Microbial Synthesis of Lactic Acid from Cotton Stalk for Polylactic Acid Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Experimental Section
2.3. Isolation of LA Producing LAB
2.4. Morphological Characterization
2.5. Fermentative Production of Lactic Acid and Screening of Isolates for Identification of Potential Isolates with Maximum Lactic Acid Production
2.6. Molecular Characterization of Potential LAB Isolates
2.7. LA and Growth Production Profiles
2.8. Cellulose Recovery from Cotton Stalk
2.9. Saccharification of Recovered Cellulose for Its Effective Conversion to Hydrolysate Containing Reducing Sugars
2.10. Microbial Synthesis of LA
2.11. Purification of LA
2.12. Synthesis of PLA from LA
2.13. Analytical Characterization
2.13.1. Quantitative Estimation of LA Using Spectrophotometric Method
2.13.2. Determination of LA through HPLC
2.13.3. FTIR Analysis
2.13.4. Nuclear Magnetic Resonance (NMR) Analysis
2.13.5. Gel Permeation Chromatography Analysis of Resultant Polymers
2.14. Statistical Analysis
3. Results
3.1. Isolation and Characterisation of LAB
3.2. Molecular Identification of Potential LAB Isolate Having Maximum LA Production
3.3. Analytical Characterization
3.3.1. Using HPLC Method for LA Production
3.3.2. Using U.V Spectrophotometric Analysis for LA Determination
3.3.3. LA Purification from Fermentation Broth
3.3.4. FTIR Analysis
3.3.5. NMR (Both 1H NMR &13C NMR) Analysis
3.3.6. Identification of Molecular Weight of PLA Using Gel Permeation Chromatography (GPC) Analysis
3.4. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tufail, T.; Ain, H.B.U.; Saeed, F.; Nasir, M.; Basharat, S.; Mahwish; Rusu, A.V.; Hussain, M.; Rocha, J.M.; Trif, M.; et al. A Retrospective on the Innovative Sustainable Valorization of Cereal Bran in the Context of Circular Bioeconomy Innovations. Sustainability 2022, 14, 14597. [Google Scholar] [CrossRef]
- Dietrich, T.; Velasco, M.V.; Echeverría, P.; Pop, B.; Rusu, A. Crop and Plant Biomass as Valuable Material for BBB. Alternatives for Valorization of Green Wastes. In Biotransformation of Agricultural Waste and By-Products; Elsevier BV: Amsterdam, The Netherlands, 2016; pp. 1–19. [Google Scholar]
- OECD; Food & Nations; Agriculture Organization of the United Nations. OECD-FAO Agricultural Outlook 2019–2028; OECD: Paris, France, 2019. [Google Scholar]
- Sołowski, G.; Konkol, I.; Shalaby, M.; Cenian, A. Rapid hydrogen generation from cotton wastes by mean of dark fermentation. SN Appl. Sci. 2020, 2, 1438. [Google Scholar] [CrossRef]
- Sasaki, C.; Kiyokawa, A.; Asada, C.; Nakamura, Y. Glucose and valuable chemicals production from cotton waste using hydrothermal method. Waste Biomass Valoriz. 2019, 10, 599–607. [Google Scholar] [CrossRef]
- Juanga-Labayen, J.P.; Labayen, I.V.; Yuan, Q. A Review on Textile Recycling Practices and Challenges. Textiles 2022, 2, 174–188. [Google Scholar] [CrossRef]
- Bangar, S.P.; Harussani, M.M.; Ilyas, R.A.; Ashogbon, A.O.; Singh, A.; Trif, M.; Jafari, S.M. Surface modifications of cellulose nanocrystals: Processes, properties, and applications. Food Hydrocoll. 2022, 130, 107689. [Google Scholar] [CrossRef]
- Isogai, A.; Atalla, R.H. Dissolution of cellulose in aqueous NaOH solutions. Cellulose 1998, 5, 309–319. [Google Scholar] [CrossRef]
- Rusu, A.V.; Criste, F.L.; Mierliţă, D.; Socol, C.T.; Trif, M. Formulation of Lipoprotein Microencapsulated Beadlets by Ionic Complexes in Algae-Based Carbohydrates. Coatings 2020, 10, 302. [Google Scholar] [CrossRef]
- Onda, A.; Ochi, T.; Kajiyoshi, K.; Yanagisawa, K. Lactic acid productionfromglucose over activated hydrotalcites as solid base catalysts in water. Catal. Commun. 2008, 9, 1050–1053. [Google Scholar] [CrossRef]
- Zhang, S.; Jin, F.; Hu, J.; Huo, Z. Improvement of lactic acid production from cellulose with the addition of Zn/Ni/C under alkaline hydrothermal conditions. Bioresour. Technol. 2011, 102, 1998–2003. [Google Scholar] [CrossRef]
- Sánchez, C.; Egüés, I.; García, A.; Llano-Ponte, R.; Labidi, J. Lactic acid production by alkaline hydrothermal treatment of corn cobs. Chem. Eng. J. 2012, 181, 655–660. [Google Scholar] [CrossRef]
- Yan, X.; Jin, F.; Tohji, K.; Kishita, A.; Enomoto, H. Hydrothermal conversion of carbohydrate biomass to lactic acid. AIChE J. 2010, 56, 2727–2733. [Google Scholar] [CrossRef]
- Perkins, E.L.; Batchelor, W.J. Water interaction in paper cellulose fibres as investigated by NMR pulsed field gradient. Carbohydr. Polym. 2012, 87, 361–367. [Google Scholar] [CrossRef]
- Cui, F.; Li, Y.; Wan, C. Lactic acid production from corn stover using mixed cultures of lactobacillus rhamnosus and lactobacillus brevis. Bioresour. Technol. 2011, 102, 1831–1836. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Q.; Wang, F.; Zhang, J.; Chen, Y.; Zhang, C.; Liu, G.; Zhang, H.; Ma, C.; Zhang, J. The Variation in the Rhizosphere Microbiome of Cotton with Soil Type, Genotype and Developmental Stage. Sci. Rep. 2017, 7, 3940. [Google Scholar] [CrossRef] [PubMed]
- Miura, S.; Arimura, T.; Itoda, N.; Dwiarti, L.; Feng, J.B.; Bin, C.H.; Okabe, M. Production of L-lactic acid from corn cob. J. Biosci. Bioeng. 2004, 97, 153–157. [Google Scholar] [CrossRef]
- Chenebault, C.; Moscoviz, R.; Trably, E.; Escudié, R.; Percheron, B. Lactic acid production from food waste using a microbial consortium: Focus on key parameters for process upscaling and fermentation residues valorization. Bioresour. Technol. 2022, 354, 127230. [Google Scholar] [CrossRef]
- Yadav, P.; Chauhan, A.K.; Singh, R.B.; Khan, S.; Halabi, G. Chapter 22—Organic acids: Microbial sources, production, and applications. In Functional Foods and Nutraceuticals in Metabolic and Non-Communicable Diseases; Singh, R.B., Watanabe, S., Isaza, A.A., Eds.; Academic Press: Cambridge, UK, 2022; pp. 325–337. [Google Scholar]
- Johnson, M.; Burgess, N.; Shi, S.; Li, J.; Blersch, D. Formulation of fish waste as a low-cost fermentative nutrient for lactic acid production by Lactobacillus pentosus. Waste Biomass Valor. 2022, 13, 2917–2925. [Google Scholar] [CrossRef]
- Byers, J.A.; Biernesser, A.B.; Delle Chiaie, K.R.; Kaur, A.; Kehl, J.A. Catalytic Systems for the Production of Poly (lactic acid). In Synthesis, Structure and Properties of Poly (lactic acid); Advances in Polymer Science; Di Lorenzo, M., Androsch, R., Eds.; Springer: Cham, Switzerland, 2019; Volume 279. [Google Scholar]
- Park, H.W.; Chang, Y.K. Economically Efficient Synthesis of Lactide Using a Solid Catalyst. Org. Process Res. Dev. 2017, 21, 1980–1984. [Google Scholar] [CrossRef]
- Singh, G.P.; Bangar, S.P.; Yang, T.; Trif, M.; Kumar, V.; Kumar, D. Effect on the Properties of Edible Starch-Based Films by the Incorporation of Additives: A Review. Polymers 2022, 14, 1987. [Google Scholar] [CrossRef]
- Choubisa, B.; Patel, M.; Dholakiya, B. Synthesis and characterization of polylactic acid (PLA) using a solid acid catalyst system in the polycondensation method. Res. Chem. Intermed. 2013, 39, 3063–3070. [Google Scholar] [CrossRef]
- Maas, R.H.; Bakker, R.R.; Eggink, G.; Weusthuis, R.A. Lactic acid production from xylose by the fungus Rhizopus oryzae. Appl. Microbiol. Biotechnol. 2006, 72, 861–868. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Z.; Duan, J.; Li, N.; Li, B.; Song, T.; Sardar, M.F.; Lv, X.; Zhu, C. Electrochemical disinfection of secondary effluent from a wastewater treatment plant: Removal efficiency of ARGs and variation of antibiotic resistance in surviving bacteria. J. Chem. Eng. 2020, 392, 123674. [Google Scholar] [CrossRef]
- Sardar, M.F.; Zhu, C.; Geng, B.; Huang, Y.; Abbasi, B.; Zhang, Z.; Song, T.; Li, H. Enhanced control of sulfonamide resistance genes and host bacteria during thermophilic aerobic composting of cow manure. Environ. Pollut. 2021, 275, 116587. [Google Scholar] [CrossRef]
- Sardar, M.F.; Li, H.; Zhu, C.; Dar, A.A.; Zhang, B.; Waqas, M.A. Differential effects of sulfamethoxazole concentrations on the enzymatic dynamics of aerobic composting. Bioresour. Technol. 2021, 336, 125330. [Google Scholar] [CrossRef]
- Chacón, M.G.; Ibenegbu, C.; Leak, D.J. Simultaneous saccharification and lactic acid fermentation of the cellulosic fraction of municipal solid waste using Bacillus smithii. Biotechnol Lett. 2021, 43, 667–675. [Google Scholar] [CrossRef]
- Borshchevskaya, L.N.; Gordeeva, T.L.; Kalinina, A.N.; Sineokii, S.P. Spectrophotometric determination of lactic acid. J. Anal. Chem. 2016, 71, 755–758. [Google Scholar] [CrossRef]
- de la Torre, I.; Ladero, M.; Santos, V.E. Production of d-lactic acid by Lactobacillus delbrueckii ssp. delbrueckii from orange peel waste: Techno-economical assessment of nitrogen sources. Appl. Microbiol. Biotechnol. 2018, 102, 10511–10521. [Google Scholar]
- Yang, P.B.; Tian, Y.; Wang, Q.; Cong, W. Effect of different types of calcium carbonate on the lactic acid fermentation performance of Lactobacillus lactis. Biochem. Eng. J. 2015, 98, 38–46. [Google Scholar] [CrossRef]
- Kumar, S.; Yadav, N.; Nain, L.; Khare, S.K. A simple downstream processing protocol for the recovery of lactic acid from the fermentation broth. Bioresour. Technol. 2020, 318, 124260. [Google Scholar] [CrossRef]
- Joglekar, H.G.; Rahman, I.; Babu, S.; Kulkarni, B.D.; Joshi, A. Comparative assessment of downstream processing options for lactic acid. Sep. Purif. Technol. 2006, 52, 1–17. [Google Scholar] [CrossRef]
- Dosuky, A.S.; Elsayed, T.R.; Yousef, E.T.; Barakat, O.S.; Nasr, N.F. Isolation, identification, and application of lactic acid-producing bacteria using salted cheese whey substrate and immobilized cells technology. J. Genet. Eng. Biotechnol. 2022, 20, 26. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, Y.; Chu, J.; Zhuang, Y.; Zhang, S. Oxygen transfer efficiency and environmental osmolarity response to neutralizing agents on l-lactic acid production efficiency by Lactobacillus paracasei. Process Biochem. 2014, 49, 2049–2054. [Google Scholar] [CrossRef]
- Levine, N.D.; Buchanan, R.E.; Gibbons, N.E. Bergey’s Manual of Determinative Bacteriology, 8th ed.; Williams & Wilkins Co.: Baltimore, MD, USA, 1975; Volume 22, p. 7. [Google Scholar]
- El-Sheshtawy, H.S.; Fahim, I.; Hosny, M.; El-Badry, M.A. Optimization of lactic acid production from agro-industrial wastes produced by Kosakonia cowanii. Curr. Res. Green Sustain. Chem. 2022, 5, 100228. [Google Scholar] [CrossRef]
- Orozco, F.G.; Valadez-González, A.; Domínguez-Maldonado, J.A.; Zuluaga, F.; Figueroa-Oyosa, L.E.; Alzate-Gaviria, L.M. Lactic Acid Yield Using Different Bacterial Strains, Its Purification, and Polymerization through Ring-Opening Reactions. Int. J. Polym. Sci. 2014, 2014, 365310. [Google Scholar] [CrossRef]
- Naser, A.Z.; Deiab, I.; Defersha, F.; Yang, S. Expanding Poly(lactic acid) (PLA) and Polyhydroxyalkanoates (PHAs) Applications: A Review on Modifications and Effects. Polymers 2021, 13, 4271. [Google Scholar] [CrossRef]
- Abedi, E.; Hashemi, S.M.B. Lactic acid production–producing microorganisms and substrates sources-state of art. Heliyon 2020, 6, e04974. [Google Scholar] [CrossRef]
- Kotzamanidis, C.; Roukas, T.; Skaracis, G. Optimization of lactic acid production from beet molasses by Lactobacillus delbrueckii NCIMB 8130. World J. Microbiol. Biotechnol. 2002, 18, 441–448. [Google Scholar] [CrossRef]
- Afzaal, M.; Saeed, F.; Shah, Y.A.; Hussain, M.; Rabail, R.; Socol, C.T.; Hassoun, A.; Pateiro, M.; Lorenzo, J.M.; Rusu, A.V.; et al. Human gut microbiota in health and disease: Unveiling the relationship. Front. Microbiol. 2022, 13, 999001. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Tashiro, Y.; Sonomoto, K. Recent advances in lactic acid production by microbial fermentation processes. Biotechnol. Adv. 2013, 31, 877–902. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Tashiro, Y.; Sonomoto, K. Lactic acid production from lignocellulose-derived sugars using lactic acid bacteria: Overview and limits. J. Biotechnol. 2011, 156, 286–301. [Google Scholar] [CrossRef]


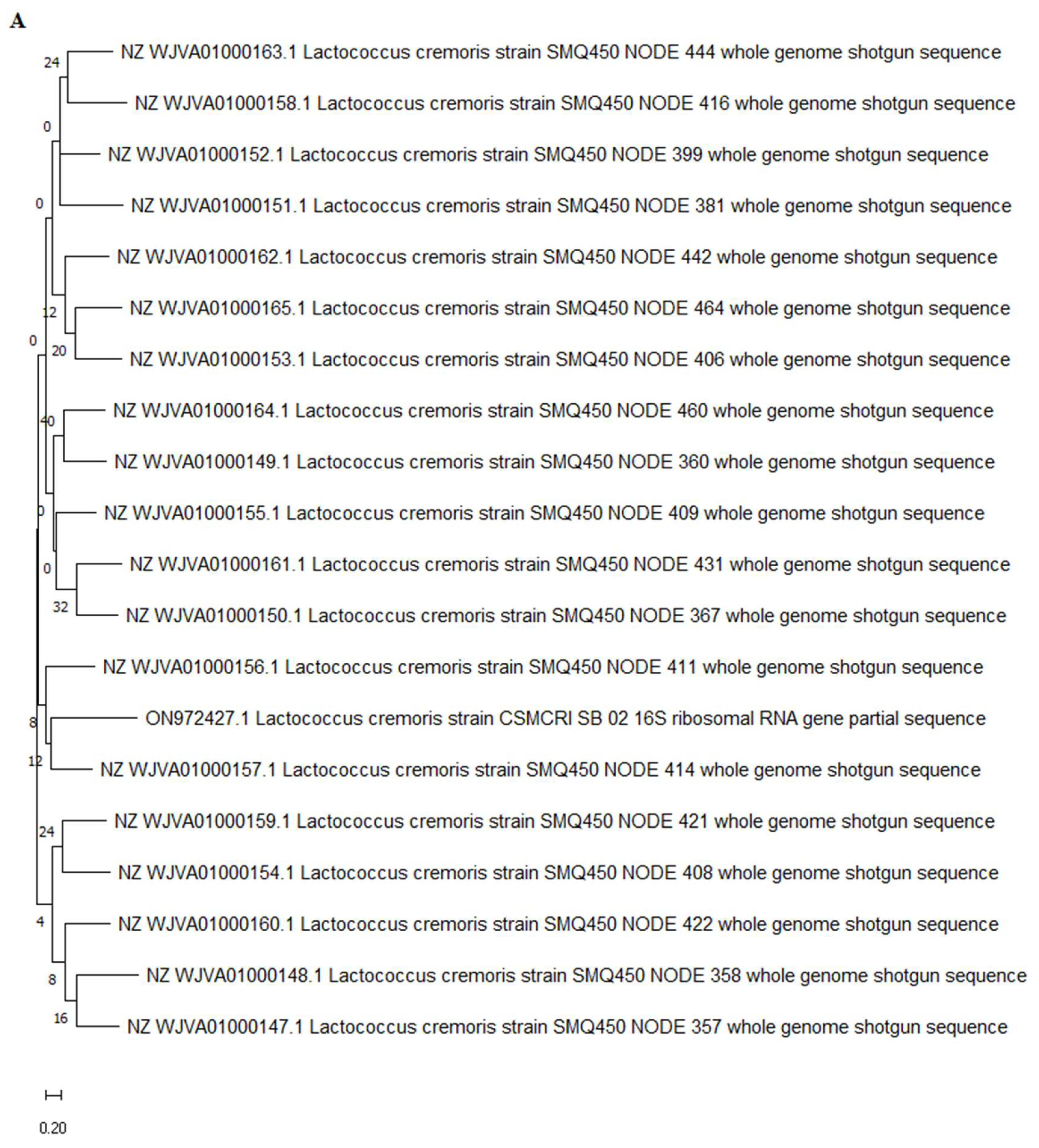


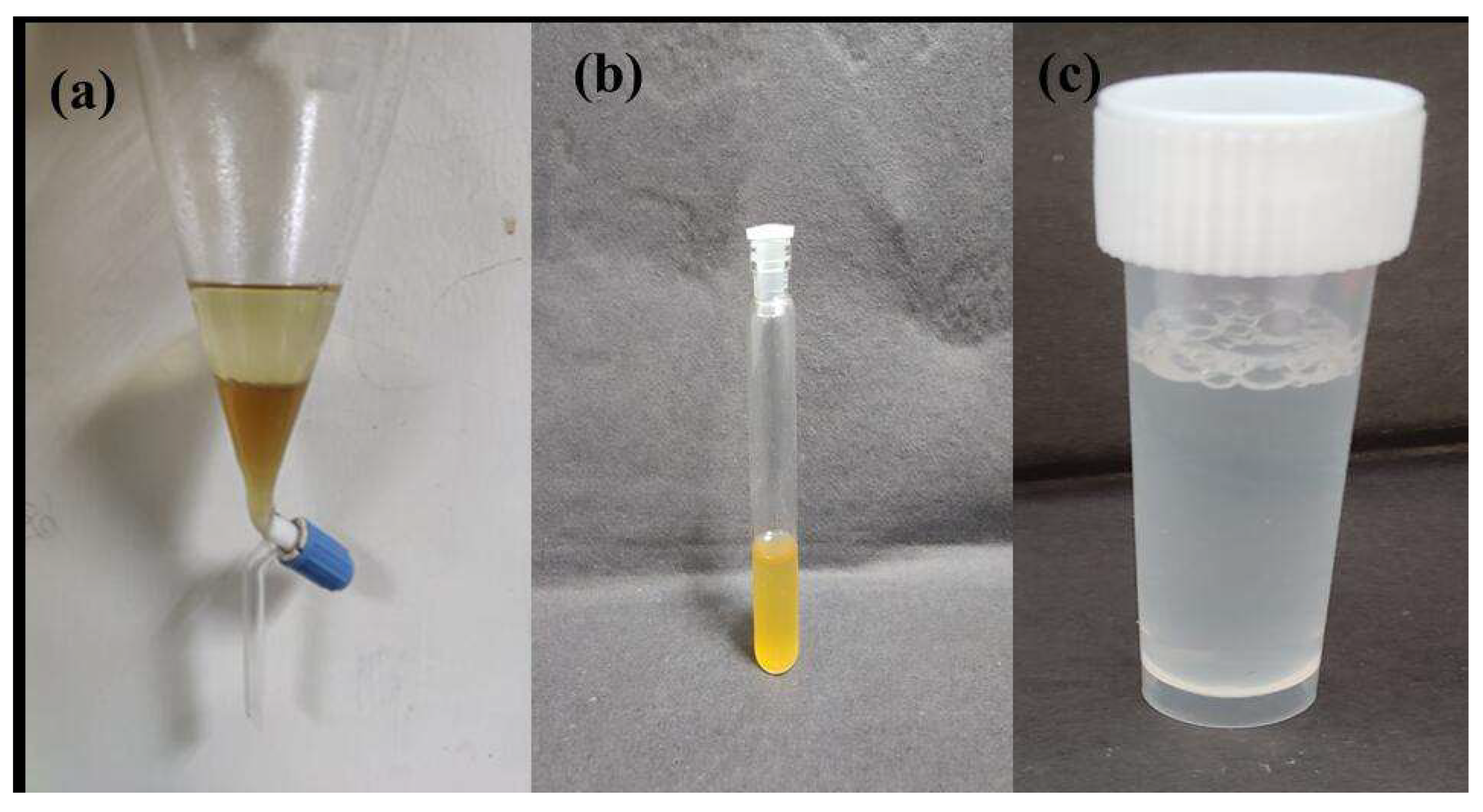

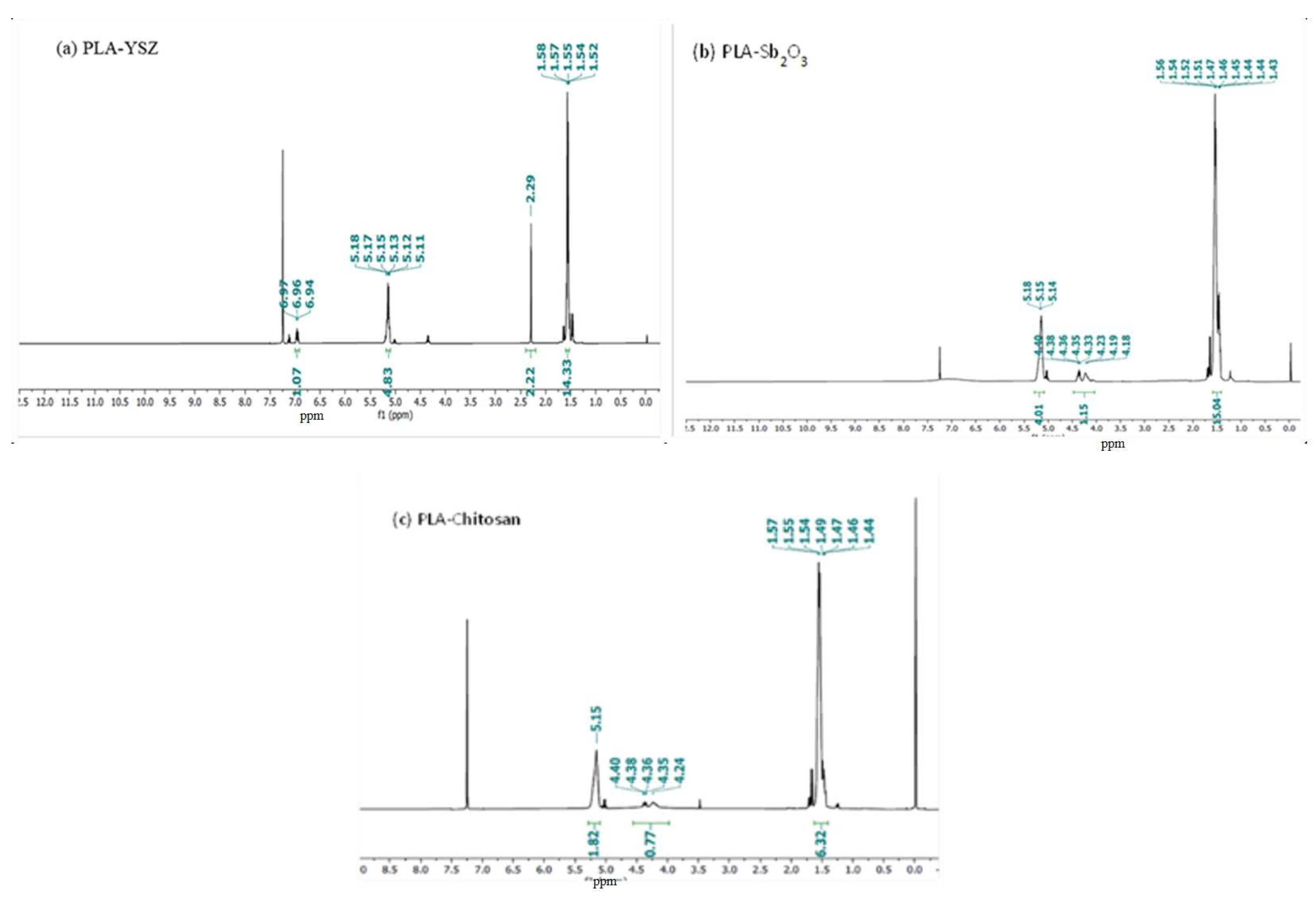
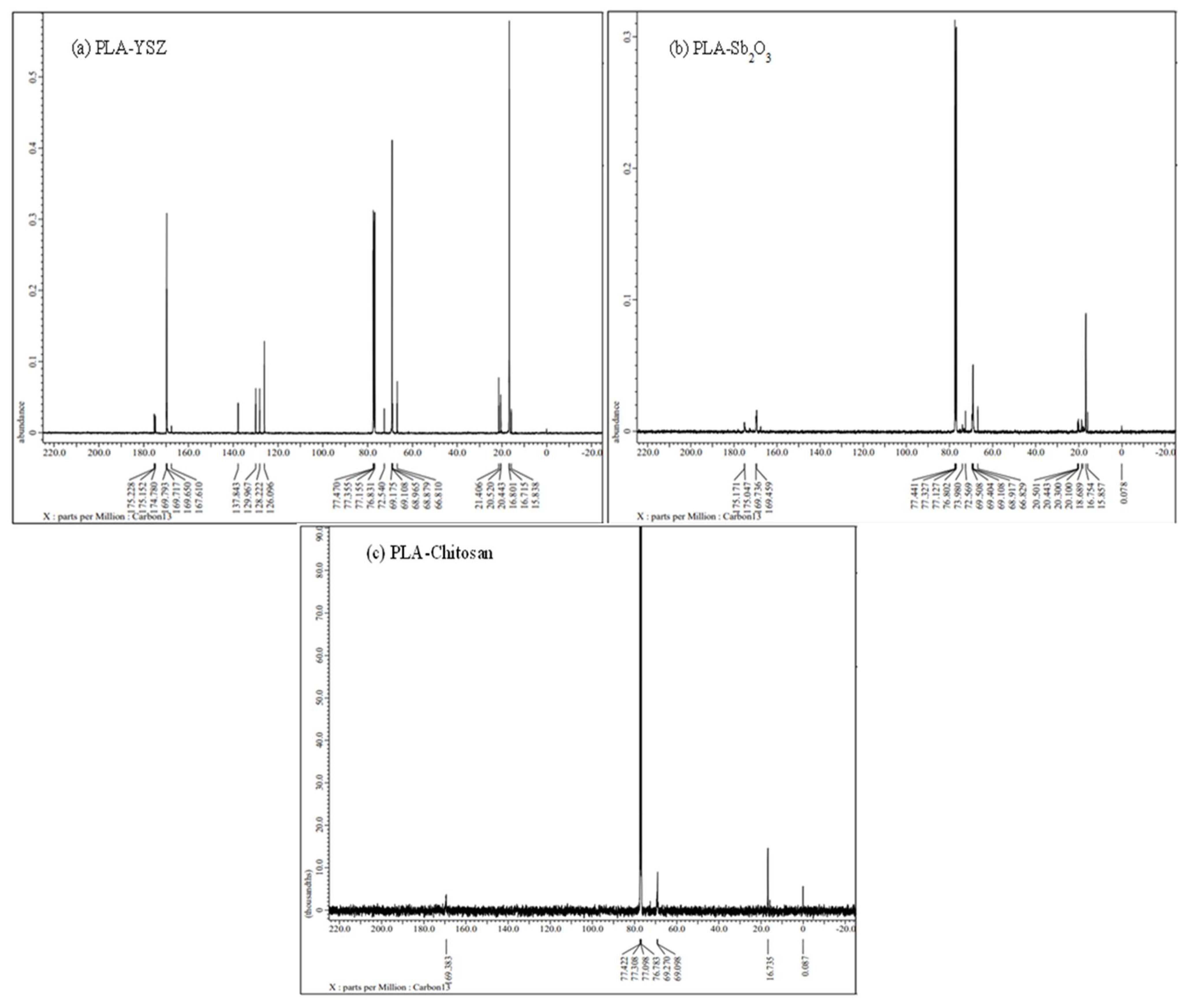
| Sample No. | Sampling Area | Latitude and Longitude | pH | Temperature | TDS (g/L) | BOD (g/L) | COD (g/L) |
|---|---|---|---|---|---|---|---|
| 1. | Kitchen waste collected from CSMCRI departmental canteen | 21°45′32″ N 72°8′39″ E | 8.4 | 42 | 150.6 | 1.2 | 0.6 |
| 2. | Dairy farm, Bhavnagar | 21°40′17″ N 72°6′25″ E | 6.1 | 40 | 73 | 1.3 | 2.5 |
| 3. | Sugarcane bagasse | 21°45′26″ N 72°7′40″ E | 7.4 | 38 | * BDL | * BDL | * BDL |
| Primer’s Name | Primer Sequence |
|---|---|
| 27F (Forward) | 5′-AGAGTTTGATCCTGGCTCAG-3′ |
| 1429R (Reversed) | 5′-GGTTACCTTGTTACGACTT-3′ |
| Isolates Name | Gram’s Staining | Growth | Source | Catalase Test | Dissolution of CaCO3 | Colony Characterization |
|---|---|---|---|---|---|---|
| LAB-1 | +ve | 24 h, 37 °C | Raw cow milk | −ve | Yes | Medium circular size, smooth texture, glossy appearance, smooth edge, opaque, whitish colony |
| LAB-2 | +ve | 24 h, 37 °C | Raw cow milk | −ve | YES | Medium circular size, smooth edge, glossy appearance, opaque, whitish colony |
| LAB-3 | +ve | 24 h, 37 °C | Raw cow milk | −ve | YES | small circular size, smooth edge, rough appearance, translucent, pale white colony |
| LAB-4 | +ve | 24 h, 37 °C | Raw cow milk | −ve | YES | Medium circular size, smooth edge, glossy appearance, opaque, whitish colony |
| LAB-5 | +ve | 24 h, 37 °C | Raw cow milk | −ve | YES | Medium circular size, smooth edge, glossy appearance, translucent, whitish colony |
| LAB-6 | +ve | 24 h, 37 °C | Kitchen waste | −ve | YES | Medium circular size, smooth edge, opaque, whitish colony |
| LAB-7 | +ve | 24 h, 37 °C | Kitchen waste | −ve | YES | Small, circular size, smooth edge, opaque, whitish colony |
| LAB-8 | −ve | 48 h, 30 °C | Kitchen waste | −ve | No | Large, circular size, wavy edge, glossy appearance, opaque, whitish colony |
| LAB-9 | +ve | 24 h, 37 °C | Kitchen waste | −ve | No | Medium circular size, smooth edge, glossy appearance, opaque, whitish colony |
| LAB-10 | +ve | 24 h, 37 °C | Kitchen waste | −ve | No | Medium circular size, smooth edge, glossy appearance, opaque, whitish colony |
| LAB-11 | −ve | 24 h, 37 °C | Kitchen waste | −ve | No | Medium circular size, smooth edge, glossy appearance, opaque, whitish colony |
| LAB-12 | +ve | 24 h, 37 °C | Sugarcane bagasse | +ve | YES | Small, circular size, rough edge, glossy appearance, transparent pale-yellow colony |
| LAB-13 | +ve | 48 h, 37 °C | Sugarcane bagasse | −ve | NO | Medium circular size, smooth edge, glossy appearance, opaque, whitish colony |
| LAB-14 | −ve | 48 h, 30 °C | Sugarcane bagasse | +ve | NO | Medium circular size, smooth edge, rough appearance, opaque, whitish colony |
| LAB-15 | −ve | 24 h, 37 °C | Soil sample | +ve | NO | Small, circular size, smooth edge, glossy appearance, opaque, whitish colony |
| LAB-16 | +ve | 24 h, 30 °C | Soil sample | +ve | NO | Medium circular size, smooth edge, glossy appearance, opaque, whitish colony |
| Isolates | LA (gm/L−1) | Dry Weight Biomass (gm/mL) | Sugar Consumed (gm/L−1) |
|---|---|---|---|
| LAB 1 | 51.3564 | 0.0624 | 41.353 |
| LAB 2 | 29.1798 | 0.0260 | 25.2038 |
| LAB 3 | 40.4100 | 0.06385 | 38.243 |
| LAB 4 | 36.908 | 0.06585 | 36.832 |
| LAB 5 | 41.3456 | 0.05784 | 38.988 |
| LAB 6 | 39.0456 | 0.07543 | 36.982 |
| LAB 11 | 46.0685 | 0.06843 | 38.654 |
| LAB 14 | 39.6854 | 0.05876 | 37.288 |
| Sample Code | Catalyst (1%) | Time (h) | Temperature (°C) | MWn | MWw | PI |
|---|---|---|---|---|---|---|
| PLA-YSZ | 1.0 | 24 h | 180 °C | 2805 | 3854 | 1.374 |
| PLA-Chitosan | 1.0 | 24 h | 180 °C | 1458 | 2305 | 1.581 |
| PLA-Sb2O3 | 1.0 | 24 h | 180 °C | 608 | 1120 | 1.843 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paswan, M.; Adhikary, S.; Salama, H.H.; Rusu, A.V.; Zuorro, A.; Dholakiya, B.Z.; Trif, M.; Bhattacharya, S. Microbial Synthesis of Lactic Acid from Cotton Stalk for Polylactic Acid Production. Microorganisms 2023, 11, 1931. https://doi.org/10.3390/microorganisms11081931
Paswan M, Adhikary S, Salama HH, Rusu AV, Zuorro A, Dholakiya BZ, Trif M, Bhattacharya S. Microbial Synthesis of Lactic Acid from Cotton Stalk for Polylactic Acid Production. Microorganisms. 2023; 11(8):1931. https://doi.org/10.3390/microorganisms11081931
Chicago/Turabian StylePaswan, Meenakshi, Sudipto Adhikary, Heba Hassan Salama, Alexandru Vasile Rusu, Antonio Zuorro, Bharatkumar Z. Dholakiya, Monica Trif, and Sourish Bhattacharya. 2023. "Microbial Synthesis of Lactic Acid from Cotton Stalk for Polylactic Acid Production" Microorganisms 11, no. 8: 1931. https://doi.org/10.3390/microorganisms11081931
APA StylePaswan, M., Adhikary, S., Salama, H. H., Rusu, A. V., Zuorro, A., Dholakiya, B. Z., Trif, M., & Bhattacharya, S. (2023). Microbial Synthesis of Lactic Acid from Cotton Stalk for Polylactic Acid Production. Microorganisms, 11(8), 1931. https://doi.org/10.3390/microorganisms11081931










