Bacterial Communities Associated with the Roots of Typha spp. and Its Relationship in Phytoremediation Processes
Abstract
1. Introduction
2. Phytoremediation
2.1. Classification of Plants Used in Phytoremediation
2.2. HM Phytoremediation Mechanisms
3. Typha Genus
Typha spp. Applications in Phytoremediation
| Species | Site/System | Heavy Metal | Metal Concentration (mg/Kg) | References | ||
|---|---|---|---|---|---|---|
| Shoots/Leaves/Stems | Roots | Rhizome | ||||
| T. latifolia | Constructed wetland | Zn | 59.29 | 177.28 | NR | [5] |
| Cu | 14.73 | 33.29 | NR | |||
| Natural wetland | Al | 38.3–48.5 | 1740–1780 | 845–1055 | [7] | |
| As | 0.08–0.12 | 1.87–2.21 | 1.21–1.65 | |||
| Cd | 0.06–0.08 | 0.39–0.46 | 0.16–0.22 | |||
| Cr | 0.95–1.01 | 5.54–6.75 | 3.24–3.85 | |||
| Cu | 4.66–5.87 | 12.8–13.1 | 9.87–11.8 | |||
| Hg | 0.49–0.63 | 2.88–3.35 | 1.55–1.83 | |||
| Mn | 29.7–41 | 132–155 | 70.1–103 | |||
| Ni | 8.42–10.3 | 35.6–41.2 | 28.5–30.2 | |||
| Pb | 0.44–0.52 | 13.5–15.2 | 4.32–6.65 | |||
| Stream | Zn | 215 | 340 | NR | [9] | |
| Ni | 40 | 55 | NR | |||
| Cu | 30 | 50 | NR | |||
| Pb | 8 | 13 | NR | |||
| Co | 10 | 24 | NR | |||
| Mn | 990 | 860 | NR | |||
| Cd | 0.21 | 0.44 | NR | |||
| Cr | 21 | 44 | NR | |||
| Artificial lagoon | Zn | 28.7–41 | 110–115 | 96.5–103 | [70] | |
| Cd | 0.1–1.85 | 0.1–25 | NR | |||
| Cr | 1–32 | 1–60 | NR | |||
| Mn | 63–1162.5 | 125–2375 | NR | |||
| Fe | 130–375 | 325–500 | NR | |||
| Pond | Fe | 178 | 8431 | 1875 | [71] | |
| Mn | 477 | 1943 | 292 | |||
| Zn | 28 | 373 | 65.6 | |||
| Cu | 3 | 8.62 | 3.97 | |||
| Cd | 0.01 | 7.28 | 2.72 | |||
| Pb | 3.0 | 12.1 | 6.33 | |||
| Ni | 3.7 | 27.8 | 8.92 | |||
| Co | 0.25 | 2.57 | 0.96 | |||
| Cr | 6 | 35.7 | 11.7 | |||
| Constructed wetland | Cd | 276–622 | 932–2339 | NR | [72] | |
| Pb | 272–927 | 1365–4867 | NR | |||
| Natural wetland | Cu | 16.00 | 13–265 | 37 | [73] | |
| Ni | 54 | 388 | 80 | |||
| Zn | 8–67 | 24–572 | 23,894 | |||
| Fe | 114–504 | 777–57,138 | 105–17,162 | |||
| Mn | 64–1734 | 16–901 | 16–552 | |||
| Mg | 564–2550 | 882–5542 | 745–2872 | |||
| Ca | 2687–16,993 | 1781–11,574 | 1209–6726 | |||
| Constructed wetland | Fe | 25–91 | 650–1250 | NR | [74] | |
| Cu | 15–49.98 | 10–31.45 | NR | |||
| Pb | 2.5–3.95 | 45,049 | NR | |||
| Hg | 2.5 | 45,082 | NR | |||
| Zn | 11,871 | 15–35 | NR | |||
| Constructed wetland | Cu | 13.52 | 32.92 | NR | [75] | |
| Cd | 11.84 | 14.68 | NR | |||
| Mn | 50.26 | 32.14 | NR | |||
| Cr | 11.46 | 10.72 | NR | |||
| Co | 8.28 | 11.1 | NR | |||
| Zn | 123.7 | 102.9 | NR | |||
| Pb | 19.38 | 24.38 | NR | |||
| Ni | 7.4 | 11.82 | NR | |||
| River | Cd | 0.89 | 1.1 | NR | [76] | |
| Ni | 1.955 | 26.9 | NR | |||
| Zn | 9.66 | 98.1 | NR | |||
| Cu | 4.885 | 30.2 | NR | |||
| Constructed wetland | Pb | NR | 65.6 | NR | [77] | |
| Cr | NR | 22.1 | NR | |||
| Mn | NR | 219 | NR | |||
| Constructed wetland | As | 0.001–0.02 | 0.008–0.03 | NR | [78] | |
| Cd | 17–118 | 185–319 | NR | |||
| Cr | 2.84 | 37–99 | NR | |||
| Lakes | Fe | 58.55 | 1252 | 125 | [79] | |
| Pb | 4.365 | 1.07 | 7.79 | |||
| Mn | 127.85 | 536 | 115 | |||
| Cd | 0.075 | 2.76 | 0.14 | |||
| Cu | 3.185 | 11.6 | 4.19 | |||
| Ni | 0.72 | 9.42 | 3.14 | |||
| Zn | 18.9 | 77.6 | 58.4 | |||
| Constructed wetland | Cd | 26.1–131 | 50.9–279 | NR | [80] | |
| T. domingensis | Constructed wetland | Fe | 63.23 | 40.6 | NR | [6] |
| Mn | 8.59 | 28.88 | NR | |||
| Ni | 4.8 | 24.3 | NR | |||
| Pb | 0.51 | 7 | NR | |||
| Cr | 8.17 | 17.6 | NR | |||
| Natural wetland | Al | 38–50.9 | 1756–1890 | 850–920 | [7] | |
| As | 0.08–0.10 | 2.78–3.21 | 1.29–1.34 | |||
| Cd | 0.05–0.08 | 0.44–0.61 | 0.15–0.18 | |||
| Cr | 1.05–1.24 | 3.67–5.88 | 3.01–4.57 | |||
| Cu | 3.50–4.67 | 15.2–18.5 | 10.4–12.7 | |||
| Hg | 0.85–0.97 | 3.21–3.67 | 2.02–2.56 | |||
| Mn | 32.1–51.2 | 138–151 | 74.2–83.8 | |||
| Ni | 10.8–10.9 | 36.6–53.3 | 29.6–38.7 | |||
| Pb | 0.65–0.71 | 10.9–13.7 | 4.21–4.33 | |||
| Zn | 35.4–38.8 | 118–122 | 97.3–103 | |||
| Natural wetland | Ba | 75.6 | 51.57 | NR | [81] | |
| Natural wetland | Cd | 1.25–21.3 | 188.62–234.10 | NR | [82] | |
| Constructed wetland | Cr | 10–90 | 50–750 | 10–300 | [83] | |
| Ni | 10–60 | 100–800 | 10–250 | |||
| Zn | 15–60 | 50–150 | 10–50 | |||
| River | Hg | 0.0506–0.5604 | 0.9785–5.474 | 0.4238–1.802 | [84] | |
| Plastic reactor | Cr | 2200–4000 | 3500–7000 | 200–1500 | [85] | |
| Ni | 1400 | 500–1000 | 200–500 | |||
| Zn | 2350–4750 | 300–3000 | 100–500 | |||
| Constructed wetland | Ba | 41.85–1398 | 303.15–3795.27 | NR | [86] | |
| Pond | Al | 187–282 | 220.82–350.55 | NR | [87] | |
| Fe | 102–173 | 307.5–582.44 | NR | |||
| Zn | 11.49–57 | 28.06–149.60 | NR | |||
| Pb | 1.7–9.0 | 1.26–20.46 | NR | |||
| Constructed wetland | Hg | 0.1785–273.3515 | NR | NR | [88] | |
| Constructed wetland | Cr | NR | 82 | NR | [89] | |
| Ni | 12 | 66 | NR | |||
| Zn | 28 | 178 | NR | |||
| T. angustifolia | Natural wetland | Al | 36.1–44.6 | 1568–1865 | 821–962 | [7] |
| As | 0.05–0.06 | 1.95–2.86 | 1.06–1.42 | |||
| Cd | 0.04 | 0.38–0.51 | 0.10–0.20 | |||
| Cr | 0.75–0.91 | 4.26–5.15 | 1.89–2.48 | |||
| Hg | 0.35–0.55 | 1.98–2.75 | 1.01–1.96 | |||
| Mn | 31.6- 36 | 95.8–126 | 77.6–103 | |||
| Ni | 8.96–12.3 | 28.8–35.7 | 20.2–21.6 | |||
| Pb | 0.52–0.75 | 8.90–10.2 | 3.25–5.23 | |||
| Constructed wetland | Zn | 33.9 | 37 | NR | [90] | |
| Cd | 7.3 | 7.2 | NR | |||
| Pb | 0.8 | 2.8 | NR | |||
| Constructed wetland | Cd | 20.3–42.3 | 241–378.3 | NR | [91] | |
| Pb | 354.9–1875.9 | 20,173.6–22,462 | NR | |||
| Constructed wetland | Cd | 0.225 | 0.82 | NR | [92] | |
| Cr | 8.345 | 59.13 | NR | |||
| Cu | 8.55 | 35.14 | NR | |||
| Fe | 701.375 | 3327 | NR | |||
| Ni | 4.025 | 21.1 | NR | |||
| Pb | 10.465 | 50.82 | NR | |||
| Zn | 100.075 | 150 | NR | |||
| Natural wetland | Cd | 0.03–0.65 | 0.1–0.8 | NR | [93] | |
| Pb | 0.3–4.5 | 1–6 | NR | |||
| Cr | 0.75–7.75 | 1.5–7.5 | NR | |||
| Ni | 1.75–16.25 | 2.5–15 | NR | |||
| Zn | 20–70 | 10–100 | NR | |||
| Cu | 0.75–25.5 | 2.5–17.5 | NR | |||
| Constructed wetland | Pb | 57.8–167.3 | 1265.2–8937.4 | 68.7–158.9 | [94] | |
| Hydroponics | Cr | 234.02–1157.28 | 287.16–4399.79 | NR | [95] | |
| T. capensis | Natural wetland | Cr | 69–3560.5 | 222–16,047 | 70–786 | [96] |
| Fe | 3176.5–8511.5 | 9413–13,833 | 2303–8970 | |||
| Zn | 21–59 | 56–162 | 24–30 | |||
| Cu | 13–31 | 35–224 | 10–56 | |||
| Co | 11–29 | 58–124 | 5–10 | |||
| Cd | 23.5–26.5 | 16–22 | 18–21 | |||
| Ni | 29–44 | 196–891 | 17–88 | |||
| Pb | 7.5–54.5 | 27–63 | 6–16 | |||
4. Bacteria Associated with the Rhizosphere of Typha spp.
4.1. Bacterial Communities Associated with Typha Roots in Natural Environments
4.2. Bacterial Communities Associated with the Roots of Typha Exposed to HMs
5. Plant Growth-Promoting Rhizobacteria Associated with HM-Tolerant Plants
5.1. Indole Acetic Acid’s Role in Plant Tolerance to HMs
5.2. Siderophore’s Role in Plant Tolerance to HMs
5.3. Phosphate Solubilization’s Role in Plant Tolerance to HMs
5.4. ACC Deaminase’s Role in Plant Tolerance to HMs
| Heavy Metal | Bacterium | Plant | PGPR Activities | Bacterial Effects on Plants | References |
|---|---|---|---|---|---|
| Cd2+ | Serratia sp. strain CP-13rif | Linum usitatissimum L. | Phosphate solubilization, IAA production, and ACC deaminase activity. | Bacterium enhances biomass accumulation and the roots and shoots growth. It increases photosynthetic pigments (Chl a, Chl b, and Chl total), proline, phenolic compounds, protein content, CAT activity and reduces H2O2 and MDA levels. | [136] |
| Cd2+ | Raoultella sp. strain X13 | Brassica chinensis L. | Phosphate solubilization, IAA, and siderophore production. | Bacteria enhance fresh and dry biomass accumulation and increase the content of soluble sugars. | [156] |
| Cd2+ | Cupriavidus necator strain GX_5 | Brassica napus | Siderophore secretion, ACC deaminase, IAA, and hydrogen cyanide (HCN) production. | Bacterium enhances dry biomass accumulation and root growth. | [157] |
| Sphingomonas sp. strain GX_15 | IAA production. | ||||
| Curtobacterium sp. strain GX_31 | ACC deaminase, IAA, and HCN production. | ||||
| Cd2+ | Kocuria rhizophila strain 14asp | Glycine max L. | Phosphate solubilization, catalase activity, ACC-deaminase, IAA, and ammonia production. | Bacterium enhances the growth of the shoots. | [158] |
| Cd2+ | Serratia marcescens strain S2I7 | Oryza sativa | Phosphate solubilization, production of siderophore, IAA, and HCN. | Bacterium increases shoot growth and root length. | [159] |
| Cd2+ | Sphingomonas sp. strain SaMR12 | Sedum alfredii | Siderophore production, phosphate solubilization, IAA production. | Bacterial inoculation increases photosynthetic pigments (Chl). It decreases H2O2 and MDA levels in roots. In shoots, it downregulates the SaZIP2 gene, whereas it upregulates SaZIP3, SaNramp6, SaHMA2, and SaHMA3 genes. In roots, the bacterium upregulates SaZIP3 and SaNramp1 genes and downregulates the SaNramp3 gene. | [133,160,161] |
| Cd2+ | Pseudomonas fluorescens strain Sasm05 | Sedum alfredii | IAA production, siderophore production, and ACC deaminase activity. | Bacterium enhances biomass accumulation, promotes shoots, and root formation and increases photosynthetic pigments (Chl). In shoots, it upregulates SaHMA2, SaHMA3, SaNramp1, SaNramp6, SaZIP2, SaZIP3, SaZIP4, and IRT1 genes, whereas in roots it upregulates SaHMA3, SaNramp6, SaZIP2, SaZIP4, SaZIP11, and IRT1 genes. | [162] |
| Cd2+ | Buttiauxella sp. strain SaSR13 | Sedum alfredii | IAA production, phosphate solubilization, siderophore production, and ACC deaminase activity. | Bacterium enhances biomass accumulation, root growth, and root-surface area, increases photosynthetic pigments (Chl), and reduces superoxide anion levels. | [135] |
| Cd2+ | Pseudomonas veronii strain E02 | Panicum virgatum | IAA production and ACC deaminase activity. | Bacterium enhances biomass accumulation and increases stem growth. | [163] |
| Cd2+ | Pseudomonas rhodesiae strains GRC065, GRC066, GRC093, GRC140 | Arabidopsis thaliana Col-0 | Phosphate solubilization, siderophore production, IAA, and ACC deaminase activity. | Bacteria promote the development of lateral roots in A. thaliana seedlings cultivated in conditions with and without cadmium. | [53] |
| Cd2+ | Enterobacter sp. strain S2 | Oryza sativa | ACC deaminase activity, IAA production, phosphate solubilization, and nitrogen fixation. | Bacterium enhances seedling growth, germination percentage, root-shoot length, fresh and dry weight, amylase, and protease activity. Furthermore, it exhibited alleviation of Cd-induced oxidative stress, reduction of stress ethylene, and decreased Cd accumulation in seedlings, conferring plant tolerance to cadmium. | [164] |
| Cd2+ | Pseudomonas fluorescens | Sedum alfredii | IAA production, siderophore production, and ACC deaminase activity. | Bacterium promotes lateral root formation, enhances biomass, Cd uptake and accumulation, increases IAA concentration, and decreases abscisic acid, brassinolide, trans-zeatin, ethylene, and jasmonic acid in roots, thereby inducing lateral root emergence. Moreover, it activates plant hormone-related genes. | [105] |
| Cd2+ | Rhodococcus ruber N7 | Sorghum bicolor | ACC deaminase activity, siderophore, and IAA production. | Bacterium increases the activity of peroxidase, laccase, and tyrosinase. Under cadmium contamination, it successfully colonizes the roots and contributes to metal accumulation in the plant roots. | [165] |
| Cd2+ | Pseudomonas rhodesiae strain GRC140 | Cucumis sativus L. | Phosphate solubilization, siderophore production, ACC deaminase activity, IAA andphenylacetic acid (PAA) synthesis. | In Cd-exposed seedlings, the bacterium improves the growth of C. sativus L. | [166] |
| Cd2+ | Enterobacter cloacae strain AS10 | Oryza sativa | Phosphate solubilization, ACC deaminase activity, nitrogen fixation, siderophore, HCN, and IAA production. | Bacterium enhances root-shoot growth at the seedling stage through Cd immobilization. It increases total sugar content and prevents the surge of ethylene and oxidative stress. | [167] |
| Cd2+, Ni2+ and Pb2+ | Citrobacter werkmanii strain WWN1 | Triticum aestivum L. | Zn, K, and PO4 solubilization, siderophore production. | Bacteria enhance plant shoot and root length, fresh and dry weight, and photosynthetic pigments (Chl a and b) under HM stress. Moreover, it improves antioxidant activity. | [168,169] |
| Enterobacter cloacaecepa strain JWM6 | |||||
| Cr6+ | Pseudomonas sp. strain NT27 | Medicago sativa | Phosphate solubilization, siderophore production, IAA and HCN production. | Bacterium increases shoot and root dry weights in the presence of Cr. Increases chlorophyll content and decreases stress markers, malondialdehyde, hydrogen peroxide, and proline levels. | [170] |
| Cr6+ | Pseudomonas sp. strain CPSB21 | Helianthus annuus L. and Solanum lycopersicum L. | Phosphate solubilization, siderophores, IAA, HCN and ammonia production. | Bacterium enhances shoot and root length, fresh and dry weight, chlorophyll, and soluble protein content. It reduces adverse effects of metal stress. | [171] |
| Cr3+ | Bacillus cereus strain B05 | Brassica nigra | Phosphate solubilization, siderophore production, ACC deaminase synthesis, phytohormones (IAA, CK, ABA). | Bacterium promotes plant growth and reduces chromium toxicity. It enhances seed germination %, shoot and root length, fresh, and dry biomass, and photosynthetic pigments. It improves phytoextraction of Cr. | [172] |
6. Other Microorganisms Associated with the Rhizosphere of Typha spp.
7. Conclusions
8. Outlooks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Inouhe, M.; Huang, H.; Chaudhary, S.K.; Gupta, D.K. Heavy Metal Bindings and Their Interactions with Thiol Peptides and Other Biological Ligands in Plant Cells. In Metal Toxicity in Plants: Perception, Signaling and Remediation; Gupta, D.K., Sandalio, L.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–21. ISBN 978-3-642-22081-4. [Google Scholar]
- Munive Cerrón, R.; Loli Figueroa, O.; Azabache Leyton, A.; Gamarra Sánchez, G. Fitorremediación Con Maíz (Zea mays L.) y Compost de Stevia En Suelos Degradados Por Contaminación Con Metales Pesados. Sci. Agropecu. 2018, 9, 551–560. [Google Scholar] [CrossRef]
- Santana-Flores, A.; Sánchez-Ayala, A.; Romero-Ramírez, Y.; Toledo-Hernández, E.; Ortega-Acosta, S.Á.; Toribio-Jiménez, J. Identification and Isolation of Heavy-Metal Tolerant and Bioaccumulator Bacteria Obtained from El Fraile Mine Tailings, Mexico. Terra Latinoam. 2020, 38, 67–75. [Google Scholar] [CrossRef]
- Nandakumar, R.; Chen, L.; Rogers, S.M.D. Agrobacterium-Mediated Transformation of the Wetland Monocot Typha latifolia L. (Broadleaf Cattail). Plant Cell Rep. 2005, 23, 744–750. [Google Scholar] [CrossRef]
- Hejna, M.; Moscatelli, A.; Stroppa, N.; Onelli, E.; Pilu, S.; Baldi, A.; Rossi, L. Bioaccumulation of Heavy Metals from Wastewater through a Typha latifolia and Thelypteris palustris Phytoremediation System. Chemosphere 2020, 241, 125018. [Google Scholar] [CrossRef]
- Shahid, M.J.; Ali, S.; Shabir, G.; Siddique, M.; Rizwan, M.; Seleiman, M.F.; Afzal, M. Comparing the Performance of Four Macrophytes in Bacterial Assisted Floating Treatment Wetlands for the Removal of Trace Metals (Fe, Mn, Ni, Pb, and Cr) from Polluted River Water. Chemosphere 2020, 243, 125353. [Google Scholar] [CrossRef]
- Bonanno, G.; Cirelli, G.L. Comparative Analysis of Element Concentrations and Translocation in Three Wetland Congener Plants: Typha domingensis, Typha latifolia and Typha angustifolia. Ecotoxicol. Environ. Saf. 2017, 143, 92–101. [Google Scholar] [CrossRef]
- Cristescu, A.C.; Covaliu, C.; Popa, L.; Dumitru, D.; Anghelet, A. Study on Use of Typha angustifolia L. in Wastewater Treatment: Promising Method in Removal of Copper Ions Present in Aquatic Solution. In Proceedings of the 17th International Scientific Conference, Jelgava, Latvia, 23–25 May 2018. [Google Scholar]
- Sasmaz, A.; Obek, E.; Hasar, H. The Accumulation of Heavy Metals in Typha latifolia L. Grown in a Stream Carrying Secondary Effluent. Ecol. Eng. 2008, 33, 278–284. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, C.; Li, S.; Lu, G.; Lu, G.; Li, S.; Zhou, Y. An Approach to Biodegradation of Chlorobenzenes: Combination of Typha angustifolia and Bacterial Effects on Hexachlorobenzene Degradation in Water. Water Sci. Technol. 2016, 74, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Zapata-Morales, A.L.; Alfaro-De la Torre, M.C.; Hernández-Morales, A.; García-De la Cruz, R.F. Isolation of Cultivable Bacteria Associated with the Root of Typha latifolia in a Constructed Wetland for the Removal of Diclofenac or Naproxen. Water Air Soil Pollut. 2020, 231, 423. [Google Scholar] [CrossRef]
- Ullah, A.; Heng, S.; Munis, M.F.H.; Fahad, S.; Yang, X. Phytoremediation of Heavy Metals Assisted by Plant Growth Promoting (PGP) Bacteria: A Review. Environ. Exp. Bot. 2015, 117, 28–40. [Google Scholar] [CrossRef]
- Etim, E. Phytoremediation and Its Mechanisms: A Review. Int. J. Environ. Bioenergy 2012, 2, 120–136. [Google Scholar]
- Reichenauer, T.G.; Germida, J.J. Phytoremediation of Organic Contaminants in Soil and Groundwater. ChemSusChem 2008, 1, 708–717. [Google Scholar] [CrossRef]
- Manahan, S.E. Introducción a la Química Ambiental; Reverte: Barcelona, Spain, 2006; ISBN 978-84-291-7907-1. [Google Scholar]
- Onweremadu, E.U. Selected Bioremediation Techniques in Polluted Tropical Soils; IntechOpen: London, UK, 2014. [Google Scholar]
- Ismail, S. Phytoremediation: A Green Technology. 2012, pp. 567–576. Available online: https://www.researchgate.net/publication/348008872_Phytoremediation_a_green_technology (accessed on 3 May 2023).
- Jadia, C.D.; Fulekar, M.H. Phytoremediation of Heavy Metals: Recent Techniques. Afr. J. Biotechnol. 2009, 8. [Google Scholar]
- Marques, A.P.G.C.; Rangel, A.O.S.S.; Castro, P.M.L. Remediation of Heavy Metal Contaminated Soils: Phytoremediation as a Potentially Promising Clean-Up Technology. Crit. Rev. Environ. Sci. Technol. 2009, 39, 622–654. [Google Scholar] [CrossRef]
- Adriano, D.C. Nickel. In Trace Elements in Terrestrial Environments; Springer: New York, NY, USA, 2001; pp. 677–705. ISBN 978-1-4684-9505-8. [Google Scholar]
- Leitenmaier, B.; Küpper, H. Compartmentation and Complexation of Metals in Hyperaccumulator Plants. Front. Plant Sci. 2013, 4, 374. [Google Scholar] [CrossRef]
- Kidd, P.S.; Castro, C.B.; Lestón, M.G.; Monterroso, C. Aplicación de plantas hiperacumuladoras de níquel en la fitoextracción natural: El género Alyssum L. Ecosistemas 2007, 16, 3. [Google Scholar]
- Llugany, M.; Tolrá, R.; Poschnrieder, C.; Barceló, J. Hiperacumulación de metales: ¿una ventaja para la planta y para el hombre? Ecosistemas 2007, 16, 4–9. [Google Scholar]
- Kopittke, P.M.; Lombi, E.; Menzies, N.W.; Naidu, R. Principles of Plant-Based Remediation of Contaminated Soils. Ind. Crops Uses 2010, 446–469. [Google Scholar] [CrossRef]
- Sanghi, R.; Singh, V. Green Chemistry for Environmental Remediation; John Wiley & Sons: Hoboken, NJ, USA, 2012; ISBN 978-1-118-28772-9. [Google Scholar]
- Neilson, S.; Rajakaruna, N. Roles of Rhizospheric Processes and Plant Physiology in Applied Phytoremediation of Contaminated Soils Using Brassica oilseeds. In The Plant Family Brassicaceae: Contribution Towards Phytoremediation; Anjum, N.A., Ahmad, I., Pereira, M.E., Duarte, A.C., Umar, S., Khan, N.A., Eds.; Environmental Pollution; Springer: Dordrecht, The Netherlands, 2012; pp. 313–330. ISBN 978-94-007-3913-0. [Google Scholar]
- Poschenrieder, C.; Tolrà, R.; Barceló, J. Can Metals Defend Plants against Biotic Stress? Trends Plant Sci. 2006, 11, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Rascio, N.; Navari-Izzo, F. Heavy Metal Hyperaccumulating Plants: How and Why Do They Do It? And What Makes Them so Interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef]
- Steinberg, C.E.W. Stress Ecology: Environmental Stress as Ecological Driving Force and Key Player in Evolution; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 978-94-007-2072-5. [Google Scholar]
- Awa, S.H.; Hadibarata, T. Removal of Heavy Metals in Contaminated Soil by Phytoremediation Mechanism: A Review. Water Air Soil Pollut. 2020, 231, 47. [Google Scholar] [CrossRef]
- Lee, B.X.Y.; Hadibarata, T.; Yuniarto, A. Phytoremediation Mechanisms in Air Pollution Control: A Review. Water Air Soil Pollut. 2020, 231, 437. [Google Scholar] [CrossRef]
- Kafle, A.; Timilsina, A.; Gautam, A.; Adhikari, K.; Bhattarai, A.; Aryal, N. Phytoremediation: Mechanisms, Plant Selection and Enhancement by Natural and Synthetic Agents. Environ. Adv. 2022, 8, 100203. [Google Scholar] [CrossRef]
- Raza, A.; Habib, M.; Kakavand, S.N.; Zahid, Z.; Zahra, N.; Sharif, R.; Hasanuzzaman, M. Phytoremediation of Cadmium: Physiological, Biochemical, and Molecular Mechanisms. Biology 2020, 9, 177. [Google Scholar] [CrossRef]
- Yan, A.; Wang, Y.; Tan, S.N.; Mohd Yusof, M.L.; Ghosh, S.; Chen, Z. Phytoremediation: A Promising Approach for Revegetation of Heavy Metal-Polluted Land. Front. Plant Sci. 2020, 11, 359. [Google Scholar] [CrossRef]
- Sharma, J.K.; Kumar, N.; Singh, N.P.; Santal, A.R. Phytoremediation Technologies and Their Mechanism for Removal of Heavy Metal from Contaminated Soil: An Approach for a Sustainable Environment. Front. Plant Sci. 2023, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Muthusaravanan, S.; Sivarajasekar, N.; Vivek, J.S.; Vasudha Priyadharshini, S.; Paramasivan, T.; Dhakal, N.; Naushad, M. Research Updates on Heavy Metal Phytoremediation: Enhancements, Efficient Post-Harvesting Strategies and Economic Opportunities. In Green Materials for Wastewater Treatment; Springer International Publishing: Cham, Switzerland, 2020; pp. 191–222. ISBN 978-3-030-17723-2. [Google Scholar]
- Gabriele, I.; Race, M.; Papirio, S.; Esposito, G. Phytoremediation of Pyrene-Contaminated Soils: A Critical Review of the Key Factors Affecting the Fate of Pyrene. J. Environ. Manag. 2021, 293, 112805. [Google Scholar] [CrossRef]
- DalCorso, G.; Fasani, E.; Manara, A.; Visioli, G.; Furini, A. Heavy Metal Pollutions: State of the Art and Innovation in Phytoremediation. Int. J. Mol. Sci. 2019, 20, 3412. [Google Scholar] [CrossRef]
- Grzegórska, A.; Rybarczyk, P.; Rogala, A.; Zabrocki, D. Phytoremediation—From Environment Cleaning to Energy Generation—Current Status and Future Perspectives. Energies 2020, 13, 2905. [Google Scholar] [CrossRef]
- Shackira, A.M.; Puthur, J.T. Phytostabilization of Heavy Metals: Understanding of Principles and Practices. In Plant-Metal Interactions; Srivastava, S., Srivastava, A.K., Suprasanna, P., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 263–282. ISBN 978-3-030-20732-8. [Google Scholar]
- Wani, Z.A.; Ahmad, Z.; Asgher, M.; Bhat, J.A.; Sharma, M.; Kumar, A.; Sharma, V.; Kumar, A.; Pant, S.; Lukatkin, A.S.; et al. Phytoremediation of Potentially Toxic Elements: Role, Status and Concerns. Plants 2023, 12, 429. [Google Scholar] [CrossRef]
- Mocek-Płóciniak, A.; Mencel, J.; Zakrzewski, W.; Roszkowski, S. Phytoremediation as an Effective Remedy for Removing Trace Elements from Ecosystems. Plants 2023, 12, 1653. [Google Scholar] [CrossRef] [PubMed]
- Sabreena; Hassan, S.; Bhat, S.A.; Kumar, V.; Ganai, B.A.; Ameen, F. Phytoremediation of Heavy Metals: An Indispensable Contrivance in Green Remediation Technology. Plants 2022, 11, 1255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yuan, X.; Xiong, T.; Wang, H.; Jiang, L. Bioremediation of Co-Contaminated Soil with Heavy Metals and Pesticides: Influence Factors, Mechanisms and Evaluation Methods. Chem. Eng. J. 2020, 398, 125657. [Google Scholar] [CrossRef]
- Khan, I.U.; Qi, S.-S.; Gul, F.; Manan, S.; Rono, J.K.; Naz, M.; Shi, X.-N.; Zhang, H.; Dai, Z.-C.; Du, D.-L. A Green Approach Used for Heavy Metals ‘Phytoremediation’ Via Invasive Plant Species to Mitigate Environmental Pollution: A Review. Plants 2023, 12, 725. [Google Scholar] [CrossRef] [PubMed]
- Raklami, A.; Meddich, A.; Oufdou, K.; Baslam, M. Plants—Microorganisms-Based Bioremediation for Heavy Metal Cleanup: Recent Developments, Phytoremediation Techniques, Regulation Mechanisms, and Molecular Responses. Int. J. Mol. Sci. 2022, 23, 5031. [Google Scholar] [CrossRef]
- Bartucca, M.L.; Cerri, M.; Del Buono, D.; Forni, C. Use of Biostimulants as a New Approach for the Improvement of Phytoremediation Performance—A Review. Plants 2022, 11, 1946. [Google Scholar] [CrossRef]
- Zhou, B.; Tu, T.; Kong, F.; Wen, J.; Xu, X. Revised Phylogeny and Historical Biogeography of the Cosmopolitan Aquatic Plant Genus Typha (Typhaceae). Sci. Rep. 2018, 8, 8813. [Google Scholar] [CrossRef]
- Baldwin, B.; Cannon, A. Typha Review; Utah State University: Logan, UT, USA, 2007. [Google Scholar]
- Sesin, V.; Davy, C.M.; Freeland, J.R. Review of Typha spp. (Cattails) as Toxicity Test Species for the Risk Assessment of Environmental Contaminants on Emergent Macrophytes. Environ. Pollut. 2021, 284, 117105. [Google Scholar] [CrossRef]
- Ciotir, C.; Freeland, J. Cryptic Intercontinental Dispersal, Commercial Retailers, and the Genetic Diversity of Native and Non-Native Cattails (Typha spp.) in North America. Hydrobiologia 2016, 768, 137–150. [Google Scholar] [CrossRef]
- Grace, J.B.; Harrison, J.S. The Biology of Canadian Weeds. Typha latifolia L., Typha angustifolia L. and Typha xglauca Godr. Can. J. Plant Sci. 1986, 66, 361–379. [Google Scholar] [CrossRef]
- Rolón-Cárdenas, G.A.; Arvizu-Gómez, J.L.; Pacheco-Aguilar, J.R.; Vázquez-Martínez, J.; Hernández-Morales, A. Cadmium-Tolerant Endophytic Pseudomonas rhodesiae Strains Isolated from Typha latifolia Modify the Root Architecture of Arabidopsis thaliana Col-0 in Presence and Absence of Cd. Braz. J. Microbiol. 2021, 52, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Ciotir, C.; Szabo, J.; Freeland, J. Genetic Characterization of Cattail Species and Hybrids (Typha spp.) in Europe. Aquat. Bot. 2017, 141, 51–59. [Google Scholar] [CrossRef]
- Eid, E.M.; Shaltout, K.H. Evaluation of Carbon Sequestration Potentiality of Lake Burullus, Egypt to Mitigate Climate Change. Egypt. J. Aquat. Res. 2013, 39, 31–38. [Google Scholar] [CrossRef]
- Mitsch, W.J.; Bernal, B.; Nahlik, A.M.; Mander, Ü.; Zhang, L.; Anderson, C.J.; Jørgensen, S.E.; Brix, H. Wetlands, Carbon, and Climate Change. Landsc. Ecol. 2013, 28, 583–597. [Google Scholar] [CrossRef]
- Valach, A.C.; Kasak, K.; Hemes, K.S.; Anthony, T.L.; Dronova, I.; Taddeo, S.; Silver, W.L.; Szutu, D.; Verfaillie, J.; Baldocchi, D.D. Productive wetlands restored for carbon sequestration quickly become net CO2 sinks with site-level factors driving uptake variability. PLoS ONE 2021, 16, e0248398. [Google Scholar] [CrossRef]
- Pal, S.; Chattopadhyay, B.; Datta, S.; Mukhopadhyay, S.K. Potential of Wetland Macrophytes to Sequester Carbon and Assessment of Seasonal Carbon Input into the East Kolkata Wetland Ecosystem. Wetlands 2017, 37, 497–512. [Google Scholar] [CrossRef]
- Hemes, K.S.; Chamberlain, S.D.; Eichelmann, E.; Knox, S.H.; Baldocchi, D.D. A Biogeochemical Compromise: The High Methane Cost of Sequestering Carbon in Restored Wetlands. Geophys. Res. Lett. 2018, 45, 6081–6091. [Google Scholar] [CrossRef]
- Günther, A.; Huth, V.; Jurasinski, G.; Glatzel, S. The Effect of Biomass Harvesting on Greenhouse Gas Emissions from a Rewetted Temperate Fen. GCB Bioenergy 2015, 7, 1092–1106. [Google Scholar] [CrossRef]
- Were, D.; Kansiime, F.; Fetahi, T.; Hein, T. Carbon Dioxide and Methane Fluxes from Various Vegetation Communities of a Natural Tropical Freshwater Wetland in Different Seasons. Environ. Process. 2021, 8, 553–571. [Google Scholar] [CrossRef]
- Vroom, R.J.E.; Xie, F.; Geurts, J.J.M.; Chojnowska, A.; Smolders, A.J.P.; Lamers, L.P.M.; Fritz, C. Typha latifolia Paludiculture Effectively Improves Water Quality and Reduces Greenhouse Gas Emissions in Rewetted Peatlands. Ecol. Eng. 2018, 124, 88–98. [Google Scholar] [CrossRef]
- Johnson, O.F.; Panda, A.; Lishawa, S.C.; Lawrence, B.A. Repeated Large-Scale Mechanical Treatment of Invasive Typha under Increasing Water Levels Promotes Floating Mat Formation and Wetland Methane Emissions. Sci. Total Environ. 2021, 790, 147920. [Google Scholar] [CrossRef]
- Saha, C.; Mukherjee, G.; Agarwal-Banka, P.; Seal, A. A Consortium of Non-Rhizobial Endophytic Microbes from Typha angustifolia Functions as Probiotic in Rice and Improves Nitrogen Metabolism. Plant Biol. 2016, 18, 938–946. [Google Scholar] [CrossRef]
- Pietrangelo, L.; Bucci, A.; Maiuro, L.; Bulgarelli, D.; Naclerio, G. Unraveling the Composition of the Root-Associated Bacterial Microbiota of Phragmites australis and Typha latifolia. Front. Microbiol. 2018, 9, 1650. [Google Scholar] [CrossRef]
- Ponce-Hernández, A.; Maldonado-Miranda, J.; Medellin-Castillo, N.; Alonso-Castro, A.; Carranza Alvarez, C. Phytoremediation Technology: Sustainable Solution for Cleaning Up of Recalcitrant Pollutants from Disturbed Environs. In Bioremediation and Biotechnology, Vol 3: Persistent and Recalcitrant Toxic Substances; Springer International Publishing: Cham, Switzerland, 2020; pp. 245–268. ISBN 978-3-030-46074-7. [Google Scholar]
- Petry, C.T.; Costa, D.T.; Droste, A. Removal of Ammoniacal Nitrogen from Municipal Landfill Leachate with Floating Typha domingensis (Typhaceae). Acta Biológica Colomb. 2020, 25, 5–13. [Google Scholar] [CrossRef]
- Ali, S.; Abbas, Z.; Rizwan, M.; Zaheer, I.E.; Yavaş, İ.; Ünay, A.; Abdel-Daim, M.M.; Bin-Jumah, M.; Hasanuzzaman, M.; Kalderis, D. Application of Floating Aquatic Plants in Phytoremediation of Heavy Metals Polluted Water: A Review. Sustainability 2020, 12, 1927. [Google Scholar] [CrossRef]
- Pacheco-Aguilar, J.R.P.; Peña-Cabriales, J.J.P.; Maldonado-Vega, M.M. Identification and Characterization of Sulfur-Oxidizing Bacteria in an Artificial Wetland That Treats Wastewater From a Tannery. Int. J. Phytoremed. 2008, 10, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Carranza-Álvarez, C.; Alonso-Castro, A.J.; Alfaro-De La Torre, M.C.; García-De La Cruz, R.F. Accumulation and Distribution of Heavy Metals in Scirpus americanus and Typha latifolia from an Artificial Lagoon in San Luis Potosí, México. Water Air Soil Pollut. 2008, 188, 297–309. [Google Scholar] [CrossRef]
- Klink, A.; Macioł, A.; Wisłocka, M.; Krawczyk, J. Metal Accumulation and Distribution in the Organs of Typha latifolia L. (Cattail) and Their Potential Use in Bioindication. Limnologica 2013, 43, 164–168. [Google Scholar] [CrossRef]
- Alonso-Castro, A.J.; Carranza-Álvarez, C.; Alfaro-De la Torre, M.C.; Chávez-Guerrero, L.; García-De la Cruz, R.F. Removal and Accumulation of Cadmium and Lead by Typha latifolia Exposed to Single and Mixed Metal Solutions. Arch. Environ. Contam. Toxicol. 2009, 57, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.J.; Crowder, A.A. Uptake and Accumulation of Heavy Metals by Typha latifolia in Wetlands of the Sudbury, Ontario Region. Can. J. Bot. 1983, 61, 63–73. [Google Scholar] [CrossRef]
- Anning, A.K.; Korsah, P.E.; Addo-Fordjour, P. Phytoremediation of Wastewater with Limnocharis flava, Thalia geniculata and Typha latifolia in Constructed Wetlands. Int. J. Phytoremed. 2013, 15, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Rana, V.; Maiti, S.K. Municipal Wastewater Treatment Potential and Metal Accumulation Strategies of Colocasia esculenta (L.) Schott and Typha latifolia L. in a Constructed Wetland. Environ. Monit. Assess. 2018, 190, 328. [Google Scholar] [CrossRef] [PubMed]
- Azizi, A.; Krika, A.; Krika, F. Heavy Metal Bioaccumulation and Distribution in Typha latifolia and Arundo donax: Implication for Phytoremediation. Casp. J. Environ. Sci. 2020, 18, 21–29. [Google Scholar] [CrossRef]
- Santos-Díaz, M.D.S.; Barrón-Cruz, M.D.C. Lead, Chromium and Manganese Removal by In Vitro Root Cultures of Two Aquatic Macrophytes Species: Typha latifolia L. and Scirpus americanus Pers. Int. J. Phytoremed. 2011, 13, 538–551. [Google Scholar] [CrossRef]
- Leura-Vicencio, A.; Alonso-Castro, A.J.; Carranza-Álvarez, C.; Loredo-Portales, R.; Alfaro-De la Torre, M.C.; García-De la Cruz, R.F. Removal and Accumulation of As, Cd and Cr by Typha latifolia. Bull. Environ. Contam. Toxicol. 2013, 90, 650–653. [Google Scholar] [CrossRef]
- Klink, A. A Comparison of Trace Metal Bioaccumulation and Distribution in Typha latifolia and Phragmites australis: Implication for Phytoremediation. Environ. Sci. Pollut. Res. 2017, 24, 3843–3852. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, Q. Phytoremediation of Cadmium-Contaminated Wetland Soil with Typha latifolia L. and the Underlying Mechanisms Involved in the Heavy-Metal Uptake and Removal. Environ. Sci. Pollut. Res. 2020, 27, 4905–4916. [Google Scholar] [CrossRef]
- de Carvalho, C.F.M.; Viana, D.G.; Pires, F.R.; Egreja Filho, F.B.; Bonomo, R.; Martins, L.F.; Cruz, L.B.S.; Nascimento, M.C.P.; Cargnelutti Filho, A.; da Rocha Júnior, P.R. Phytoremediation of Barium-Affected Flooded Soils Using Single and Intercropping Cultivation of Aquatic Macrophytes. Chemosphere 2019, 214, 10–16. [Google Scholar] [CrossRef]
- Oliveira, J.P.V.; Pereira, M.P.; Duarte, V.P.; Corrêa, F.F.; Castro, E.M.; Pereira, F.J. Cadmium Tolerance of Typha domingensis Pers. (Typhaceae) as Related to Growth and Leaf Morphophysiology. Braz. J. Biol. 2018, 78, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Hadad, H.R.; Mufarrege, M.D.L.M.; Di Luca, G.A.; Maine, M.A. Long-Term Study of Cr, Ni, Zn, and P Distribution in Typha domingensis Growing in a Constructed Wetland. Environ. Sci. Pollut. Res. 2018, 25, 18130–18137. [Google Scholar] [CrossRef] [PubMed]
- Lominchar, M.Á.; Sierra, M.J.; Jiménez-Moreno, M.; Guirado, M.; Martín-Doimeadios, R.C.R.; Millán, R. Mercury Species Accumulation and Distribution in Typha domingensis under Real Field Conditions (Almadén, Spain). Environ. Sci. Pollut. Res. 2019, 26, 3138–3144. [Google Scholar] [CrossRef]
- Mufarrege, M.M.; Hadad, H.R.; Di Luca, G.A.; Maine, M.A. The Ability of Typha domingensis to Accumulate and Tolerate High Concentrations of Cr, Ni, and Zn. Environ. Sci. Pollut. Res. 2015, 22, 286–292. [Google Scholar] [CrossRef]
- de Castro Ribeiro, P.R.C.; Viana, D.G.; Pires, F.R.; Egreja Filho, F.B.; Bonomo, R.; Cargnelutti Filho, A.; Martins, L.F.; Cruz, L.B.S.; Nascimento, M.C.P. Selection of Plants for Phytoremediation of Barium-Polluted Flooded Soils. Chemosphere 2018, 206, 522–530. [Google Scholar] [CrossRef]
- Hegazy, A.K.; Abdel-Ghani, N.T.; El-Chaghaby, G.A. Phytoremediation of Industrial Wastewater Potentiality by Typha domingensis. Int. J. Environ. Sci. Technol. 2011, 8, 639–648. [Google Scholar] [CrossRef]
- Gomes, M.V.T.; de Souza, R.R.; Teles, V.S.; Araújo Mendes, É. Phytoremediation of Water Contaminated with Mercury Using Typha domingensis in Constructed Wetland. Chemosphere 2014, 103, 228–233. [Google Scholar] [CrossRef]
- Maine, M.A.; Hadad, H.R.; Camaño Silvestrini, N.E.; Nocetti, E.; Sanchez, G.C.; Campagnoli, M.A. Cr, Ni, and Zn Removal from Landfill Leachate Using Vertical Flow Wetlands Planted with Typha domingensis and Canna indica. Int. J. Phytoremed. 2022, 24, 66–75. [Google Scholar] [CrossRef]
- Sricoth, T.; Meeinkuirt, W.; Pichtel, J.; Taeprayoon, P.; Saengwilai, P. Synergistic Phytoremediation of Wastewater by Two Aquatic Plants (Typha angustifolia and Eichhornia crassipes) and Potential as Biomass Fuel. Environ. Sci. Pollut. Res. 2018, 25, 5344–5358. [Google Scholar] [CrossRef]
- Panich-Pat, T.; Upatham, S.; Pokethitiyook, P.; Kruatrachue, M.; Lanza, G.R. Phytoextraction of Metal Contaminants by Typha angustifolia: Interaction of Lead and Cadmium in Soil-Water Microcosms. J. Environ. Prot. 2010, 1, 431–437. [Google Scholar] [CrossRef]
- Chandra, R.; Yadav, S. Phytoremediation of Cd, Cr, Cu, Mn, Fe, Ni, Pb and Zn from Aqueous Solution Using Phragmites cummunis, Typha angustifolia and Cyperus Esculentus. Int. J. Phytoremed. 2011, 13, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Demırezen, D.; Aksoy, A. Accumulation of Heavy Metals in Typha angustifolia (L.) and Potamogeton pectinatus (L.) Living in Sultan Marsh (Kayseri, Turkey). Chemosphere 2004, 56, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Panich-Pat, T.; Pokethitiyook, P.; Kruatrachue, M.; Upatham, E.S.; Srinives, P.; Lanza, G.R. Removal of Lead from Contaminated Soils by Typha angustifolia. Water Air Soil Pollut. 2004, 155, 159–171. [Google Scholar] [CrossRef]
- Vidayanti, V.; Choesin, D.N.; Iriawati, I. Phytoremediation of Chromium: Distribution and Speciation of Chromium in Typha angustifolia. Int. J. Plant Biol. 2017, 8, 6870. [Google Scholar] [CrossRef]
- Zaranyika, M.F.; Nyati, W. Uptake of Heavy Metals by Typha capensis from Wetland Sites Polluted by Effluent from Mineral Processing Plants: Implications of Metal–Metal Interactions. 3 Biotech 2017, 7, 286. [Google Scholar] [CrossRef]
- Gao, T.; Shi, X.-Y. Taxonomic Structure and Function of Seed-Inhabiting Bacterial Microbiota from Common Reed (Phragmites australis) and Narrowleaf Cattail (Typha angustifolia L.). Arch. Microbiol. 2018, 200, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Zhu, J.N.; Liu, Q.F.; Liu, Y.; Liu, M.; Liu, L.; Zhang, Q. Comparison of the Diversity of Root-Associated Bacteria in Phragmites australis and Typha angustifolia L. in Artificial Wetlands. World J. Microbiol. Biotechnol. 2013, 29, 1499–1508. [Google Scholar] [CrossRef]
- Arroyo, P.; de Miera, L.E.S.; Ansola, G. Influence of Environmental Variables on the Structure and Composition of Soil Bacterial Communities in Natural and Constructed Wetlands. Sci. Total Environ. 2015, 506–507, 380–390. [Google Scholar] [CrossRef]
- Kennedy, A.C.; de Luna, L.Z. Rhizosphere. In Encyclopedia of Soils in the Environment; Hillel, D., Ed.; Elsevier: Oxford, UK, 2005; pp. 399–406. ISBN 978-0-12-348530-4. [Google Scholar]
- Jha, P.N.; Kumar, A. Endophytic Colonization of Typha australis by a Plant Growth-promoting Bacterium Klebsiella oxytoca Strain GR-3. J. Appl. Microbiol. 2007, 103, 1311–1320. [Google Scholar] [CrossRef]
- Ashkan, M.F.; Bleakley, B. Isolation, Characterization and Identification of Putative Bacterial Endophytes from Some Plants in Hot Springs, South Dakota. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 756–767. [Google Scholar] [CrossRef]
- Lagos, L.; Maruyama, F.; Nannipieri, P.; Mora, M.L.; Ogram, A.; Jorquera, M.A. Current Overview on the Study of Bacteria in the Rhizosphere by Modern Molecular Techniques: A Mini-review. J. Soil Sci. Plant Nutr. 2015, 15, 504–523. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going Back to the Roots: The Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, L.; Liu, Q.; Vestergård, M.; Topalovic, O.; Wang, Q.; Zhou, Q.; Huang, L.; Yang, X.; Feng, Y. The Plant-Growth Promoting Bacteria Promote Cadmium Uptake by Inducing a Hormonal Crosstalk and Lateral Root Formation in a Hyperaccumulator Plant Sedum alfredii. J. Hazard. Mater. 2020, 395, 122661. [Google Scholar] [CrossRef] [PubMed]
- Shehzadi, M.; Fatima, K.; Imran, A.; Mirza, M.S.; Khan, Q.M.; Afzal, M. Ecology of Bacterial Endophytes Associated with Wetland Plants Growing in Textile Effluent for Pollutant-Degradation and Plant Growth-Promotion Potentials. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2016, 150, 1261–1270. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, J.; Wen, C. Community Composition and Abundance of Anammox Bacteria in Cattail Rhizosphere Sediments at Three Phenological Stages. Curr. Microbiol. 2017, 74, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Chellaiah, E.R. Cadmium (Heavy Metals) Bioremediation by Pseudomonas Aeruginosa: A Minireview. Appl. Water Sci. 2018, 8, 154. [Google Scholar] [CrossRef]
- Fakhar, A.; Gul, B.; Gurmani, A.R.; Khan, S.M.; Ali, S.; Sultan, T.; Chaudhary, H.J.; Rafique, M.; Rizwan, M. Heavy Metal Remediation and Resistance Mechanism of Aeromonas, Bacillus, and Pseudomonas: A Review. Crit. Rev. Environ. Sci. Technol. 2022, 52, 1868–1914. [Google Scholar] [CrossRef]
- Renu, S.; Sarim, K.M.; Singh, D.P.; Sahu, U.; Bhoyar, M.S.; Sahu, A.; Kaur, B.; Gupta, A.; Mandal, A.; Thakur, J.K.; et al. Deciphering Cadmium (Cd) Tolerance in Newly Isolated Bacterial Strain, Ochrobactrum intermedium BB12, and Its Role in Alleviation of Cd Stress in Spinach Plant (Spinacia oleracea L.). Front. Microbiol. 2022, 12, 758144. [Google Scholar] [CrossRef]
- Pandey, S.; Ghosh, P.K.; Ghosh, S.; De, T.K.; Maiti, T.K. Role of Heavy Metal Resistant Ochrobactrum sp. and Bacillus spp. Strains in Bioremediation of a Rice Cultivar and Their PGPR like Activities. J. Microbiol. 2013, 51, 11–17. [Google Scholar] [CrossRef]
- Faisal, M.; Hasnain, S. Beneficial Role of Hydrophytes in Removing Cr(VI) from Wastewater in Association with Chromate-Reducing Bacterial Strains Ochrobactrum intermedium and Brevibacterium. Int. J. Phytoremed. 2005, 7, 271–277. [Google Scholar] [CrossRef]
- Kavita, B.; Keharia, H. Reduction of Hexavalent Chromium by Ochrobactrum intermedium BCR400 Isolated from a Chromium-Contaminated Soil. 3 Biotech 2012, 2, 79. [Google Scholar] [CrossRef]
- Ozdemir, G.; Ozturk, T.; Ceyhan, N.; Isler, R.; Cosar, T. Heavy Metal Biosorption by Biomass of Ochrobactrum anthropi Producing Exopolysaccharide in Activated Sludge. Bioresour. Technol. 2003, 90, 71–74. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Gupta, A. Evaluation of Acinetobacter sp. B9 for Cr (VI) Resistance and Detoxification with Potential Application in Bioremediation of Heavy-Metals-Rich Industrial Wastewater. Environ. Sci. Pollut. Res. 2013, 20, 6628–6637. [Google Scholar] [CrossRef]
- Pang, B.; Lv, L.; Pang, C.; Ye, F.; Shang, C. Optimization of Growth Conditions of Acinetobacter sp. Cr1 for Removal of Heavy Metal Cr Using Central Composite Design. Curr. Microbiol. 2021, 78, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, Z.A.; Zakaria, Z.; Surif, S.; Ahmad, W.A. Hexavalent Chromium Reduction by Acinetobacter haemolyticus Isolated from Heavy-Metal Contaminated Wastewater. J. Hazard. Mater. 2007, 146, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Ndeddy Aka, R.J.; Babalola, O.O. Effect of Bacterial Inoculation of Strains of Pseudomonas aeruginosa, Alcaligenes feacalis and Bacillus subtilis on Germination, Growth and Heavy Metal (Cd, Cr, and Ni) Uptake of Brassica juncea. Int. J. Phytoremed. 2016, 18, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Sodhi, K.K.; Kumar, M.; Singh, D.K. Multi-Metal Resistance and Potential of Alcaligenes sp. MMA for the Removal of Heavy Metals. SN Appl. Sci. 2020, 2, 1885. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Q.; Wang, B.; Hou, J.; Luo, Y.; Tang, C.; Franks, A.E. Plant Growth-Promoting Rhizobacteria Enhance the Growth and Cd Uptake of Sedum plumbizincicola in a Cd-Contaminated Soil. J. Soils Sediments 2015, 15, 1191–1199. [Google Scholar] [CrossRef]
- Salam, L.B.; Shomope, H.; Ummi, Z.; Bukar, F. Mercury Contamination Imposes Structural Shift on the Microbial Community of an Agricultural Soil. Bull. Natl. Res. Cent. 2019, 43, 163. [Google Scholar] [CrossRef]
- Li, D.; Chen, J.; Zhang, X.; Shi, W.; Li, J. Structural and Functional Characteristics of Soil Microbial Communities in Response to Different Ecological Risk Levels of Heavy Metals. Front. Microbiol. 2022, 13, 1072389. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, Q.; Fan, S.; Zhang, Y.; Zhang, M.; Zhang, J. Distinction between Cr and Other Heavy-Metal-Resistant Bacteria Involved in C/N Cycling in Contaminated Soils of Copper Producing Sites. J. Hazard. Mater. 2021, 402, 123454. [Google Scholar] [CrossRef]
- Rolón-Cárdenas, G.A.; Martínez-Martínez, J.G.; Arvizu-Gómez, J.L.; Soria-Guerra, R.E.; Alfaro-De la Torre, M.C.; Alatorre-Cobos, F.; Rubio-Santiago, J.; González-Balderas, R.D.M.; Carranza-Álvarez, C.; Macías-Pérez, J.R.; et al. Enhanced Cd-Accumulation in Typha latifolia by Interaction with Pseudomonas rhodesiae GRC140 under Axenic Hydroponic Conditions. Plants 2022, 11, 1447. [Google Scholar] [CrossRef]
- Rubio-Santiago, J.; Hernández-Morales, A.; Rolón-Cárdenas, G.A.; Arvizu-Gómez, J.L.; Soria-Guerra, R.E.; Carranza-Álvarez, C.; Rubio-Salazar, J.E.; Rosales-Loredo, S.; Pacheco-Aguilar, J.R.; Macías-Pérez, J.R.; et al. Characterization of Endophytic Bacteria Isolated from Typha latifolia and Their Effect in Plants Exposed to Either Pb or Cd. Plants 2023, 12, 498. [Google Scholar] [CrossRef]
- Barillot, C.D.C.; Sarde, C.-O.; Bert, V.; Tarnaud, E.; Cochet, N. A Standardized Method for the Sampling of Rhizosphere and Rhizoplan Soil Bacteria Associated to a Herbaceous Root System. Ann. Microbiol. 2013, 63, 471–476. [Google Scholar] [CrossRef]
- Singh, A.K.; Varaprasad, K.S. Criteria for Identification and Assessment of Agro-Biodiversity Heritage Sites: Evolving Sustainable Agriculture. Curr. Sci. 2008, 94, 1131–1138. [Google Scholar]
- Khanna, K.; Ohri, P.; Bhardwaj, R.; Ahmad, P. Unsnarling Plausible Role of Plant Growth-Promoting Rhizobacteria for Mitigating Cd-Toxicity from Plants: An Environmental Safety Aspect. J. Plant Growth Regul. 2021, 41, 2514–2542. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- He, W.; Megharaj, M.; Wu, C.-Y.; Subashchandrabose, S.R.; Dai, C.-C. Endophyte-Assisted Phytoremediation: Mechanisms and Current Application Strategies for Soil Mixed Pollutants. Crit. Rev. Biotechnol. 2020, 40, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Patten, C.L.; Blakney, A.J.C.; Coulson, T.J.D. Activity, Distribution and Function of Indole-3-Acetic Acid Biosynthetic Pathways in Bacteria. Crit. Rev. Microbiol. 2013, 39, 395–415. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Hasnain, S. Auxins as One of the Factors of Plant Growth Improvement by Plant Growth Promoting Rhizobacteria. Pol. J. Microbiol. 2014, 63, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Luo, S.; Shen, J.; Wang, Q.; Ye, J.; Meng, Q.; Wu, Y.; Chen, B.; Cao, X.; Yang, X.; et al. The Effects of Endophytic Bacterium SaMR12 on Sedum alfredii Hance Metal Ion Uptake and the Expression of Three Transporter Family Genes after Cadmium Exposure. Environ. Sci. Pollut. Res. Int. 2017, 24, 9350–9360. [Google Scholar] [CrossRef]
- Chen, B.; Luo, S.; Wu, Y.; Ye, J.; Wang, Q.; Xu, X.; Pan, F.; Khan, K.Y.; Feng, Y.; Yang, X. The Effects of the Endophytic Bacterium Pseudomonas fluorescens Sasm05 and IAA on the Plant Growth and Cadmium Uptake of Sedum alfredii Hance. Front. Microbiol. 2017, 8, 2538. [Google Scholar] [CrossRef]
- Wu, K.; Luo, J.; Li, J.; An, Q.; Yang, X.; Liang, Y.; Li, T. Endophytic Bacterium buttiauxella sp. SaSR13 Improves Plant Growth and Cadmium Accumulation of Hyperaccumulator Sedum alfredii. Environ. Sci. Pollut. Res. 2018, 25, 21844–21854. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Javed, M.T.; Masood, S.; Akram, M.S.; Azeem, M.; Ali, Q.; Gilani, R.; Basit, F.; Abid, A.; Lindberg, S. Serratia sp. CP-13 Augments the Growth of Cadmium (Cd)-Stressed Linum usitatissimum L. by Limited Cd Uptake, Enhanced Nutrient Acquisition and Antioxidative Potential. J. Appl. Microbiol. 2019, 126, 1708–1721. [Google Scholar] [CrossRef] [PubMed]
- Tamariz-Angeles, C.; Huamán, G.D.; Palacios-Robles, E.; Olivera-Gonzales, P.; Castañeda-Barreto, A. Characterization of Siderophore-Producing Microorganisms Associated to Plants from High-Andean Heavy Metal Polluted Soil from Callejón de Huaylas (Ancash, Perú). Microbiol. Res. 2021, 250, 126811. [Google Scholar] [CrossRef] [PubMed]
- Asad, S.A.; Farooq, M.; Afzal, A.; West, H. Integrated Phytobial Heavy Metal Remediation Strategies for a Sustainable Clean Environment—A Review. Chemosphere 2019, 217, 925–941. [Google Scholar] [CrossRef]
- Rathaur, P.; Ramteke, P.W.; Raja, W.; John, S.A. Isolation and Characterization of Nickel and Cadmium Tolerant Plant Growth Promoting Rhizobacteria from Rhizosphere of Withania somnifera. J. Biol. Environ. Sci. 2012, 6, 253–261. [Google Scholar]
- Xinxian, L.; Xuemei, C.; Yagang, C.; Woon-Chung, W.J.; Zebin, W.; Qitang, W. Isolation and Characterization Endophytic Bacteria from Hyperaccumulator Sedum alfredii Hance and Their Potential to Promote Phytoextraction of Zinc Polluted Soil. World J. Microbiol. Biotechnol. 2011, 27, 1197–1207. [Google Scholar] [CrossRef]
- Sinha, S.; Mukherjee, S.K. Cadmium–Induced Siderophore Production by a High Cd-Resistant Bacterial Strain Relieved Cd Toxicity in Plants Through Root Colonization. Curr. Microbiol. 2008, 56, 55–60. [Google Scholar] [CrossRef]
- Dimkpa, C.O.; Svatoš, A.; Dabrowska, P.; Schmidt, A.; Boland, W.; Kothe, E. Involvement of Siderophores in the Reduction of Metal-Induced Inhibition of Auxin Synthesis in Streptomyces spp. Chemosphere 2008, 74, 19–25. [Google Scholar] [CrossRef]
- Nair, A.; Juwarkar, A.A.; Singh, S.K. Production and Characterization of Siderophores and Its Application in Arsenic Removal from Contaminated Soil. Water Air Soil Pollut. 2007, 180, 199–212. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Freitas, H. Endophytic Bacteria and Their Potential to Enhance Heavy Metal Phytoextraction. Chemosphere 2009, 77, 153–160. [Google Scholar] [CrossRef]
- Perea-Vélez, Y.S.; Carrillo-González, R.; González-Chávez, M.C.A. Fitorremediación Asistida Por Microorganismos: Enfásis En Bacterias Promotoras Del Crecimiento De Plantas. Agro Product. 2017, 10, 34–40. [Google Scholar]
- Saha, M.; Sarkar, S.; Sarkar, B.; Sharma, B.K.; Bhattacharjee, S.; Tribedi, P. Microbial Siderophores and Their Potential Applications: A Review. Environ. Sci. Pollut. Res. 2016, 23, 3984–3999. [Google Scholar] [CrossRef]
- Paredes-Mendoza, M.; Espinosa-Victoria, D. Ácidos orgánicos producidos por rizobacterias que solubilizan fosfato: Una revisión crítica. Terra Latinoam. 2010, 28, 61–70. [Google Scholar]
- Ahemad, M. Phosphate-Solubilizing Bacteria-Assisted Phytoremediation of Metalliferous Soils: A Review. 3 Biotech 2015, 5, 111–121. [Google Scholar] [CrossRef]
- Teng, Z.; Shao, W.; Zhang, K.; Huo, Y.; Li, M. Characterization of Phosphate Solubilizing Bacteria Isolated from Heavy Metal Contaminated Soils and Their Potential for Lead Immobilization. J. Environ. Manag. 2019, 231, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Camelo, M.; Vera, S.P.; Bonilla, R.R. Mecanismos de acción de las rizobacterias promotoras del crecimiento vegetal. Cienc. Tecnol. Agropecu. 2011, 12, 159–166. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Plant and Endophyte Relationships. In Comprehensive Biotechnology; Elsevier: Amsterdam, The Netherlands, 2011; pp. 713–727. ISBN 978-0-08-088504-9. [Google Scholar]
- Glick, B.R. Modulation of Plant Ethylene Levels by the Bacterial Enzyme ACC Deaminase. FEMS Microbiol. Lett. 2005, 251, 1–7. [Google Scholar] [CrossRef]
- Saleem, M.; Arshad, M.; Hussain, S.; Bhatti, A.S. Perspective of Plant Growth Promoting Rhizobacteria (PGPR) Containing ACC Deaminase in Stress Agriculture. J. Ind. Microbiol. Biotechnol. 2007, 34, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Grichko, V.P.; Filby, B.; Glick, B.R. Increased Ability of Transgenic Plants Expressing the Bacterial Enzyme ACC Deaminase to Accumulate Cd, Co, Cu, Ni, Pb, and Zn. J. Biotechnol. 2000, 81, 45–53. [Google Scholar] [CrossRef]
- Chandwani, S.; Amaresan, N. Role of ACC Deaminase Producing Bacteria for Abiotic Stress Management and Sustainable Agriculture Production. Environ. Sci. Pollut. Res. Int. 2022, 29, 22843–22859. [Google Scholar] [CrossRef]
- Xu, S.; Xing, Y.; Liu, S.; Huang, Q.; Chen, W. Role of Novel Bacterial Raoultella sp. Strain X13 in Plant Growth Promotion and Cadmium Bioremediation in Soil. Appl. Microbiol. Biotechnol. 2019, 103, 3887–3897. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yan, Z.; Gu, D.; Li, D.; Tao, Y.; Zhang, D.; Su, L.; Ao, Y. Characterization of Cadmium-Resistant Rhizobacteria and Their Promotion Effects on Brassica napus Growth and Cadmium Uptake. J. Basic Microbiol. 2019, 59, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Amna; Kamran, M.A.; Javed, M.T.; Hayat, K.; Farooq, M.A.; Ali, N.; Ali, M.; Manghwar, H.; Jan, F.; et al. Individual and Combinatorial Application of Kocuria rhizophila and Citric Acid on Phytoextraction of Multi-Metal Contaminated Soils by Glycine max L. Environ. Exp. Bot. 2019, 159, 23–33. [Google Scholar] [CrossRef]
- Kotoky, R.; Nath, S.; Kumar Maheshwari, D.; Pandey, P. Cadmium Resistant Plant Growth Promoting Rhizobacteria Serratia marcescens S2I7 Associated with the Growth Promotion of Rice Plant. Environ. Sustain. 2019, 2, 135–144. [Google Scholar] [CrossRef]
- Pan, F.; Meng, Q.; Wang, Q.; Luo, S.; Chen, B.; Khan, K.Y.; Yang, X.; Feng, Y. Endophytic Bacterium Sphingomonas SaMR12 Promotes Cadmium Accumulation by Increasing Glutathione Biosynthesis in Sedum alfredii Hance. Chemosphere 2016, 154, 358–366. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, L.; Zhu, Z.; Yang, X.; Wang, Y.; An, Q. Colonization and Modulation of Host Growth and Metal Uptake by Endophytic Bacteria of Sedum alfredii. Int. J. Phytoremed. 2013, 15, 51–64. [Google Scholar] [CrossRef]
- Chen, S.; Han, X.; Fang, J.; Lu, Z.; Qiu, W.; Liu, M.; Sang, J.; Jiang, J.; Zhuo, R. Sedum alfredii SaNramp6 Metal Transporter Contributes to Cadmium Accumulation in Transgenic Arabidopsis thaliana. Sci. Rep. 2017, 7, 13318. [Google Scholar] [CrossRef]
- Begum, N.; Afzal, S.; Zhao, H.; Lou, L.; Cai, Q. Shoot Endophytic Plant Growth-Promoting Bacteria Reduce Cadmium Toxicity and Enhance Switchgrass (Panicum virgatum L.) Biomass. Acta Physiol. Plant 2018, 40, 170. [Google Scholar] [CrossRef]
- Mitra, S.; Pramanik, K.; Sarkar, A.; Ghosh, P.K.; Soren, T.; Maiti, T.K. Bioaccumulation of Cadmium by Enterobacter sp. and Enhancement of Rice Seedling Growth under Cadmium Stress. Ecotoxicol. Environ. Saf. 2018, 156, 183–196. [Google Scholar] [CrossRef]
- Muratova, A.; Lyubun, Y.; German, K.; Turkovskaya, O. Effect of Cadmium Stress and Inoculation with a Heavy-Metal-Resistant Bacterium on the Growth and Enzyme Activity of Sorghum bicolor. Environ. Sci. Pollut. Res. 2015, 22, 16098–16109. [Google Scholar] [CrossRef] [PubMed]
- Rolón-Cárdenas, G. Effect of Pseudomonas rhodesiae GRC140 on Cucumis sativus L. Seedlings with and without Cadmium. J. Nat. Resour. Life Sci. Educ. 2020, 7, 14–20. [Google Scholar] [CrossRef]
- Ghosh, A.; Pramanik, K.; Bhattacharya, S.; Mondal, S.; Ghosh, S.K.; Maiti, T.K. A Potent Cadmium Bioaccumulating Enterobacter Cloacae Strain Displays Phytobeneficial Property in Cd-Exposed Rice Seedlings. Curr. Res. Microb. Sci. 2021, 3, 100101. [Google Scholar] [CrossRef]
- Ajmal, A.W.; Yasmin, H.; Hassan, M.N.; Khan, N.; Jan, B.L.; Mumtaz, S. Heavy Metal–Resistant Plant Growth–Promoting Citrobacter werkmanii Strain WWN1 and Enterobacter cloacae Strain JWM6 Enhance Wheat (Triticum aestivum L.) Growth by Modulating Physiological Attributes and Some Key Antioxidants Under Multi-Metal Stress. Front. Microbiol. 2022, 13, 815704. [Google Scholar] [CrossRef]
- Ajmal, A.W.; Saroosh, S.; Mulk, S.; Hassan, M.N.; Yasmin, H.; Jabeen, Z.; Nosheen, A.; Shah, S.M.U.; Naz, R.; Hasnain, Z.; et al. Bacteria Isolated from Wastewater Irrigated Agricultural Soils Adapt to Heavy Metal Toxicity While Maintaining Their Plant Growth Promoting Traits. Sustainability 2021, 13, 7792. [Google Scholar] [CrossRef]
- Tirry, N.; Kouchou, A.; El Omari, B.; Ferioun, M.; El Ghachtouli, N. Improved Chromium Tolerance of Medicago sativa by Plant Growth-Promoting Rhizobacteria (PGPR). J. Genet. Eng. Biotechnol. 2021, 19, 149. [Google Scholar] [CrossRef]
- Gupta, P.; Rani, R.; Chandra, A.; Kumar, V. Potential Applications of Pseudomonas sp. (Strain CPSB21) to Ameliorate Cr6+ Stress and Phytoremediation of Tannery Effluent Contaminated Agricultural Soils. Sci. Rep. 2018, 8, 4860. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Ilyas, N.; Yasmin, H.; Sayyed, R.Z.; Hasnain, Z.; Elsayed, E.A.; El Enshasy, H.A. Role of Bacillus cereus in Improving the Growth and Phytoextractability of Brassica nigra (L.) K. Koch in Chromium Contaminated Soil. Molecules 2021, 26, 1569. [Google Scholar] [CrossRef]
- Khan, A.A.H. Endophytic Fungi and Their Impact on Agroecosystems. In Medicinal Plants: Biodiversity, Sustainable Utilization and Conservation; Khasim, S.M., Long, C., Thammasiri, K., Lutken, H., Eds.; Springer: Singapore, 2020; pp. 443–499. ISBN 9789811516368. [Google Scholar]
- Yadav, A.; Goyal, D.; Prasad, M.; Singh, T.B.; Shrivastav, P.; Ali, A.; Dantu, P.K. Bioremediation of Toxic Pollutants: Features, Strategies, and Applications. In Contaminants in Agriculture: Sources, Impacts and Management; Naeem, M., Ansari, A.A., Gill, S.S., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 361–383. ISBN 978-3-030-41552-5. [Google Scholar]
- Guan, M.; Pan, X.-C.; Wang, S.; Wei, X.-L.; Zhang, C.-B.; Wang, J.; Liu, W.-L.; Liu, S.-Y.; Chang, J. Comparison of Fungal Communities among Ten Macrophyte Rhizospheres. Fungal Biol. 2018, 122, 867–874. [Google Scholar] [CrossRef]
- Cheng, S. Effects of Heavy Metals on Plants and Resistance Mechanisms. Environ. Sci. Pollut. Res. 2003, 10, 256–264. [Google Scholar] [CrossRef]
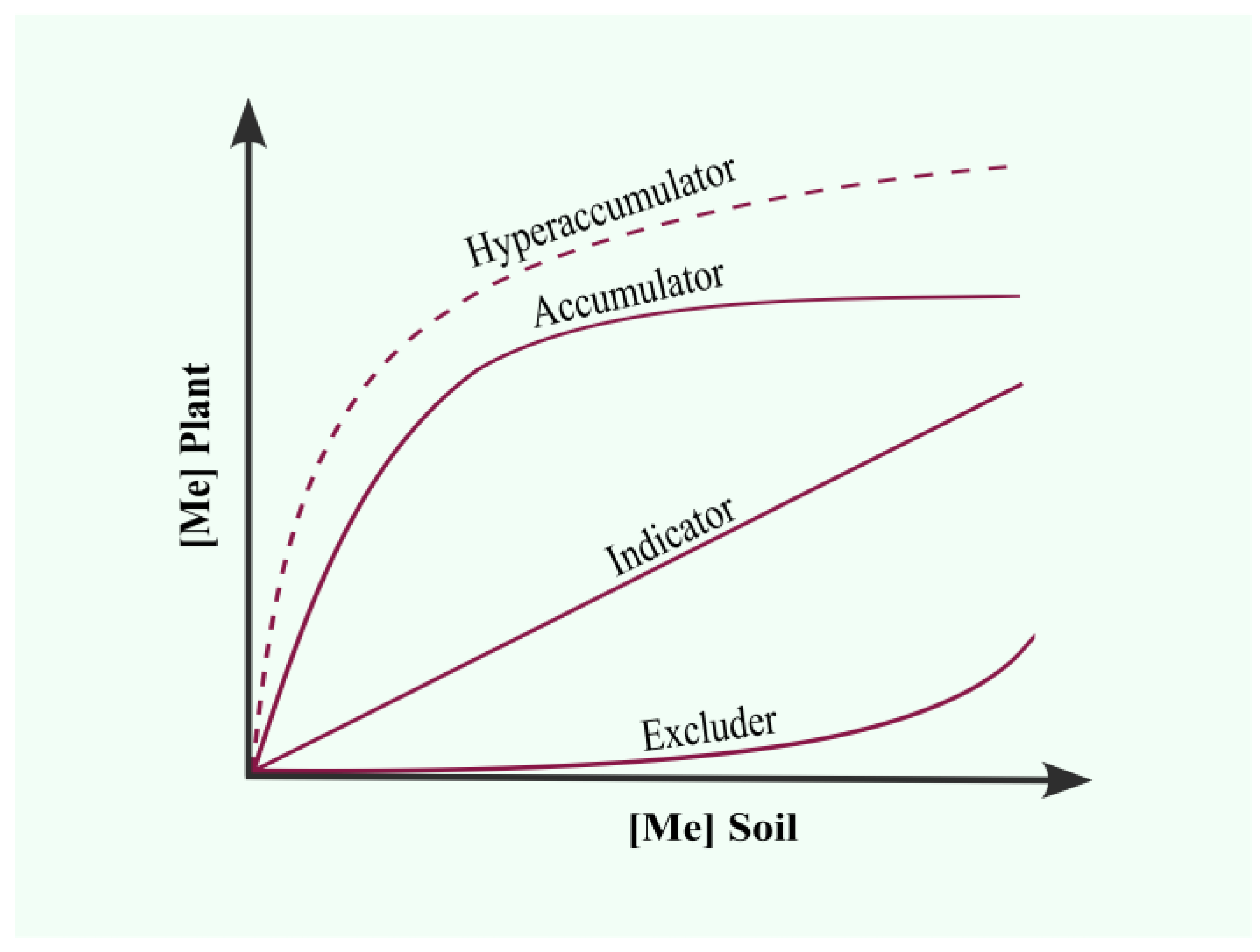
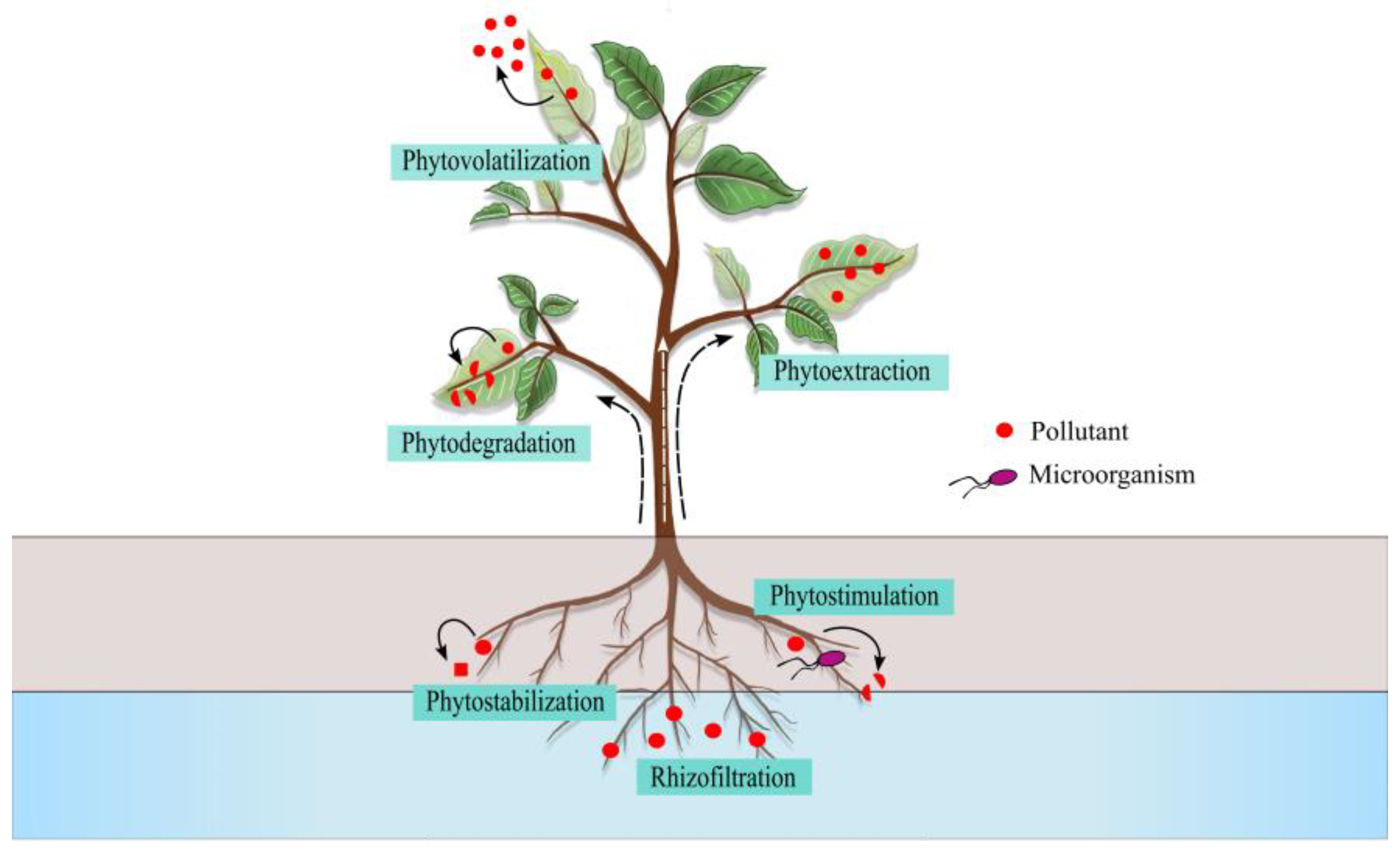



| Process | Mechanism | Contaminant |
|---|---|---|
| Phytoextraction | Hyperaccumulation | HMs, organic compounds, and radioisotopes |
| Phytovolatilization | Leaf volatilization | Organic compounds and Hg, As, and Se. |
| Phytostabilization | Precipitation, formation of insoluble complexes, valence reduction, and adsorption | HMs |
| Rhizofiltration | Accumulation in the rhizosphere | HMs and organic compounds |
| Phytodegradation | Enzymatic degradation | Organic pollutants |
| Phytostimulation | Microbial growth by stimulation | Organic pollutants |
| Specie | Phylum | Metal | Site | References |
|---|---|---|---|---|
| Typha sp. | Proteobacteria | Cr | Wetland | [69] |
| T. domingensis | Proteobacteria Firmicutes Actinobacteria Bacteroidetes | Cr, Ni, Fe | Pond and stream | [106] |
| T. angustifolia | Firmicutes Proteobacteria Actinobacteria | Fe | Wetland | [64] |
| T. orientalis | Planctomycetes Uncultured bacterium | Cu, Zn, Pb | Lake | [107] |
| T. latifolia | Proteobacteria | Cd | Contaminated site | [53] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Martínez, J.G.; Rosales-Loredo, S.; Hernández-Morales, A.; Arvizu-Gómez, J.L.; Carranza-Álvarez, C.; Macías-Pérez, J.R.; Rolón-Cárdenas, G.A.; Pacheco-Aguilar, J.R. Bacterial Communities Associated with the Roots of Typha spp. and Its Relationship in Phytoremediation Processes. Microorganisms 2023, 11, 1587. https://doi.org/10.3390/microorganisms11061587
Martínez-Martínez JG, Rosales-Loredo S, Hernández-Morales A, Arvizu-Gómez JL, Carranza-Álvarez C, Macías-Pérez JR, Rolón-Cárdenas GA, Pacheco-Aguilar JR. Bacterial Communities Associated with the Roots of Typha spp. and Its Relationship in Phytoremediation Processes. Microorganisms. 2023; 11(6):1587. https://doi.org/10.3390/microorganisms11061587
Chicago/Turabian StyleMartínez-Martínez, Joana Guadalupe, Stephanie Rosales-Loredo, Alejandro Hernández-Morales, Jackeline Lizzeta Arvizu-Gómez, Candy Carranza-Álvarez, José Roberto Macías-Pérez, Gisela Adelina Rolón-Cárdenas, and Juan Ramiro Pacheco-Aguilar. 2023. "Bacterial Communities Associated with the Roots of Typha spp. and Its Relationship in Phytoremediation Processes" Microorganisms 11, no. 6: 1587. https://doi.org/10.3390/microorganisms11061587
APA StyleMartínez-Martínez, J. G., Rosales-Loredo, S., Hernández-Morales, A., Arvizu-Gómez, J. L., Carranza-Álvarez, C., Macías-Pérez, J. R., Rolón-Cárdenas, G. A., & Pacheco-Aguilar, J. R. (2023). Bacterial Communities Associated with the Roots of Typha spp. and Its Relationship in Phytoremediation Processes. Microorganisms, 11(6), 1587. https://doi.org/10.3390/microorganisms11061587






