M.D.; Schembri, M.; Monteleone, M.; Montague, C.R.; et al. Mycobacterium tuberculosis requires glyoxylate shunt and reverse methylcitrate cycle for lactate and pyruvate metabolism. Mol. Microbiol. 2019, 112, 1284–1307. [Google Scholar] [CrossRef]
- Daniel, J.; Deb, C.; Dubey, V.S.; Sirakova, T.D.; Abomoelak, B.; Morbidoni, H.R.; Kolattukudy, P.E. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 2004, 186, 5017–5030. [Google Scholar] [CrossRef]
- Zhang, L.; Hendrickson, R.C.; Meikle, V.; Lefkowitz, E.J.; Ioerger, T.R.; Niederweis, M. Comprehensive analysis of iron utilization by Mycobacterium tuberculosis. PLoS Pathog. 2020, 16, e1008337. [Google Scholar] [CrossRef]
- Martinez-Olivares, C.E.; Hernández-Pando, R.; Mixcoha, E. In silico EsxG· EsxH rational epitope selection: Candidate epitopes for vaccine design against pulmonary tuberculosis. bioRxiv 2022. [Google Scholar] [CrossRef]
- Wang, L.; Asare, E.; Shetty, A.C.; Sanchez-Tumbaco, F.; Edwards, M.R.; Saranathan, R.; Weinrick, B.; Xu, J.; Chen, B.; Bénard, A.; et al. Multiple genetic paths including massive gene amplification allow Mycobacterium tuberculosis to overcome loss of ESX-3 secretion system substrates. Proc. Natl. Acad. Sci. USA 2022, 119, e2112608119. [Google Scholar] [CrossRef] [PubMed]
- Tufariello, J.M.; Chapman, J.R.; Kerantzas, C.A.; Wong, K.W.; Vilchèze, C.; Jones, C.M.; Cole, L.E.; Tinaztepe, E.; Thompson, V.; Fenyö, D.; et al. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc. Natl. Acad. Sci. USA 2016, 113, E348–E357. [Google Scholar] [CrossRef]
- Martini, M.C.; Hicks, N.D.; Xiao, J.; Alonso, M.N.; Barbier, T.; Sixsmith, J.; Fortune, S.M.; Shell, S.S. Loss of RNase J leads to multi-drug tolerance and accumulation of highly structured mRNA fragments in Mycobacterium tuberculosis. PLoS Pathog. 2022, 18, e1010705. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Chaiprasert, A.; Ngamphiw, C.; Tongsima, S.; Regmi, S.M.; Clark, T.G.; Ong, R.T.; Teo, Y.Y.; Prammananan, T.; Palittapongarnpim, P. Genetic signatures of Mycobacterium tuberculosis Nonthaburi genotype revealed by whole genome analysis of isolates from tuberculous meningitis patients in Thailand. PeerJ 2016, 4, e1905. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.R.; Wernisch, L.; Stabler, R.; Mangan, J.A.; Hinds, J.; Laing, K.G.; Young, D.B.; Butcher, P.D. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 2002, 148, 3129–3138. [Google Scholar] [CrossRef]
- Hakiem, O.R.; Batra, J.K. Role of HrcA in stress management in Mycobacterium tuberculosis. J. Appl. Microbiol. 2022, 132, 3315–3326. [Google Scholar] [CrossRef]
- Deep, A.; Kaundal, S.; Agarwal, S.; Singh, R.; Thakur, K.G. Crystal structure of Mycobacterium tuberculosis VapC20 toxin and its interactions with cognate antitoxin, VapB20, suggest a model for toxin–antitoxin assembly. FEBS J. 2017, 284, 4066–4082. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, D.; Kong, Y.; Zhang, L.; Marrs, C.F.; Foxman, B.; Bates, J.H.; Wilson, F.; Cave, M.D. Clinical relevance of Mycobacterium tuberculosis plcD gene mutations. Am. J. Respir. Crit. Care Med. 2005, 171, 1436–1442. [Google Scholar] [CrossRef]
- Shafipour, M.; Shirzad-Aski, H.; Kochaksaraii, M.B.; Sohrabi, A.; Taziki, M.; Mahghani, G.A.; Ghaemi, E.A. The prevalence of plcD gene and evaluation of IS6110 insertion status in this gene in some clinical Mycobacterium tuberculosis isolates. Mol. Genet. Microbiol. Virol. 2021, 36, 111–118. [Google Scholar] [CrossRef]
- Kohli, S.; Singh, Y.; Sharma, K.; Mittal, A.; Ehtesham, N.Z.; Hasnain, S.E. Comparative genomic and proteomic analyses of PE/PPE multigene family of Mycobacterium tuberculosis H₃₇Rv and H₃₇Ra reveal novel and interesting differences with implications in virulence. Nucleic Acids Res. 2012, 40, 7113–7122. [Google Scholar] [CrossRef]
- Ahmad, J.; Khubaib, M.; Sheikh, J.A.; Pancsa, R.; Kumar, S.; Srinivasan, A.; Babu, M.M.; Hasnain, S.E.; Ehtesham, N.Z. Disorder-to-order transition in PE–PPE proteins of Mycobacterium tuberculosis augments the pro-pathogen immune response. FEBS Open Bio. 2020, 10, 70–85. [Google Scholar] [CrossRef]
- Campuzano, J.; Aguilar, D.; Arriaga, K.; León, J.C.; Salas-Rangel, L.P.; González-y-Merchand, J.; Hernández-Pando, R.; Espitia, C. The PGRS domain of Mycobacterium tuberculosis PE_PGRS Rv1759c antigen is an efficient subunit vaccine to prevent reactivation in a murine model of chronic tuberculosis. Vaccine 2007, 25, 3722–3729. [Google Scholar] [CrossRef]
- De Maio, F.; Berisio, R.; Manganelli, R.; Delogu, G. PE_PGRS proteins of Mycobacterium tuberculosis: A specialized molecular task force at the forefront of host–pathogen interaction. Virulence 2020, 11, 898–915. [Google Scholar] [CrossRef]
- Sharma, T.; Grover, S.; Arora, N.; Manjunath, P.; Ehtesham, N.Z.; Hasnain, S.E. PGRS domain of Rv0297 of Mycobacterium tuberculosis is involved in modulation of macrophage functions to favor bacterial persistence. Front. Cell Infect. Microbiol. 2020, 10, 451. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Zhou, Q.; Li, L.; Zhou, Y.; Sha, W. Epidemiological characteristics of extrapulmonary tuberculosis patients with or without pulmonary tuberculosis. Epidemiol. Infect. 2022, 150, e158. [Google Scholar] [CrossRef] [PubMed]
- Yates, T.A.; Khan, P.Y.; Knight, G.M.; Taylor, J.G.; McHugh, T.D.; Lipman, M.; White, R.G.; Cohen, T.; Cobelens, F.G.; Wood, R.; et al. The transmission of Mycobacterium tuberculosis in high burden settings. Lancet Infect. Dis. 2016, 16, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.D.; Chiu, C.; Churchyard, G.J.; Esmail, H.; Lewinsohn, D.M.; Gandhi, N.R.; Fennelly, K.P. Tuberculosis infectiousness and host susceptibility. J. Infect. Dis. 2017, 216 (Suppl. S6), S636–S643. [Google Scholar] [CrossRef]
- Chandra, P.; Grigsby, S.J.; Philips, J.A. Immune evasion and provocation by Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2022, 20, 750–766. [Google Scholar] [CrossRef] [PubMed]

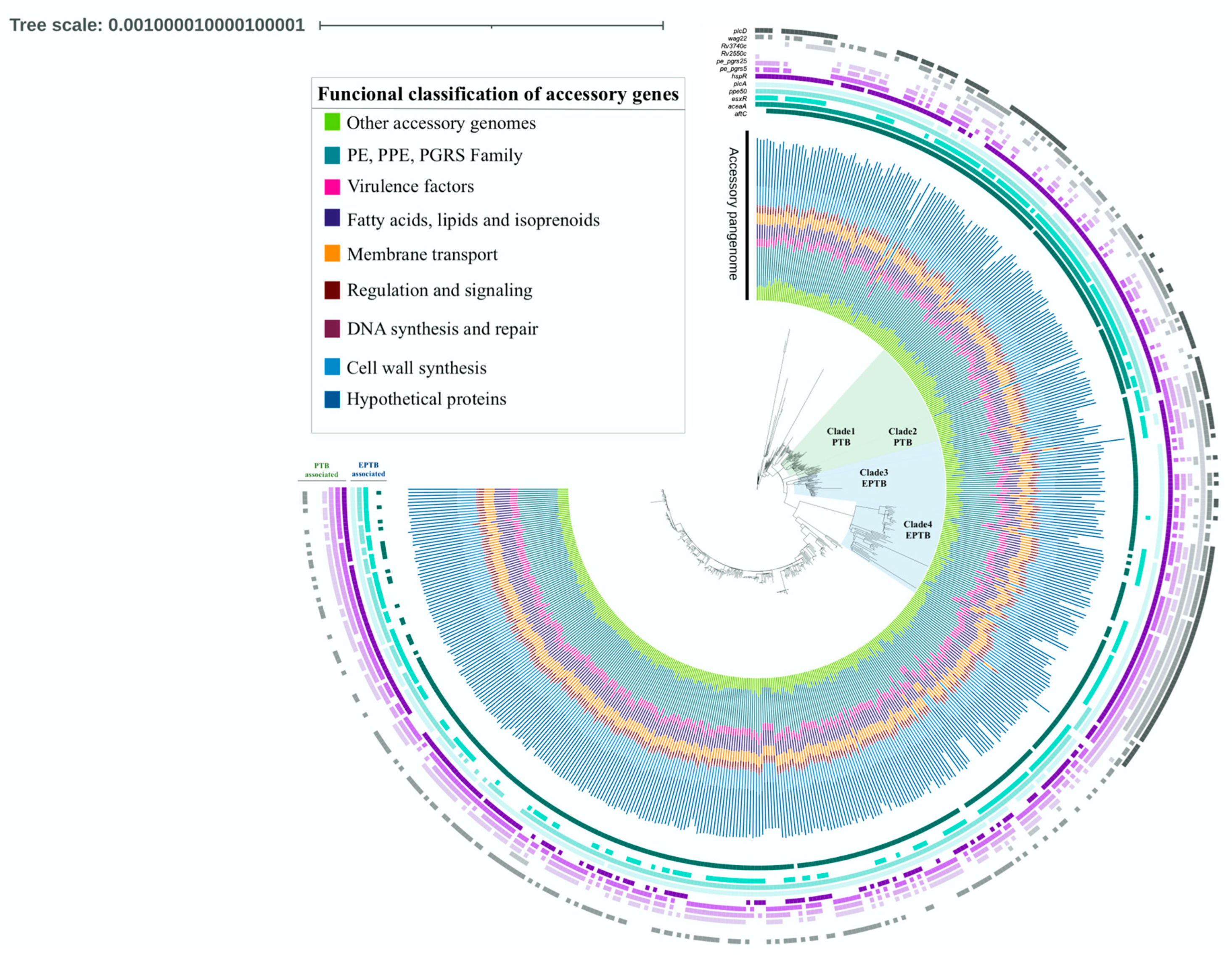

| Gene | Product | Functional Classification | p-Value | OR* | CI* 95% | Association | |
|---|---|---|---|---|---|---|---|
| Virulence | Rv0353/hspR | Heat shock protein transcriptional repressor HspR | Virulence | 0.0483 | 1.5 | 1.0034 to 2.4059 | PTB |
| Rv1755c/plcD | Phospholipase C | Virulence | 0.0006 | 2 | 1.3487 to 2.9849 | PTB | |
| Rv1915/aceA | Isocitrate lyase AceAa | Virulence | 0.0005 | 2.03 | 1.3606 to 3.0357 | EPTB | |
| Rv2351c/plcA | Phospholipase C | Virulence | 0.007 | 2.78 | 1.3136 to 5.9075 | EPTB | |
| Rv3019c/esxR | Secreted ESAT-6 like protein EsxR | Virulence | 0.0132 | 1.6 | 1.1010 to 2.2783 | EPTB | |
| Rv2550c | Antitoxin VapB20 | Virulence | 0.001 | 2.8 | 1.5405 to 5.5661 | PTB | |
| Other | Rv3135/ppe50 | PPE family protein PPE50 | Pe/ppe/pgrs | <0.0001 | 6.4 | 1.6896 to 4.5763 | EPTB |
| Rv0297/pe_pgrs5 | PE-PGRS family protein PE PGRS5 | Pe/ppe/pgrs | 0.0303 | 1.5 | 1.0419 to 2.2758 | PTB | |
| Rv1396c/pe_pgrs25 | PE-PGRS family protein PE PGRS25 | Pe/ppe/pgrs | 0.0255 | 1.6 | 1.0553 to 2.2802 | PTB | |
| Rv1759c/wag22 | PE-PGRS family protein Wag22 | Pe/ppe/pgrs | <0.0001 | 2.4 | 1.6427 to 3.4220 | PTB | |
| Rv3514/pe_pgrs57 | PE-PGRS family protein PE PGRS57 | Pe/ppe/pgrs | 0.0201 | 1.5 | 1.0679 to 2.1640 | PTB | |
| Rv3740c | Triacylglycerol synthase (diacylglycerol acyltransferase) | Lipid metabolism | 0.0266 | 1.5 | 1.0516 to 2.2586 | PTB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Negrete-Paz, A.M.; Vázquez-Marrufo, G.; Gutiérrez-Moraga, A.; Vázquez-Garcidueñas, M.S. Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype. Microorganisms 2023, 11, 1495. https://doi.org/10.3390/microorganisms11061495
Negrete-Paz AM, Vázquez-Marrufo G, Gutiérrez-Moraga A, Vázquez-Garcidueñas MS. Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype. Microorganisms. 2023; 11(6):1495. https://doi.org/10.3390/microorganisms11061495
Chicago/Turabian StyleNegrete-Paz, Andrea Monserrat, Gerardo Vázquez-Marrufo, Ana Gutiérrez-Moraga, and Ma. Soledad Vázquez-Garcidueñas. 2023. "Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype" Microorganisms 11, no. 6: 1495. https://doi.org/10.3390/microorganisms11061495
APA StyleNegrete-Paz, A. M., Vázquez-Marrufo, G., Gutiérrez-Moraga, A., & Vázquez-Garcidueñas, M. S. (2023). Pangenome Reconstruction of Mycobacterium tuberculosis as a Guide to Reveal Genomic Features Associated with Strain Clinical Phenotype. Microorganisms, 11(6), 1495. https://doi.org/10.3390/microorganisms11061495






