Microbial Growth under Limiting Conditions-Future Perspectives
Abstract
:1. Introduction
2. Microbial Growth
3. Growth at Optimum (Maximum) Rate
4. Growth at Slow (Limited) Rate
5. Growth at Near-Zero Rate (Severe Growth Limitation)
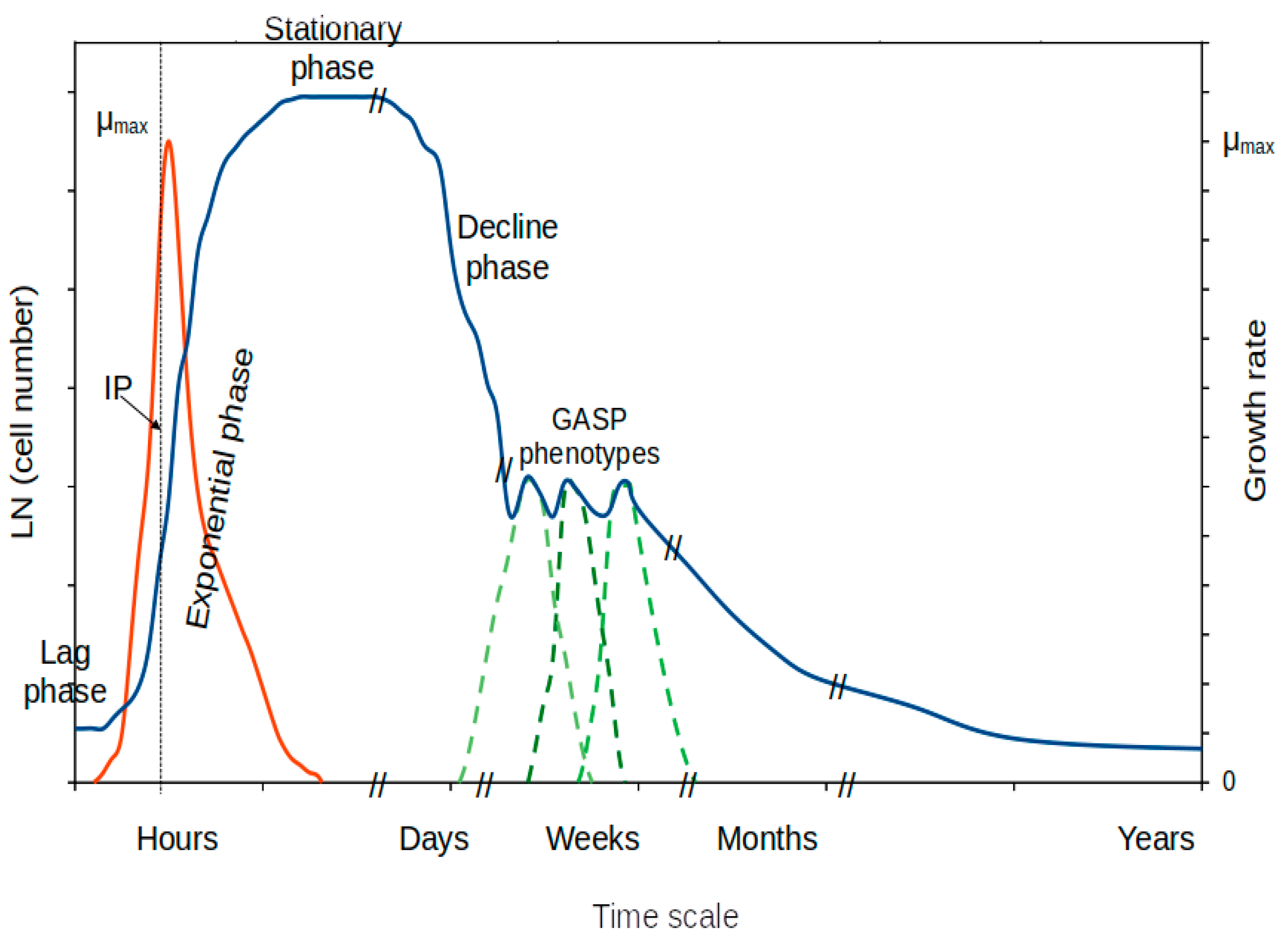
6. Non-Growing Stages
6.1. Stationary Phase of Growth
6.2. Decline (Decay) Phase of the Growth Curve
6.3. Late Stages of the Stationary Phase of Growth
6.4. Microbial Decay and Survival
6.5. Maintenance Metabolism
7. Physiological Stages and Methods
8. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uroz, S.; Calvaruso, C.; Turpault, M.-P.; Frey-Klett, P. Mineral weathering by bacteria: Ecology, actors and mechanisms. Trends Microbiol. 2009, 17, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Conant, R.T.; Ryan, M.G.; Ågren, G.I.; Birge, H.E.; Davidson, E.A.; Eliasson, P.E.; Evans, S.E.; Frey, S.D.; Giardina, C.P.; Hopkins, F.; et al. Temperature and soil organic matter decomposition rates—Synthesis of current knowledge and a way forward. Glob. Change Biol. 2011, 17, 3392–3404. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Zhao, Y.; Gillings, M.; Penuelas, J.; Ok, Y.S.; Capon, A.; Banwart, S. Soil biota, antimicrobial resistance and planetary health. Environ. Int. 2019, 131, 105059. [Google Scholar] [CrossRef]
- Santana, M.M.; Gonzalez, J.M.; Cruz, C. Nitric oxide accumulation: The evolutionary trigger for phytopathogenesis. Front. Microbiol. 2017, 8, 1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowther, T.W.; van den Hoogen, J.; Wan, J.; Mayes, M.A.; Keiser, A.D.; Mo, L.; Averill, C.; Maynard, D.S. The global soil community and its influence on biogeochemistry. Science 2019, 365, eaav0550. [Google Scholar] [CrossRef]
- Davidson, E.A.; Janssens, I.A. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 2006, 440, 165–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.; Fang, C.M.; Dawson, J.J.C.; Moncrieff, J.B. Impact of global warming on soil organic carbon. Adv. Agron. 2008, 97, 1–43. [Google Scholar] [CrossRef]
- Lladó, S.; López-Mondéjar, R.; Baldrian, P. Forest Soil Bacteria: Diversity, Involvement in Ecosystem Processes, and Response to Global Change. Microbiol. Mol. Biol. Rev. 2017, 81, e00063–16. [Google Scholar] [CrossRef] [Green Version]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientist’s warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef] [Green Version]
- Cernava, T.; Berg, G. The emergence of disease-preventing bacteria within the plant microbiota. Environ. Microbiol. 2022, 8, 3259–3263. [Google Scholar] [CrossRef] [PubMed]
- Wabrick, I.; Heke, D.; Breed, M. Indigenous Knowledge and the Microbiome—Bridging the Disconnect between Colonized Places, Peoples, and the Unseen Influences That Shape Our Health and Well-Being. Msystems 2023, 8, e008722. [Google Scholar] [CrossRef]
- Antwis, R.E.; Griffiths, S.M.; Harrison, X.A.; Aranega-Bou, P.; Arce, A.; Bettridge, A.S.; Brailsford, F.L.; de Menezes, A.; Devaynes, A.; Forbes, K.M.; et al. Fifty important research questions in microbial ecology. FEMS Microbiol. Ecol. 2017, 93, fix044. [Google Scholar] [CrossRef]
- Radjabzadeh, D.; Bosch, J.A.; Uitterlinden, A.G.; Zwinderman, A.H.; Ikram, M.A.; van Meurs, J.B.J.; Luik, A.l.; Nieuwdorp, M.; Lok, A.; van Duijn, C.M.; et al. Gut microbiome-wide association study of depressive symptoms. Nat. Commun. 2022, 13, 7128. [Google Scholar] [CrossRef]
- Santana, M.M.; González, J.M. High temperature microbial activity in upper soil layers. FEMS Microbiol. Lett. 2015, 362, fnv182. [Google Scholar] [CrossRef]
- Gibbons, S.M.; Gilbert, J.A. Microbial diversity—Exploration of natural ecosystems and microbiomes. Curr. Opin. Genet. Dev. 2015, 35, 66–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtis, T.P.; Sloan, W.T.; Scannell, J.W. Estimating prokaryotic diversity and its limits. Proc Natl. Acad. Sci. USA 2002, 99, 10494–10499. [Google Scholar] [CrossRef] [Green Version]
- Tecon, R.; Or, D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 2017, 41, 599–623. [Google Scholar] [CrossRef] [Green Version]
- Grundmann, G.L. Spatial scales of soil bacterial diversity- the size of a clone. FEMS Microiol. Ecol. 2004, 48, 119–127. [Google Scholar] [CrossRef]
- Dubey, S.K.; Tripathi, A.K.; Upadhyay, S.N. Exploration of soil bacterial communities for their potential as bioresource. Bioresour. Technol. 2006, 97, 2217–2224. [Google Scholar] [CrossRef] [PubMed]
- Pedrós-Alió, C. Marine microbial diversity: Can it be determined? Trends Microbiol. 2006, 14, 257–263. [Google Scholar] [CrossRef] [PubMed]
- González, J.M.; Portillo, M.C.; Belda-Ferre, P.; Mira, A. Amplification by PCR artificially reduces the proportion of the rare biosphere in microbial communities. PLoS ONE 2012, 7, e29973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roesch, L.F.; Fulthorpe, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.; Kent, A.D.; Daroub, S.H.; Camargo, F.A.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 4, 283–290. [Google Scholar] [CrossRef]
- Lempp, M.; Lubrano, P.; Bange, G.; Link, H. Metabolism of non-growing bacteria. Biol. Chem. 2020, 401, 1479–1485. [Google Scholar] [CrossRef]
- Goelzer, A.; Fromion, V. Bacterial growth rate reflects a bottleneck in resource allocation. Biochim. Biophys. Acta 2011, 10, 978–988. [Google Scholar] [CrossRef]
- Dunlap, V.P. Microbial Diversity. In Encyclopedia of Biodiversity, 2nd ed.; Levin, A.S., Ed.; Academic Press: Waltham, MA, USA, 2001; Volume 5, pp. 280–291. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Martiny, C.A.; Brodie, E.; Chase, B.A.; Verdugo-Rodríguez, A.; Treseder, K.K.; Allison, D.S. Investigating the eco-evolutionary response of microbiomes to environmental change. Ecol. Lett. 2023, 00, 1–10. [Google Scholar] [CrossRef]
- Himeoka, Y.; Mitarai, N. Dynamics of bacterial populations under the feast-famine cycles. Phys. Rev. Res. 2020, 2, 013372. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Dai, X. Stringent response ensures the timely adaptation of bacterial growth to nutrient downshift. Nat. Commun. 2023, 14, 467. [Google Scholar] [CrossRef]
- Morita, R.Y. Bacteria in Oligotrophic Environments: Starvation-Survival Lifestyle; Chapman & Hall: New York, NY, USA, 1997. [Google Scholar]
- Wang, J.D.; Levin, P.A. Metabolism, cell growth and the bacterial cell cycle. Nat. Rev. Microbiol. 2009, 7, 822–827. [Google Scholar] [CrossRef]
- Cuecas, A.; Portillo, M.C.; Kanoksilapatham, W.; González, J.M. Bacterial distribution along a 50 °C temperature gradient reveals a parceled out hot spring environment. Microb. Ecol. 2014, 68, 729–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to Environmental Stresses. Plant. Cell. 1995, 7, 1099–1111. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Shen, L.; Wen, C.; Yan, Q.; Ning, D.; Qin, Y.; Xue, K.; Wu, L.; He, Z.; et al. Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 2016, 7, 12083. [Google Scholar] [CrossRef] [Green Version]
- Rath, K.M.; Fierer, N.; Murphy, D.V.; Rousk, J. Linking bacterial community composition to soil salinity along environmental gradients. ISME J. 2019, 3, 836–846. [Google Scholar] [CrossRef] [Green Version]
- Naz, M.; Dai, Z.; Hussain, S.; Tariq, M.; Danish, S.; Khan, I.U.; Qi, S.; Du, D. The soil pH and heavy metals revealed their impact on soil microbial community. J. Environ. Manag. 2022, 321, 115770. [Google Scholar] [CrossRef]
- Horner-Devine, M.C.; Carney, K.M.; Bohannan, B.J. An ecological perspective on bacterial biodiversity. Proc. Biol. Sci. 2004, 271, 113–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martiny, J.B.; Bohannan, B.J.; Brown, J.H.; Colwell, R.K.; Furhman, J.A.; Green, J.L.; Horner-Devine, M.C.; Kane, M.; Krumins, J.A.; Kuske, C.R.; et al. Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 2006, 2, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Stetter, K.O. Extremophiles and their adaptation to hot environments. FEBS Lett. 1999, 452, 22–25. [Google Scholar] [CrossRef]
- Oren, A. Halophilic archaea on Earth and in space: Growth and survival under extreme conditions. Philos. Trans. A Math. Phys. Eng. Sci. 2014, 372, 20140194. [Google Scholar] [CrossRef] [PubMed]
- Vásquez-Dean, J.; Maza, F.; Morel, I.; Pulgar, R.; González, M. Microbial communities from arid environments on a global scale. A systematic review. Biol. Res. 2020, 53, 29. [Google Scholar] [CrossRef]
- Huber, C.; Loy, A.; Nickel, M.; Arnosti, C.; Baranyi, C.; Bruchert, V.; Ferdelman, T.; Finster, K.; Christensen, F.M.; Rosa de Rezende, J.; et al. A constant flux of diverse thermophilic bacteria into the cold Arctic seabed. Science 2009, 325, 1541–1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, J.; Lara-Gutiérrez, J.; Stocker, R. Environmental fluctuations and their effects on microbial communities, populations and individuals. FEMS Mirobiol. Rev. 2021, 45, fuaa068. [Google Scholar] [CrossRef] [PubMed]
- Ai, D.; Li, X.; Liu, G.; Liang, X.; Xia, L.C. Constructing th emicrobial association network from large-scale time series data using Granger causality. Genes 2019, 10, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridenhour, B.J.; Brooker, S.L.; Williams, J.E.; van Leuven, J.T.; Miller, A.W.; Dearing, M.D.; Remien, C.H. Modeling time-series data from microbial communities. ISME J. 2017, 11, 2526–2537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darmon, E.; Leach, D.R. Bacterial genome instability. Microbiol. Mol. Biol. Rev. 2014, 78, 1–39. [Google Scholar] [CrossRef] [Green Version]
- Cuecas, A.; Kanoksilapatham, W.; González, J.M. Evidence of horizontal gene transfer by transposase gene analyses in Fervidobacterium species. PLoS ONE 2017, 12, e0173961. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, K.; Kawasaki, H. Genetic variation during long-term preservation of bacteria in public culture collections. Int. J. Syst. Evol. Microbiol. 2018, 68, 1815–1821. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Bauchinger, F.; Laroche, B.; Buyl, S.; Lahti, L.; Washburne, A.D.; Gonze, D.; Widder, S. Signatures of ecological processes in microbial community time series. Microbiome 2018, 6, 120. [Google Scholar] [CrossRef] [Green Version]
- Finkel, S.E. Long-term survival during stationary phase: Evolution and GASP phenotype. Nat. Rev. Microbiol. 2006, 4, 113–120. [Google Scholar] [CrossRef]
- Foster, P.L. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 373–397. [Google Scholar] [CrossRef]
- Jaishankar, J.; Srivastava, P. Molecular Basis of Stationary Phase Survival and Applications. Front. Microbiol. 2017, 8, 2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS System: A Complex and Tightly Regulated Response to DNA Damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, K. Regulation Systems of Bacteria such as Escherichia coli in Response to Nutrient Limitation and Environmental Stresses. Metabolites 2014, 4, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Baharoglu, Z.; Mazel, D. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 2014, 38, 1126–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zambrano, M.M.; Siggiele, D.A.; Almirón, M.; Tormo, A.; Kolter, R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 1999, 259, 1757–1760. [Google Scholar] [CrossRef] [Green Version]
- Zambrano, M.M.; Kolter, R. GASPing for life in stationary phase. Cell 1996, 86, 181–184. [Google Scholar] [CrossRef] [Green Version]
- Bacun-Druzina, V.; Cagalj, Z.; Gjuracic, K. The growth advantage in stationary-phase (GASP) phenomenon in mixed cultures of enterobacteria. FEMS Microbiol. Lett. 2007, 266, 119–127. [Google Scholar] [CrossRef] [Green Version]
- Lemonnier, M.; Levin, B.R.; Romeo, T.; Garner, K.; Baquero, M.R.; Mercante, J.; Lemichez, E.; Baquero, F.; Blázquez, J. The evolution of contact-dependent inhibition in non-growing populations of Escherichia coli. Proc. Biol. Sci. 2008, 275, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Llorens, J.M.; Tormo, A.; Martínez-García, E. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 2010, 34, 476–495. [Google Scholar] [CrossRef] [Green Version]
- D’Ari, R. The SOS system. Biochimie 1985, 67, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Roszak, D.B.; Colwell, R.R. Survival Strategies of Bacteria in the Natural Environment. Microbiol. Rev. 1987, 51, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Ercan, O.; Bisschops, M.M.; Overkamp, W.; Jorgensen, T.R.; Ram, A.; Smid, E.J.; Pronk, J.; Kuipers, O.; Daran-Lapujade, P.; Kleerebezem, M. Physiological and Transcripcional Responses of Different Industrial Microbes at Near-Zero Specific Growth Rates. Appl. Environ. Microbiol. 2015, 81, 17. [Google Scholar] [CrossRef] [Green Version]
- Demain, A.L.; Fang, A. The natural functions of secondary metabolites. Adv. Biochem. Eng. Biotechnol. 2000, 69, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Post, W.M. A theorethical reassessment of microbial maintenance and implications for microbial ecology modeling. FEMS Microbiol. Ecol. 2012, 81, 610–617. [Google Scholar] [CrossRef] [Green Version]
- Bergkessel, M.; Basta, D.W.; Newman, D.K. The physiology of growth arrest: Uniting molecular and environmental microbiology. Nat. Rev. Microbiol. 2016, 14, 549–562. [Google Scholar] [CrossRef]
- Tyc, O.; Song, C.; Dickschat, J.S.; Vos, M.; Garbeva, P. The Ecological Role of Volatile and Soluble Secondary Metabolites Produced by Soil Bacteria. Trends Microbiol. 2017, 25, 280–292. [Google Scholar] [CrossRef]
- Seyedsayamdost, M.R. Toward a global picture of bacterial secondary metabolism. J. Ind. Microbiol. Biotechnol. 2019, 46, 301–311. [Google Scholar] [CrossRef]
- Joergensen, R.G.; Wichern, F. Alive and kicking: Why dormant soil microorganisms matter. Soil. Biol. Biochem. 2018, 116, 419–430. [Google Scholar] [CrossRef]
- Akin, E.; Pelen, N.N.; Tiryaki, I.U.; Yalcin, F. Parameter identification for gompertz and logistic dynamic equations. PLoS ONE 2020, 15, e0230582. [Google Scholar] [CrossRef] [Green Version]
- Asadi, M.; Di Crescenzo, A.; Sajadi, F.A.; Spina, S. A generalized Gompertz growth model with applications and related birth-death processes. Ric. Mat. 2020, 1–36. [Google Scholar] [CrossRef]
- El-Shaarawi, A.H.; Pipes, W.O. Enumeration and statistical inference. In Bacterial Indicators of Pollution; Pipes, W.O., Ed.; CRC Press: Boca Raton, FL, USA, 1982. [Google Scholar]
- Zwietering, M.H.; Jongenburguer, I.; Rombouts, F.M.; van’t Riet, K. Modelling of the bacterial growth curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjørve, K.M.C.; Tjørve, E. The use of Gompertz models in growth analyses, and new Gompertz-model approach: An addition to the Unified-Richards family. PLoS ONE 2017, 12, e0178691. [Google Scholar] [CrossRef] [PubMed]
- Steen, A.D.; Christoph-Crits, A.; Carini, P.; DeAngelis, M.K.; Fierer, N.; Lloyd, G.K.; Trash-Cameron, J. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019, 13, 3126–3130. [Google Scholar] [CrossRef] [Green Version]
- Martiny, A.C. The ‘1% culturability paradigm’ needs to be carefully defined. ISME J. 2020, 14, 10–11. [Google Scholar] [CrossRef]
- Lee, K.C.; Herbold, C.W.; Dunfield, P.F.; Morgan, X.C.; McDOnald, I.R.; Stott, M.B. Phylogenetic delineation of the novel phylum Armanimonadetes (former candidate division OP10) and definition of two novel candidate divisions. Appl. Environ. Microbiol. 2013, 79, 2484–2487. [Google Scholar] [CrossRef] [Green Version]
- Murugkar, P.P.; Collins, A.J.; Chen, T.; Dewhirst, F.E. Isolation and cultivation of candidate phyla radiation Saccaribacteria (TM7) bacteria in coculture with bacterial hosts. J. Oral Microbiol. 2020, 12, 1814666. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, F. Single-cell metagenomics: Challenges and applications. Protein Cell 2018, 9, 501–510. [Google Scholar] [CrossRef] [Green Version]
- Gao, D.; Huang, X.; Tao, Y. A critical review of NanoSIMS in analysis of microbial metabolic activities at single-cell level. Crit. Rev. Biotechnol. 2016, 36, 884–890. [Google Scholar] [CrossRef]
- Adamberg, K.; Valgepea, K.; Vilu, R. Advanced continuous cultivation methods for systems microbiology. Microbiology 2015, 161, 1707–1719. [Google Scholar] [CrossRef]
- Greenman, J.; Mendis, B.A.; Gajda, I.; Ieropoulos, I.A. Microbial fuel cell compared to a chemostat. Chemosphere 2022, 296, 133967. [Google Scholar] [CrossRef]
- Novick, A.; Szilard, L. Description of the chemostat. Science 1950, 112, 715–716. [Google Scholar] [CrossRef]
- Gresham, D.; Hong, J. The functional basis of adaptive evolution in chemostats. FEMS Microbiol. Rev. 2015, 39, 2–16. [Google Scholar] [CrossRef] [Green Version]
- Boender, L.G.; van Maris, J.; de Hulster, E.A.; Almering, M.J.; van der Klei, I.J.; Veenhuis, M.; de Winde, J.H.; Pronk, J.T.; Daran-Lapujade, J. Cellular responses of Saccharomyces cerevisiae at near-zero growth rates: Transcriptome analysis of anaerobic retentostat cultures. FEMS Yeast Res. 2011, 11, 603–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overkamp, W.; Ercan, O.; Herber, M.; van Maris, A.J.; Kleerebezem, M.; Kuipers, O.P. Physiological and cell morphology adaptation of Bacillus subtilis at near-zero specific growth rates: A transcriptome analysis. Environ. Microbiol. 2015, 17, 346–363. [Google Scholar] [CrossRef]
- Mastrigt, O.; Egas, R.A.; Abee, T.; Smid, E.J. Aroma formation in retentostat co-cultures of Lactococcus lactis and Leuconostoc mesenteroides. Food Microbiol. 2019, 82, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Ercan, O.; den Besten, M.W.H.; Smid, J.E.; Kleerebezem, M. The growth-survival trade-off is hard-wired in the Lactococcus lactis gene regulation network. Environ. Microbiol. Rep. 2022, 14, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Parkes, R.J.; Cragg, B.A.; Fry, J.C.; Herbert, R.A.; Wimpenny, J.W.T. Bacterial biomass and activity in deep sediment layers from the Peru margin. Philos. Trans. A Math. Phys. Eng. Sci. 1990, 331, 139–153. [Google Scholar] [CrossRef]
- Hoehler, T.M.; Jorgensen, B.B. Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 2013, 11, 83–94. [Google Scholar] [CrossRef]
- Lever, M.A.; Rogers, K.L.; Lloyd, K.G.; Overmann, J.; Schink, B.; Thauer, R.K.; Hoehler, T.M.; Jorgensen, B.B. Life under extreme energy limitation: A synthesis of laboratory-and field-based investigations. FEMS Microbiol. Rev. 2015, 39, 688–728. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xu, J.; Kong, L.; Liu, T.; Yi, L.; Wang, H.; Huang, W.E.; Zheng, C. Raman-deuterium isotope probing to study metabolic activities of single bacterial cells in human intestinal microbiota. Microb. Biotechnol. 2020, 13, 572–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleiner, M.; Kouris, A.; Violette, M.; D’Angelo, G.; Liu, Y.; Korenek, A.; Tolić, N.; Sachsenberg, T.; McCalder, J.; Lipton, M.S.; et al. Ultra-sensitive isotope probing to quantify activity and substrate assimilation in microbiomes. Microbiome 2023, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Escudero, C.; Oggerin, M.; Amils, R. The deep continental subsurface: The dark biosphere. Int. Microbiol. 2018, 21, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, J.M.; Puerta-Fernández, E.; Santana, M.M.; Rekadwad, B. On a non-discrete concept of prokaryotic species. Microorganisms 2020, 8, 1723. [Google Scholar] [CrossRef] [PubMed]
- Madigan, M.T.; Bender, K.S.; Buckley, D.H.; Sattley, W.M.; Stahl, D.A. Brock Biology of Microorganisms, 16th ed.; Pearson Education: London, UK, 2021. [Google Scholar]
- Neuenschwander, S.M.; Ghai, R.; Pernthaler, J.; Salcher, M.M. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J. 2017, 12, 185–198. [Google Scholar] [CrossRef] [Green Version]
- García-Pastor, L.; Sánchez-Romero, M.A.; Gutiérrez, G.; Puerta-Fernández, E.; Casadesús, J. Formation of phenotypic lineages in Salmonella enterica by a pleiotropic fimbrial switch. PLoS Genet. 2018, 14, e1007677. [Google Scholar] [CrossRef] [Green Version]
- Chaillou, S.; Daty, M.; Bariage, F.; Dudez, A.M.; Anglade, P.; Jones, R.; Alpert, C.A.; Chanpomier-Vergès, M.C.; Zagorec, M. Intraspecies genomic diversity and natural population structure of the meat-borne lactic acid bacterium Lactobacillus sakei. Appl. Environ. Microbiol. 2009, 75, 970–980. [Google Scholar] [CrossRef] [Green Version]
- Lapujade-Daran, P.; Daran, M.J. Chemostat-Based Micro-Array Analysis in Baker’s Yeast. Adv. Microb. Physiol. 2009, 54, 257–417. [Google Scholar] [CrossRef]
- Reyes-Ruiz, L.M.; Williams, C.L.; Tamayo, R. Enhancing bacterial survival through phenotypic heterogeneity. PLoS Pathog. 2020, 16, e1008439. [Google Scholar] [CrossRef]
- Tappe, W.; Laverman, A.; Bohland, M.; Braster, M.; Rittershaus, S.; Groeneweg, J.; van Verseveld, H.W. Maintenance energy demand and starvation recovery dynamics of Nitrosomonas europaea and Nitrobacter winogradskyi cultivated in a retentostat with complete biomass retention. Appl. Environ. Microbiol. 1999, 65, 2471–2477. [Google Scholar] [CrossRef] [Green Version]
- Cases, I.; Lorenzo, V. Transcription regulation and environmetal adaptation in bacteria. Trends Microbiol. 2003, 11, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Aertsen, A.; Michiels, C. Stress and How Bacteria Cope with Death and Survival. Crit. Rev. Microbiol. 2008, 30, 4–263. [Google Scholar] [CrossRef]
- Okabe, S.; Kamigaito, A.; Kobayashi, K. Maintenance power requirements of anammox bacteria “Candidatus Brocadia sinica” and “Candidatus scalindua sp”. ISME J. 2021, 15, 3566–3575. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D. The viable but non-culturable state in bacteria. J. Microbiol. 2005, 43, 93–100. [Google Scholar]
- Marchant, R.; Franzetti, A.; Pavlostathis, S.G.; Tas, D.O.; Erdbrugger, I.; Unyayar, A.; Mazmanci, M.A.; Banat, I.M. Thermophilic bacteria in cool temperate soils: Are they metabolically active or continually added by global atmospheric transport? Appl. Microbiol. Biotechnol. 2008, 78, 841–852. [Google Scholar] [CrossRef]
- Portillo, M.C.; Santana, M.; González, J.M. Presence and potencial role of thermophilic bacteria in temperate terrestrial environments. Naturwissenschaften 2012, 99, 43–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, J.M.; Portillo, M.C.; Piñeiro-Vidal, M. Latitude-dependent underestimation of microbial extracellular enzyme activity in soils. Int. J. Environ. Sci. Technol. 2015, 12, 2427–2434. [Google Scholar] [CrossRef] [Green Version]
- DePoy, A.N.; King, G.M. Distribution and diversity of anaerobic thermophiles and putative anaerobic nickel-dependent carbon monoxide-oxidizing thermophiles in mesothermal soils and sediments. Front. Microbiol. 2023, 13, 1096186. [Google Scholar] [CrossRef] [PubMed]
- Zeigler, D.R. The Geobacillus paradox: Why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology 2014, 160, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.L.; An, D.; Caffrey, S.M.; Soh, J.; Dong, X.; Sensen, C.W.; Oldenburg, T.B.; Larter, S.R.; Voordouw, G. Roles of thermophiles and fungi in bitumen degradation in mostly cold oil sands outcrops. Appl. Environ. Microbiol. 2015, 81, 6825–6838. [Google Scholar] [CrossRef] [Green Version]
- Panikov, N.S.; Mandalakis, M.; Dai, S.; Mulcahy, L.R.; Fowle, W.; Garret, W.S.; Karger, B.L. Near-zero growth kinetics of Pseudomonas putida deduced from proteomic analysis. Environ. Microbiol. 2015, 17, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Jõers, A.; Tenson, T. Growth resumption from stationary phase reveals memory in Escherichia coli cultures. Sci. Rep. 2016, 6, 24055. [Google Scholar] [CrossRef] [Green Version]
- Gefen, O.; Fridman, O.; Ronin, I.; Balaban, N.Q. Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc. Natl. Acad. Sci. USA 2014, 111, 556–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolter, R. Growth in studying the cessation of growth. J. Bacteriol. 1999, 181, 697–699. [Google Scholar] [CrossRef] [Green Version]
- Schürch, A.C.; Arredondo-Alonso, S.; Willems, R.J.L.; Goering, R.V. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphisms versus gene-by-gene-based approaches. Clin. Microbiol. Infect. 2018, 24, 350–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, W.-I.; Chung, M.-S. Bacillus spores: A review of their properties and inactivation processing technologies. Food Sci. Biotechnol. 2020, 29, 1447–1461. [Google Scholar] [CrossRef]
- Zhang, X.-H.; Ahmad, W.; Zhu, X.-Y.; Chen, J.; Austin, B. Viable but nonculturable bacteria and their resuscitation: Implications for cultivating uncultured marine microorganisms. Mar. Life Sci. Technol. 2021, 3, 189–203. [Google Scholar] [CrossRef]
- Maalej, S.; Gdoura, R.; Dunkan, D.; Hammami, A.; Bouain, A. Maintenance of pathogenicity during entry into and resuscitation from viable but nonculturable state in Aeromonas hydrophila exposed to natural seawater at low temperature. J. Appl. Microbiol. 2004, 97, 557–565. [Google Scholar] [CrossRef]
- Grigorian, A.V.; Lustig, R.B.; Guzmán, E.C.; Mahaffy, J.M.; Zyskind, J.W. Escherichia coli cells with increased levels of DnaA and deficient in recombinational repair have decreased viability. J. Bacteriol. 2003, 185, 630–644. [Google Scholar] [CrossRef] [Green Version]
- Barcina, I.; González, J.M.; Iriberri, J.; Egea, L. Effect of visible light on progressive dormancy of E. coli cells during the survival process in natural fresh waters. Appl. Environ. Microbiol. 1989, 55, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Pirt, S.J. The maintenance energy of bacteria in growing cultures. Proc. R. Soc. Lond. B Biol. Sci. 1965, 163, 224–231. [Google Scholar] [PubMed]
- Pirt, S.J. The energetics of microbes at slow growth rates: Maintenance energies and dormant organisms. J. Ferment. Technol. 1987, 65, 173–177. [Google Scholar] [CrossRef]
- Kempes, C.P.; van Bodegom, P.M.; Wolpert, D.; Libby, E.; Amend, J.; Hoehler, T. Drivers of bacterial maintenance and minimal energy requirements. Front. Microbiol. 2017, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Bodegom, P. Microbial maintenance: A critical review on its quantification. Microb. Ecol. 2007, 53, 513–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booth, J.A.; Špírek, M.; Lobie, T.A.; Skarstad, K.; Krejci, L.; Bjørås, M. Antibiotic-induced DNA damage results in a controlled loss of pH homeostasis and genome instability. Sci. Rep. 2020, 10, 19422. [Google Scholar] [CrossRef]
- Hudson, M.A.; Siegele, D.A.; Lockless, S.W. Use of fluorescence-based assay to measure Escherichia coli membrane potential changes in high throughput. Antimicrob. Agents Chemother. 2020, 64, e00910–e00920. [Google Scholar] [CrossRef] [PubMed]
- Hoefel, D.; Grooby, W.L.; Monis, P.T.; Andrews, S.; Saint, C.P. A comparative study of carboxyfluorescein diacetate and carboxyfluorescein diacetate succinimidyl ester as indicators of bacterial activity. J. Microbiol. Methods 2003, 52, 379–388. [Google Scholar] [CrossRef]
- Kirchman, D. Measuring bacterial biomass production and growth rates from leucine incorporation in natural aquatic environments. Methods Microbiol. 2001, 30, 227–237. [Google Scholar]
- Gonzalez, J.M. Special Issue: New Methods in Microbial Research 2.0: Editorial. Microorganisms 2023, 11, 718. [Google Scholar] [CrossRef]


| Characteristics /Relevance | Optimum Growth | Slow Growth | Near-Zero Growth | Stationary Phase | Maintenance Metabolism |
|---|---|---|---|---|---|
| Conditions | Optimum/ Laboratory | Restricted growth | Severely limited growth | Nutrient depletion/growth inhibition | Maintenance /Persistence |
| Expected net growth | Optimum/maximum | ≥0.025 h−1 | <0.025 h−1 | 0–Decline | 0 |
| Degree of understanding | High | Medium/Low | Low | Low | Low |
| Environmental relevance | Poor | High | High | high | high |
| Value in taxonomy * | High | Poor | Poor | Poor | Poor |
| Discovering cell capabilities | Limited | Enhanced | High | High | High |
| Expected variability | Low | High | Very high | Very high | Medium |
| Secondary metabolite discovery | Low | High | Very high | High | Medium |
| Genome understanding | Medium | High | High | High | High |
| New gene annotation | Low/Medium | Medium/High | High | High | High |
| Microbial behavior understanding | Basic | Medium | High | High | High |
| Microbiome analysis value | Poor | Medium | High | High | Medium |
| Understanding adversity | Null | Medium | High | High | High |
| Microbial interaction potential | Poor | Medium | High | High | High |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez, J.M.; Aranda, B. Microbial Growth under Limiting Conditions-Future Perspectives. Microorganisms 2023, 11, 1641. https://doi.org/10.3390/microorganisms11071641
Gonzalez JM, Aranda B. Microbial Growth under Limiting Conditions-Future Perspectives. Microorganisms. 2023; 11(7):1641. https://doi.org/10.3390/microorganisms11071641
Chicago/Turabian StyleGonzalez, Juan M., and Beatriz Aranda. 2023. "Microbial Growth under Limiting Conditions-Future Perspectives" Microorganisms 11, no. 7: 1641. https://doi.org/10.3390/microorganisms11071641
APA StyleGonzalez, J. M., & Aranda, B. (2023). Microbial Growth under Limiting Conditions-Future Perspectives. Microorganisms, 11(7), 1641. https://doi.org/10.3390/microorganisms11071641






