The Isolation, Genetic Analysis and Biofilm Characteristics of Listeria spp. from the Marine Environment in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sand Sample Collection and Listeria spp. Isolation
2.2. DNA Extraction, Sequencing, Assembly, and Species Identification
2.3. Core, Pan-Genome Analyses, and Phylogenetic Analyses Based on Single-Nucleotide Polymorphism (SNP)
2.4. Analyses of Virulence Genes, Resistance Genes, and Plasmid
2.5. Biofilm Formation Assessment
3. Results
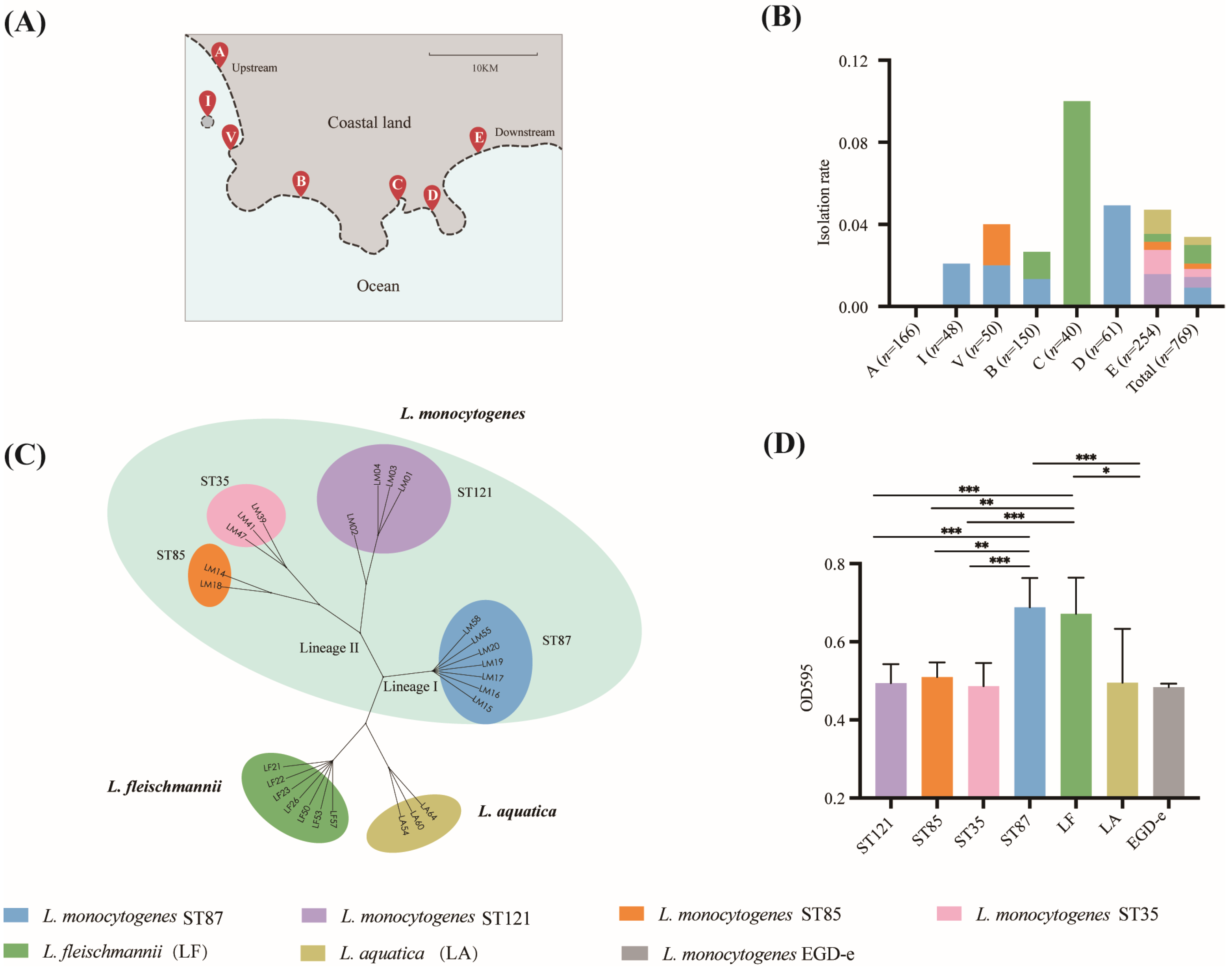
3.1. Occurrence of Listeria Isolates and Molecular Subtyping of L. monocytogenes
3.2. The Biofilm Formation Ability of Listeria from Beach Sand
3.3. Virulence Genes, Resistance Genes, and Plasmids of L. monocytogenes Isolates
3.4. The Genetic Relationship of L. monocytogenes Isolates
3.5. The General Genomic Features of L. fleischmannii and L. aquatica Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parte, A.C. LPSN—List of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014, 42, D613–D616. [Google Scholar] [CrossRef] [PubMed]
- Freitag, N.E.; Port, G.C.; Miner, M.D. Listeria monocytogenes—From saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 2009, 7, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Mao, P.; Jiang, H.; Zhang, L.; Liu, D.; Cao, X.; Wang, Y.; Wang, Y.; Sun, H.; Huang, Y.; et al. Two Prevalent Listeria ivanovii subsp. ivanovii Clonal Strains with Different Virulence Exist in Wild Rodents and Pikas of China. Front. Vet. Sci. 2020, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Orsi, R.H.; Wiedmann, M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 2016, 100, 5273–5287. [Google Scholar] [CrossRef]
- De Roin, M.A.; Foong, S.C.; Dixon, P.M.; Dickson, J.S. Survival and recovery of Listeria monocytogenes on ready-to-eat meats inoculated with a desiccated and nutritionally depleted dustlike vector. J. Food Prot. 2003, 66, 962–969. [Google Scholar] [CrossRef]
- Bertsch, D.; Rau, J.; Eugster, M.R.; Haug, M.C.; Lawson, P.A.; Lacroix, C.; Meile, L. Listeria fleischmannii sp. nov., isolated from cheese. Int. J. Syst. Evol. Microbiol. 2013, 63, 526–532. [Google Scholar] [CrossRef]
- Terentjeva, M.; Steingolde, Z.; Meistere, I.; Elferts, D.; Avsejenko, J.; Streikisa, M.; Gradovska, S.; Alksne, L.; Kibilds, J.; Berzins, A. Prevalence, Genetic Diversity and Factors Associated with Distribution of Listeria monocytogenes and Other Listeria spp. in Cattle Farms in Latvia. Pathogens 2021, 10, 851. [Google Scholar] [CrossRef]
- den Bakker, H.C.; Warchocki, S.; Wright, E.M.; Allred, A.F.; Ahlstrom, C.; Manuel, C.S.; Stasiewicz, M.J.; Burrell, A.; Roof, S.; Strawn, L.K.; et al. Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov. and Listeria grandensis sp. nov., from agricultural and natural environments. Int. J. Syst. Evol. Microbiol. 2014, 64, 1882–1889. [Google Scholar] [CrossRef]
- Denis, M.; Ziebal, C.; Boscher, E.; Picard, S.; Perrot, M.; Nova, M.V.; Roussel, S.; Diara, A.; Pourcher, A.M. Occurrence and Diversity of Listeria monocytogenes Isolated from Two Pig Manure Treatment Plants in France. Microbes Environ. 2022, 37, ME22019. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Bjorkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- Muchaamba, F.; Eshwar, A.K.; Stevens, M.J.A.; Stephan, R.; Tasara, T. Different Shades of Listeria monocytogenes: Strain, Serotype, and Lineage-Based Variability in Virulence and Stress Tolerance Profiles. Front. Microbiol. 2021, 12, 792162. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiao, Y.; Lan, R.; Xu, X.; Liu, G.; Wang, X.; Zhang, L.; Pang, H.; Jin, D.; Dai, H.; et al. Characterization of Listeria monocytogenes isolated from human Listeriosis cases in China. Emerg. Microbes Infect. 2015, 4, e50. [Google Scholar] [CrossRef] [PubMed]
- Lauer, J.R.; Simsek, S.; Bergholz, T.M. Fate of Salmonella and Enterohemorrhagic Escherichia coli on Wheat Grain. J. Food Prot. 2021, 84, 2109–2115. [Google Scholar] [CrossRef]
- Selvaganapathi, R.; Jeyasekaran, G.; Shakila, R.J.; Sukumar, D.; Kumar, M.P.; Sivaraman, B. Occurrence of Listeria monocytogenes on the seafood contact surfaces of Tuticorin Coast of India. J. Food Sci. Technol. 2018, 55, 2808–2812. [Google Scholar] [CrossRef]
- Huss, H.H.; Jorgensen, L.V.; Vogel, B.F. Control options for Listeria monocytogenes in seafoods. Int. J. Food Microbiol. 2000, 62, 267–274. [Google Scholar] [CrossRef]
- Iwu, C.D.; Iwu-Jaja, C.J.; Elhadi, R.; Semerjian, L.; Okoh, A.I. Modelling the Potential Risk of Infection Associated with Listeria monocytogenes in Irrigation Water and Agricultural Soil in Two District Municipalities in South Africa. Microorganisms 2022, 10, 181. [Google Scholar] [CrossRef]
- O’Driscoll, J.; Nnadi, C.; McLauchlin, J. Listeria monocytogenes septic arthritis in an immunocompetent adult. Clin. Microbiol. Infect. 1999, 5, 234–235. [Google Scholar] [CrossRef]
- Gan, L.; Cao, X.; Wang, Y.; Wang, Y.; Jiang, H.; Lan, R.; Xu, J.; Ye, C. Carriage and potential long distance transmission of Listeria monocytogenes by migratory black-headed gulls in Dianchi Lake, Kunming. Emerg. Microbes Infect. 2019, 8, 1195–1204. [Google Scholar] [CrossRef]
- Cooley, M.B.; Quinones, B.; Oryang, D.; Mandrell, R.E.; Gorski, L. Prevalence of shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front. Cell. Infect. Microbiol. 2014, 4, 30. [Google Scholar] [CrossRef]
- Colburn, K.G.; Kaysner, C.A.; Abeyta, C., Jr.; Wekell, M.M. Listeria species in a California coast estuarine environment. Appl. Environ. Microbiol. 1990, 56, 2007–2011. [Google Scholar] [CrossRef]
- Gorski, L.; Cooley, M.B.; Oryang, D.; Carychao, D.; Nguyen, K.; Luo, Y.; Weinstein, L.; Brown, E.; Allard, M.; Mandrell, R.E.; et al. Prevalence and Clonal Diversity of over 1200 Listeria monocytogenes Isolates Collected from Public Access Waters near Produce Production Areas on the Central California Coast during 2011 to 2016. Appl. Environ. Microbiol. 2022, 88, e0035722. [Google Scholar] [CrossRef]
- Bou-m’handi, N.; Jacquet, C.; El Marrakchi, A.; Martin, P. Phenotypic and molecular characterization of Listeria monocytogenes strains isolated from a marine environment in Morocco. Foodborne Pathog. Dis. 2007, 4, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Raschle, S.; Stephan, R.; Stevens, M.J.A.; Cernela, N.; Zurfluh, K.; Muchaamba, F.; Nuesch-Inderbinen, M. Environmental dissemination of pathogenic Listeria monocytogenes in flowing surface waters in Switzerland. Sci. Rep. 2021, 11, 9066. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- Pasquali, F.; Palma, F.; Guillier, L.; Lucchi, A.; De Cesare, A.; Manfreda, G. Listeria monocytogenes Sequence Types 121 and 14 Repeatedly Isolated Within One Year of Sampling in a Rabbit Meat Processing Plant: Persistence and Ecophysiology. Front. Microbiol. 2018, 9, 596. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.; Lee, S.; Kathariou, S. Heavy Metal Resistance Determinants of the Foodborne Pathogen Listeria monocytogenes. Genes 2018, 10, 11. [Google Scholar] [CrossRef]
- Mao, P.; Wang, Y.; Gan, L.; Sun, H.; Wang, Y.; Li, L.; Ji, S.; Song, Z.; Jiang, H.; Ye, C. Function and distribution of the conjugative plasmid pLM1686 in foodborne Listeria monocytogenes in China. Int. J. Food Microbiol. 2021, 352, 109261. [Google Scholar] [CrossRef]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 2013, 14, 47. [Google Scholar] [CrossRef]
- Dwight, R.H.; Fernandez, L.M.; Baker, D.B.; Semenza, J.C.; Olson, B.H. Estimating the economic burden from illnesses associated with recreational coastal water pollution—A case study in Orange County, California. J. Environ. Manag. 2005, 76, 95–103. [Google Scholar] [CrossRef]
- Bompangue Nkoko, D.; Giraudoux, P.; Plisnier, P.D.; Tinda, A.M.; Piarroux, M.; Sudre, B.; Horion, S.; Tamfum, J.J.; Ilunga, B.K.; Piarroux, R. Dynamics of cholera outbreaks in Great Lakes region of Africa, 1978–2008. Emerg. Infect. Dis. 2011, 17, 2026–2034. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.P.; Avery, L.M.; Killham, K.; Jones, D.L. Persistence, dissipation, and activity of Escherichia coli O157:H7 within sand and seawater environments. FEMS Microbiol. Ecol. 2007, 60, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Gholipour, S.; Nikaeen, M.; Farhadkhani, M.; Nikmanesh, B. Survey of Listeria monocytogenes contamination of various environmental samples and associated health risks. Food Control 2020, 108, 106843. [Google Scholar] [CrossRef]
- Wagner, M.; Melzner, D.; Bago, Z.; Winter, P.; Egerbacher, M.; Schilcher, F.; Zangana, A.; Schoder, D. Outbreak of clinical listeriosis in sheep: Evaluation from possible contamination routes from feed to raw produce and humans. J. Vet. Med. B Infect. Dis. Vet. Public Health 2005, 52, 278–283. [Google Scholar] [CrossRef]
- Shuval, H. Estimating the global burden of thalassogenic diseases: Human infectious diseases caused by wastewater pollution of the marine environment. J. Water Health 2003, 1, 53–64. [Google Scholar] [CrossRef]
- Halliday, E.; Gast, R.J. Bacteria in beach sands: An emerging challenge in protecting coastal water quality and bather health. Environ. Sci. Technol. 2011, 45, 370–379. [Google Scholar] [CrossRef]
- Rubini, S.; Baruffaldi, M.; Taddei, R.; D’Annunzio, G.; Scaltriti, E.; Tambassi, M.; Menozzi, I.; Bondesan, G.; Mazzariol, S.; Centelleghe, C.; et al. Loggerhead Sea Turtle as Possible Source of Transmission for Zoonotic Listeriosis in the Marine Environment. Vet. Sci. 2023, 10, 344. [Google Scholar] [CrossRef]
- Chen, M.; Cheng, J.; Wu, Q.; Zhang, J.; Chen, Y.; Xue, L.; Lei, T.; Zeng, H.; Wu, S.; Ye, Q.; et al. Occurrence, Antibiotic Resistance, and Population Diversity of Listeria monocytogenes Isolated from Fresh Aquatic Products in China. Front. Microbiol. 2018, 9, 2215. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, W.; Wang, J.; Xu, B.; Liu, H.; Dong, Q.; Zhang, X. 10-Year Molecular Surveillance of Listeria monocytogenes Using Whole-Genome Sequencing in Shanghai, China, 2009–2019. Front. Microbiol. 2020, 11, 551020. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, L.; Li, Q.; Wang, H.; Wang, Y.; Sun, H.; Xu, J.; Lan, R.; Ye, C. Genomic dissection of the most prevalent Listeria monocytogenes clone, sequence type ST87, in China. BMC Genom. 2019, 20, 1014. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, L.; Ji, S.; Li, Q.; Wang, H.; Zhang, Z.; Mao, P.; Sun, H.; Li, L.; Wang, Y.; et al. Dissecting Listeria monocytogenes Persistent Contamination in a Retail Market Using Whole-Genome Sequencing. Microbiol. Spectr. 2022, 10, e0018522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Zhang, P.; Niu, Y.; Chen, Q.; Ma, X. Genomic Characterization of Clinical Listeria monocytogenes Isolates in Beijing, China. Front. Microbiol. 2021, 12, 751003. [Google Scholar] [CrossRef]
- Rychli, K.; Wagner, E.M.; Ciolacu, L.; Zaiser, A.; Tasara, T.; Wagner, M.; Schmitz-Esser, S. Comparative genomics of human and non-human Listeria monocytogenes sequence type 121 strains. PLoS ONE 2017, 12, e0176857. [Google Scholar] [CrossRef] [PubMed]
- Kropac, A.C.; Eshwar, A.K.; Stephan, R.; Tasara, T. New Insights on the Role of the pLMST6 Plasmid in Listeria monocytogenes Biocide Tolerance and Virulence. Front. Microbiol. 2019, 10, 1538. [Google Scholar] [CrossRef] [PubMed]
- Naditz, A.L.; Dzieciol, M.; Wagner, M.; Schmitz-Esser, S. Plasmids contribute to food processing environment-associated stress survival in three Listeria monocytogenes ST121, ST8, and ST5 strains. Int. J. Food Microbiol. 2019, 299, 39–46. [Google Scholar] [CrossRef]
- Anast, J.M.; Schmitz-Esser, S. Certain Listeria monocytogenes plasmids contribute to increased UVC ultraviolet light stress. FEMS Microbiol. Lett. 2021, 368, fnab123. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.E.; Fricker, A.D.; Kapili, B.J.; Petassi, M.T. Heteromeric transposase elements: Generators of genomic islands across diverse bacteria. Mol. Microbiol. 2014, 93, 1084–1092. [Google Scholar] [CrossRef]
- Li, F.; Lu, G.; Chan, B.P.L.; Zheng, X.; ZHOU, Z.; MO, Y. Status of wintering waterbirds on Hainan Island: Results of annual waterbird surveys between 2008–2020. Forktail 2020, 36, 79–89. [Google Scholar]
- Xue, N.; Wang, L.; Li, W.; Wang, S.; Pan, X.; Zhang, D. Increased inheritance of structure and function of bacterial communities and pathogen propagation in plastisphere along a river with increasing antibiotics pollution gradient. Environ. Pollut. 2020, 265, 114641. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, C.; Zhang, W.; Di, P.; Yi, N.; Chen, C. Vertical and horizontal assemblage patterns of bacterial communities in a eutrophic river receiving domestic wastewater in southeast China. Environ. Pollut. 2017, 230, 469–478. [Google Scholar] [CrossRef]
- Hansen, C.H.; Vogel, B.F.; Gram, L. Prevalence and survival of Listeria monocytogenes in Danish aquatic and fish-processing environments. J. Food Prot. 2006, 69, 2113–2122. [Google Scholar] [CrossRef] [PubMed]
- Qian, P.-Y.; Cheng, A.; Wang, R.; Zhang, R. Marine biofilms: Diversity, interactions and biofouling. Nat. Rev. Microbiol. 2022, 20, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, F.; Perez-Rodriguez, F.; Servadio, J.L.; Gummalla, S.; Hedberg, C.W. Quantitative risk assessment model to investigate the public health impact of varying Listeria monocytogenes allowable levels in different food commodities: A retrospective analysis. Int. J. Food Microbiol. 2022, 383, 109932. [Google Scholar] [CrossRef] [PubMed]
- Gorski, L.; Parker, C.T.; Liang, A.S.; Walker, S.; Romanolo, K.F. The Majority of Genotypes of the Virulence Gene inlA Are Intact among Natural Watershed Isolates of Listeria monocytogenes from the Central California Coast. PLoS ONE 2016, 11, e0167566. [Google Scholar] [CrossRef] [PubMed]
- Mohiuddin, M.M.; Salama, Y.; Schellhorn, H.E.; Golding, G.B. Shotgun metagenomic sequencing reveals freshwater beach sands as reservoir of bacterial pathogens. Water Res. 2017, 115, 360–369. [Google Scholar] [CrossRef]
- Wen, R.; Fu, Z.; Fu, P.; Wang, S. Quick detection of Listeria in food with the VIDAS automatic Immunoassay system. Chin. J. Health Lab Technol. 1999, 50–51. (In Chinese) [Google Scholar]
- Liu, X.; He, J.; Xing, K.; Feng, L. Analysis of Microbial Pollution in Catering and Other Food Products in Hainan Province from 2014 to 2015. Pract. Prev. Healthc. 2018, 25, 261–263. (In Chinese) [Google Scholar] [CrossRef]


| Accession No. of Plasmid | Species | ST | Accession No. of Strains | Geographic Location | Isolation Source | Collection Date | Assemble Level |
|---|---|---|---|---|---|---|---|
| FR667692.1 | * LM | 66 | GCA_000197755.2 | unknown | unknown | unknown | ** |
| CP006595.1 | * LM | 3 | GCA_000438585.1 | USA | food | 1994 | ** |
| CP015985.1 | * LM | 7 | GCA_001596775.2 | Italy | blood (human) | 2015 | ** |
| CP025561.1 | * LM | 199 | GCA_003031975.1 | unknown | unknown | unknown | ** |
| CP041214.1 | * LM | 3 | GCA_004142705.2 | USA | chocolate milk | 2018 | ** |
| CP045973.1 | * LM | 122 | GCA_009664775.1 | Australia | human | 2009 | ** |
| CP090056.1 | * LM | 31 | GCA_021403065.1 | USA | Salmon processing facility | 1998 | ** |
| MZ127848.1 | * LIN | Poland | food contact surface swab | 2009 | ** | ||
| CP045744.1 | * LIN | GCA_009648575.1 | Italy | minced meat | 2005 | ** | |
| MZ869809.1 | * LW | Russia | environmental surface, meat processing plant | 2020 | ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, P.; Wang, Y.; Li, L.; Ji, S.; Li, P.; Liu, L.; Chen, J.; Sun, H.; Luo, X.; Ye, C. The Isolation, Genetic Analysis and Biofilm Characteristics of Listeria spp. from the Marine Environment in China. Microorganisms 2023, 11, 2166. https://doi.org/10.3390/microorganisms11092166
Mao P, Wang Y, Li L, Ji S, Li P, Liu L, Chen J, Sun H, Luo X, Ye C. The Isolation, Genetic Analysis and Biofilm Characteristics of Listeria spp. from the Marine Environment in China. Microorganisms. 2023; 11(9):2166. https://doi.org/10.3390/microorganisms11092166
Chicago/Turabian StyleMao, Pan, Yan Wang, Lingling Li, Shunshi Ji, Peijing Li, Lingyun Liu, Jinni Chen, Hui Sun, Xia Luo, and Changyun Ye. 2023. "The Isolation, Genetic Analysis and Biofilm Characteristics of Listeria spp. from the Marine Environment in China" Microorganisms 11, no. 9: 2166. https://doi.org/10.3390/microorganisms11092166
APA StyleMao, P., Wang, Y., Li, L., Ji, S., Li, P., Liu, L., Chen, J., Sun, H., Luo, X., & Ye, C. (2023). The Isolation, Genetic Analysis and Biofilm Characteristics of Listeria spp. from the Marine Environment in China. Microorganisms, 11(9), 2166. https://doi.org/10.3390/microorganisms11092166






