Feed Regime Slightly Modifies the Bacterial but Not the Fungal Communities in the Intestinal Mucosal Microbiota of Cobia Fish (Rachycentron canadum)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish and Experimental Design
2.2. Intestinal Samples
2.3. DNA Extraction and Amplicon Sequencing
2.4. Identification of Microbial Communities Using Bioinformatic Analysis
2.5. Sample Size and Statistical Analysis
3. Results
3.1. Bacterial Community Composition
3.1.1. Effect of Diet on Alpha Diversity (Primary Outcome)
3.1.2. Effect of Diet on Beta Diversity
3.1.3. Differential Abundances of Bacterial Communities
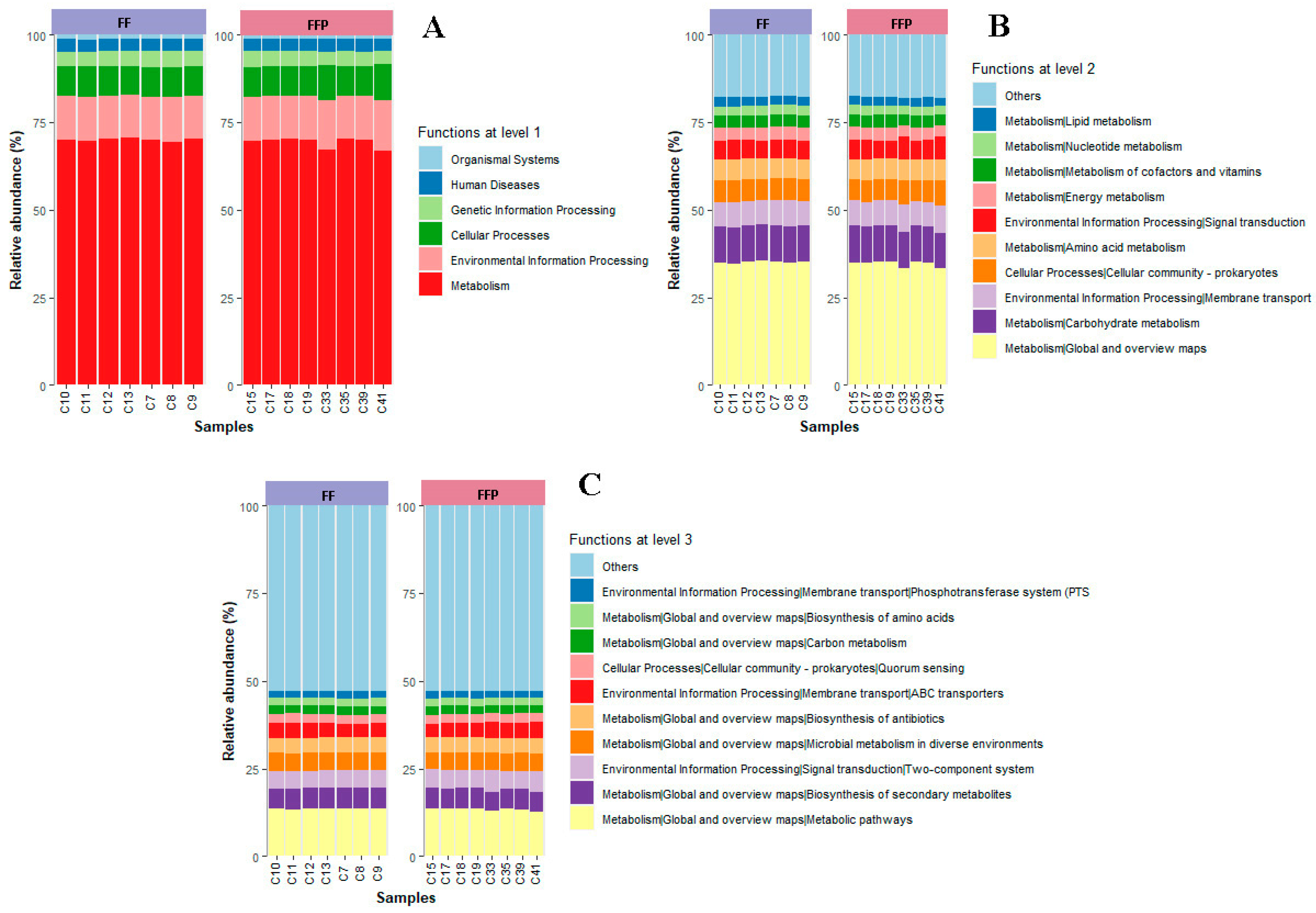
3.1.4. Effect of Diet on Inferred Bacterial Functions
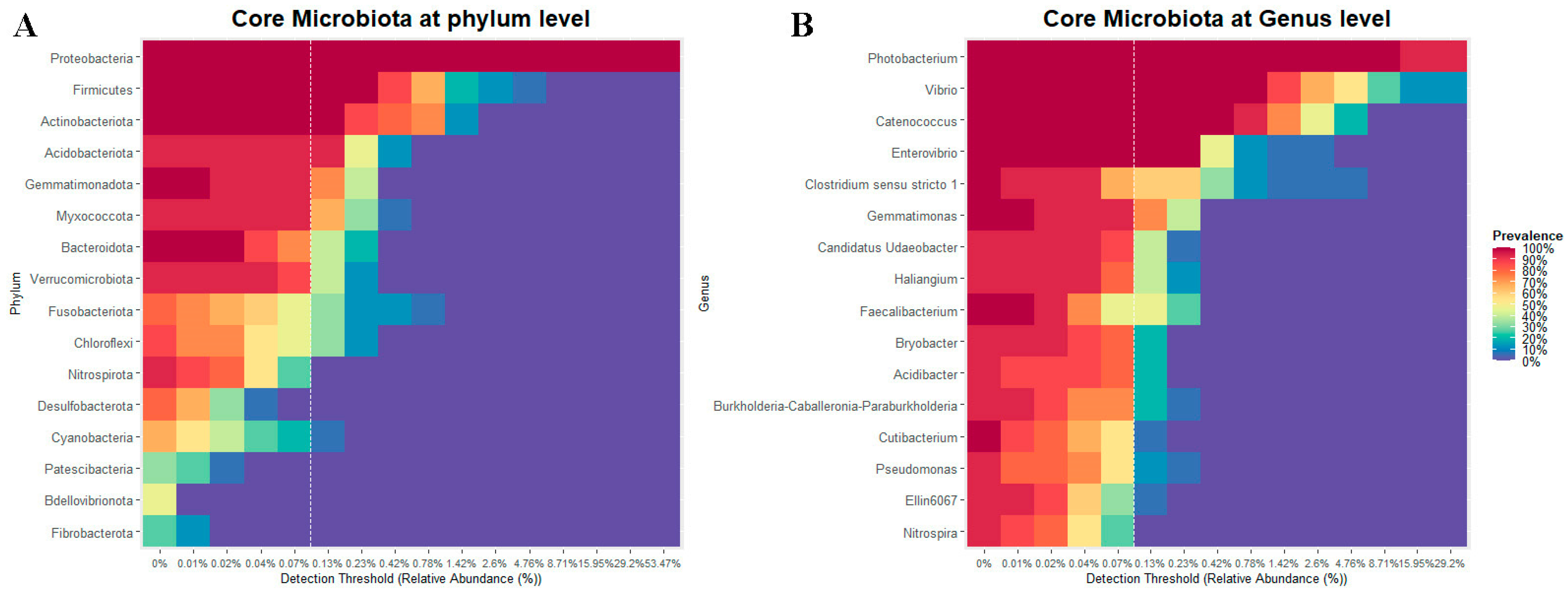
3.1.5. Core of the Bacterial Community
3.2. Fungal Community Composition
3.2.1. Effect of Diet on Alpha Diversity (Primary Outcome)
3.2.2. Effect of Diet on Beta Diversity
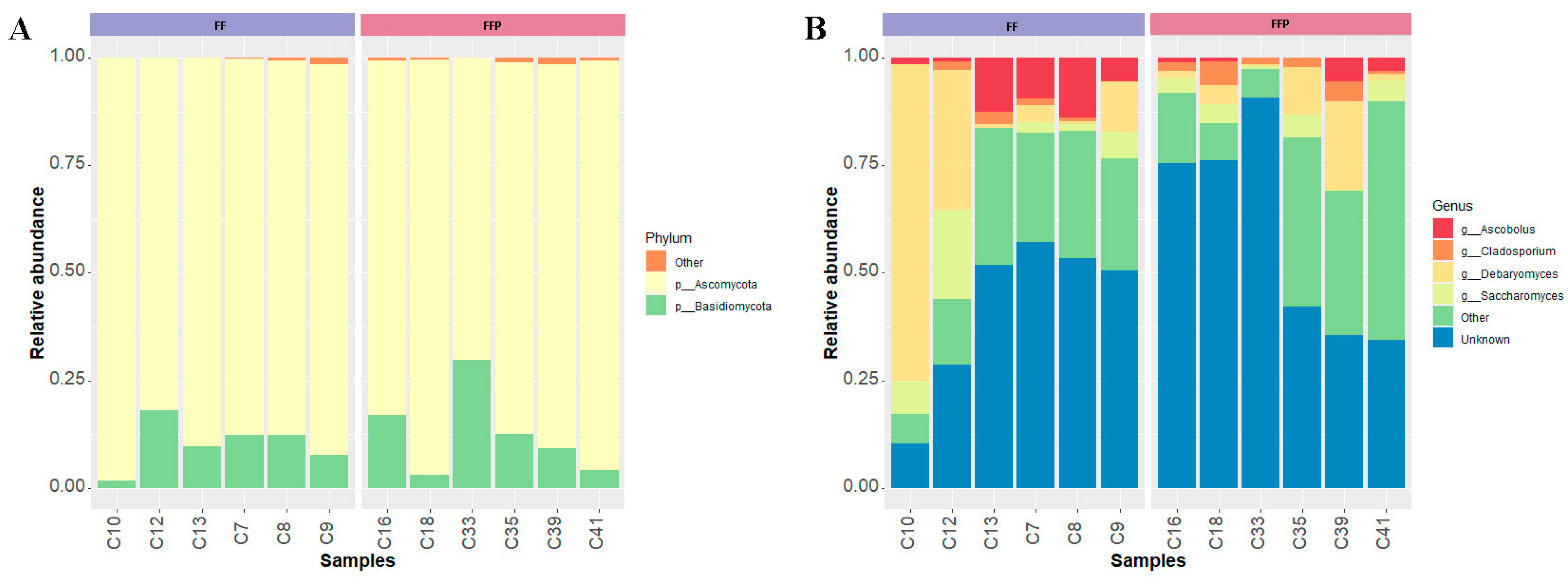
3.2.3. Differential Abundances of Fungal Communities
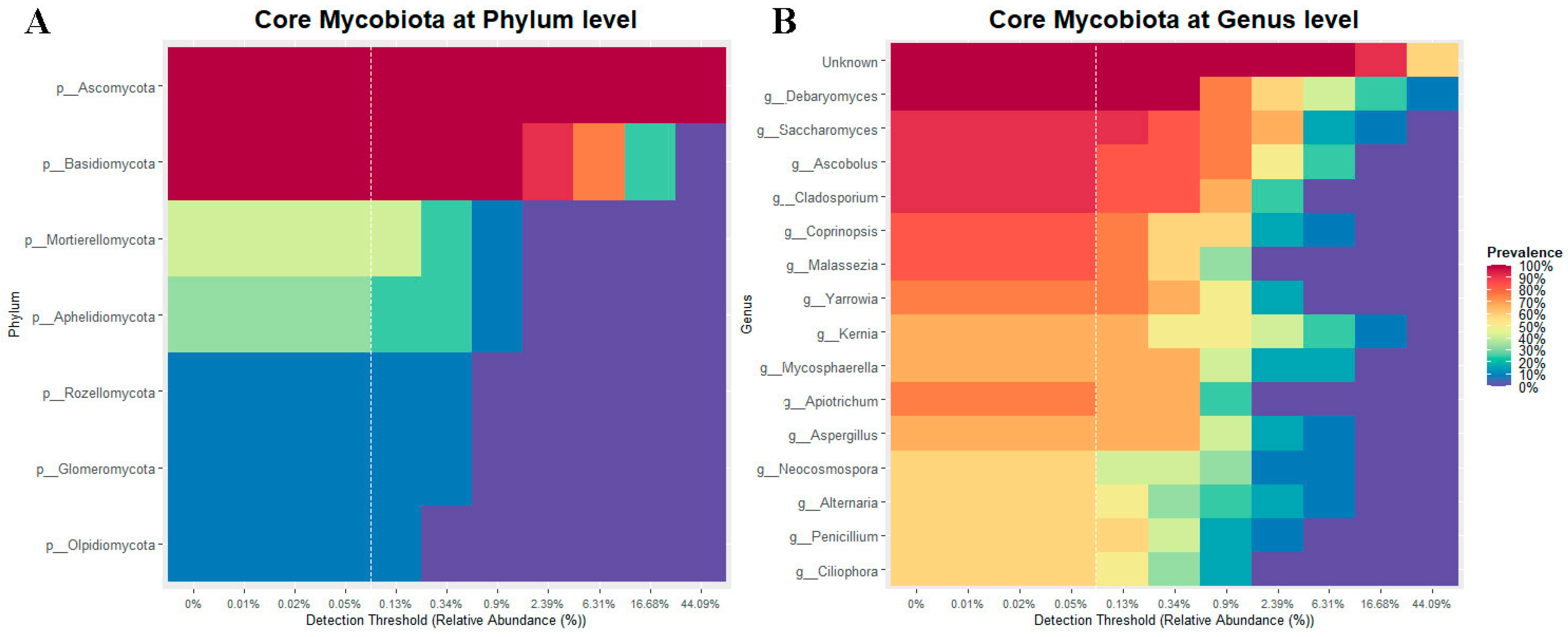
3.2.4. Core of the Mycobiota
3.3. Correlation between Bacterial and Fungal Communities
4. Discussion
4.1. Bacterial Community
4.2. Fungal Community
4.3. Correlation between Bacterial and Fungal Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Merrifield, D.L.; Rodiles, A. The Fish Microbiome and Its Interactions with Mucosal Tissues. In Mucosal Health in Aquaculture; Elsevier Inc.: Amsterdam, The Netherlands, 2015; pp. 273–295. ISBN 9780124171862. [Google Scholar]
- Nayak, S.K. Role of Gastrointestinal Microbiota in Fish. Aquac. Res. 2010, 41, 1553–1573. [Google Scholar] [CrossRef]
- Romero, J.; Ringø, E.; Merrifield, D.L. The Gut Microbiota of Fish. In Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics; Merrifield, D.L., Ringø, E., Eds.; Aquaculture Nutrition: West Sussex, UK, 2014; pp. 75–100. ISBN 9780470672716. [Google Scholar]
- Gómez, G.D.; Balcázar, J.L. A Review on the Interactions between Gut Microbiota and Innate Immunity of Fish. FEMS Immunol. Med. Microbiol. 2008, 52, 145–154. [Google Scholar] [CrossRef]
- Rawls, J.F.; Samuel, B.S.; Gordon, J.I. Gnotobiotic Zebrafish Reveal Evolutionarily Conserved Responses to the Gut Microbiota. Proc. Natl. Acad. Sci. USA 2004, 101, 4596–4601. [Google Scholar] [CrossRef] [PubMed]
- Zoetendal, E.G.; Von Wright, A.; Vilpponen-Salmela, T.; Ben-Amor, K.; Akkermans, A.D.L.; De Vos, W.M. Mucosa-Associated Bacteria in the Human Gastrointestinal Tract Are Uniformly Distributed along the Colon and Differ from the Community Recovered from Feces. Appl. Environ. Microbiol. 2002, 68, 3401–3407. [Google Scholar] [CrossRef]
- Nelson, J.S. Fishes of the World, 4th ed.; Nelson, J.S., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; ISBN 978-0-471-25031-9. [Google Scholar]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’Connor, M.P.; Rosen, G.L.; Knight, R.; Kilham, S.S.; Russell, J.A. Environmental and Ecological Factors That Shape the Gut Bacterial Communities of Fish: A Meta-Analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef] [PubMed]
- Benetti, D.; Sardenberg, B.; Welch, A.; Hoenig, R.; Orhun, M.R.; Zink, I. Intensive Larval Husbandry and Fingerling Production of Cobia Rachycentron canadum. Aquaculture 2008, 281, 22–27. [Google Scholar] [CrossRef]
- Rasheeda, M.K.; Rangamaran, V.R.; Srinivasan, S.; Ramaiah, S.K.; Gunasekaran, R.; Jaypal, S.; Gopal, D.; Ramalingam, K. Comparative Profiling of Microbial Community of Three Economically Important Fishes Reared in Sea Cages under Tropical Offshore Environment. Mar. Genom. 2017, 34, 57–65. [Google Scholar] [CrossRef]
- Sumithra, T.G.; Gayathri, S.; Krupesha Sharma, S.R.; Ebeneezar, S.; Anikuttan, K.K.; Sajina, K.A.; Iyyapparaja Narasimapallavan, G.; Reshma, K.J.; Vishnu, R.; Tamilmani, G.; et al. Metagenomic Signatures of Transportation Stress in the Early Life Stages of Cobia (Rachycentron canadum) to Aid in Mitigation Strategies. Aquaculture 2022, 559, 738407. [Google Scholar] [CrossRef]
- Villegas-Plazas, M.; Villamil, L.; Angélica Martínez-Silva, M.; González-Jiménez, T.; Salazar, M.; Güiza, L.; Mendoza, M.; Junca, H.; Org, M. Microbiome Composition and Autochthonous Probiotics from Contrasting Probiosis/Dysbiosis States in Cobia (Rachycentron canadum) Fish Epitheliocystis. Access Microbiol. 2022, 4, 405. [Google Scholar] [CrossRef]
- Wang, W.Z.; Huang, J.S.; Zhang, J.D.; Wang, Z.L.; Li, H.J.; Amenyogbe, E.; Chen, G. Effects of Hypoxia Stress on the Intestinal Microflora of Juvenile of Cobia (Rachycentron canadum). Aquaculture 2021, 536, 736419. [Google Scholar] [CrossRef]
- Raggi, P.; Lopez, P.; Diaz, A.; Carrasco, D.; Silva, A.; Velez, A.; Opazo, R.; Magne, F.; Navarrete, P. Debaryomyces hansenii and Rhodotorula mucilaginosa Comprised the Yeast Core Gut Microbiota of Wild and Reared Carnivorous Salmonids, Croaker and Yellowtail. Environ. Microbiol. 2014, 16, 2791–2803. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, B.; Ruiz, J.J.; Gutiérrez, M.S.; Alveal, K.; Caruffo, M.; Oliva, M.; Flores, H.; Silva, A.; Toro, M.; Reyes-Jara, A.; et al. Cultivable Yeast Microbiota from the Marine Fish Species Genypterus chilensis and Seriolella violacea. J. Fungi 2021, 7, 515. [Google Scholar] [CrossRef] [PubMed]
- Reinoso, S.; Gutiérrez, M.S.; Domínguez-Borbor, C.; Argüello-Guevara, W.; Bohórquez-Cruz, M.; Sonnenholzner, S.; Nova-Baza, D.; Mardones, C.; Navarrete, P. Selection of Autochthonous Yeasts Isolated from the Intestinal Tracts of Cobia Fish (Rachycentron canadum) with Probiotic Potential. J. Fungi 2023, 9, 274. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Bolchacova, E.; Voigt, K.; Crous, P.W.; et al. Nuclear Ribosomal Internal Transcribed Spacer (ITS) Region as a Universal DNA Barcode Marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Hoggard, M.; Vesty, A.; Wong, G.; Montgomery, J.M.; Fourie, C.; Douglas, R.G.; Biswas, K.; Taylor, M.W. Characterizing the Human Mycobiota: A Comparison of Small Subunit RRNA, ITS1, ITS2, and Large Subunit RRNA Genomic Targets. Front. Microbiol. 2018, 9, 2208. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome Diversity: High-Throughput Sequencing and Identification of Fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef]
- Marden, C.L.; McDonald, R.; Schreier, H.J.; Watts, J.E. Investigation into the Fungal Diversity within Different Regions of the Gastrointestinal Tract of Panaque Nigrolineatus, a Wood-Eating Fish. AIMS Microbiol. 2017, 3, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Siriyappagouder, P.; Kiron, V.; Lokesh, J.; Rajeish, M.; Kopp, M.; Fernandes, J. The Intestinal Mycobiota in Wild Zebrafish Comprises Mainly Dothideomycetes While Saccharomycetes Predominate in Their Laboratory-Reared Counterparts. Front. Microbiol. 2018, 9, 387. [Google Scholar] [CrossRef]
- Ghori, I.; Tabassum, M.; Ahmad, T.; Zuberi, A.; Imra, M. Geotrichum Candidum Enhanced the Enterococcus Faecium Impact in Improving Physiology, and Health of Labeo Rohita (Hamilton, 1822) by Modulating Gut Microbiome Under Mimic Aquaculture Conditions. Turkish J. Fish. Aquat. Sci. 2018, 18, 1255–1267. [Google Scholar] [CrossRef]
- Zhou, L.; Han, Y.; Wang, D.; Li, Y.; Huang, X.; He, A. Comparison of Fungal Community Composition within Different Intestinal Segments of Tilapia and Bighead Carp. J. Oceanol. Limnol. 2021, 39, 1961–1971. [Google Scholar] [CrossRef]
- Caruffo, M.; Navarrete, N.; Salgado, O.; Díaz, A.; López, P.; García, K.; Feijóo, C.G.; Navarrete, P. Potential Probiotic Yeasts Isolated from the Fish Gut Protect Zebrafish (Danio rerio) from a Vibrio Anguillarum Challenge. Front. Microbiol. 2015, 6, 1093. [Google Scholar] [CrossRef] [PubMed]
- Caruffo, M.; Navarrete, N.C.; Salgado, O.A.; Faúndez, N.B.; Gajardo, M.C.; Feijóo, C.G.; Reyes-Jara, A.; García, K.; Navarrete, P. Protective Yeasts Control V. anguillarum Pathogenicity and Modulate the Innate Immune Response of Challenged Zebrafish (Danio rerio) Larvae. Front. Cell. Infect. Microbiol. 2016, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Siriyappagouder, P.; Galindo-Villegas, J.; Lokesh, J.; Mulero, V.; Fernandes, J.M.O.; Kiron, V. Exposure to Yeast Shapes the Intestinal Bacterial Community Assembly in Zebrafish Larvae. Front. Microbiol. 2018, 9, 1868. [Google Scholar] [CrossRef] [PubMed]
- Vargas, O.; Gutiérrez, M.S.; Caruffo, M.; Valderrama, B.; Medina, D.A.; García, K.; Reyes-Jara, A.; Toro, M.; Feijóo, C.G.; Navarrete, P. Probiotic Yeasts and Vibrio anguillarum Infection Modify the Microbiome of Zebrafish Larvae. Front. Microbiol. 2021, 12, 1639. [Google Scholar] [CrossRef]
- Butt, R.L.; Volkoff, H. Gut Microbiota and Energy Homeostasis in Fish. Front. Endocrinol. 2019, 10, 6–8. [Google Scholar] [CrossRef]
- Gorokhova, E. Effects of Preservation and Storage of Microcrustaceans in RNAlater on RNA and DNA Degradation. Limnol. Oceanogr. Methods 2005, 3, 143–148. [Google Scholar] [CrossRef]
- RNAlater Solutions for RNA Stabilization and Storage. Available online: https://www.thermofisher.com/ec/en/home/brands/product-brand/rnalater.html (accessed on 30 January 2023).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Barnett, D.J.M.; Arts, I.C.W.; Penders, J. MicroViz: An R Package for Microbiome Data Visualization and Statistics. J. Open Source Softw. 2021, 6, 3201. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE Database for Molecular Identification of Fungi: Handling Dark Taxa and Parallel Taxonomic Classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Gao, J.; Peng, C.; Song, J.; Xie, Z.; Jia, J.; Li, H.; Zhao, S.; Liang, Y.; Gong, B. The Effect of the Microalgae Chlorella vulgaris on the Gut Microbiota of Juvenile Nile Tilapia (Oreochromis niloticus) Is Feeding-Time Dependent. Microorganisms 2023, 11, 1002. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.J.; Qiang, J.; Tao, Y.F.; Ngoepe, T.K.; Bao, J.W.; Chen, D.J.; Xu, P. Physiological and Gut Microbiome Changes Associated with Low Dietary Protein Level in Genetically Improved Farmed Tilapia (GIFT, Oreochromis niloticus) Determined by 16S RRNA Sequence Analysis. Microbiologyopen 2020, 9, e1000. [Google Scholar] [CrossRef]
- SHAPIRO, S.S.; WILK, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package 2022. Available online: https://github.com/vegandevs/vegan (accessed on 21 February 2023).
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R Package for Data Mining in Microbial Community Ecology. FEMS Microbiol. Ecol. 2021, 97, 255. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of Habitat-Specific Functional Profiles and Functional Redundancy Based on 16S RRNA Gene Sequences. Environ. Microbiomes 2020, 15, 11. [Google Scholar] [CrossRef]
- Larios-Soriano, E.; Zavala, R.C.; López, L.M.; Gómez-Gil, B.; Ramírez, D.T.; Sanchez, S.; Canales, K.; Galaviz, M.A. Soy Protein Concentrate Effects on Gut Microbiota Structure and Digestive Physiology of Totoaba Macdonaldi. J. Appl. Microbiol. 2022, 132, 1384–1396. [Google Scholar] [CrossRef]
- Dam, C.T.M.; Booth, M.; Pirozzi, I.; Salini, M.; Smullen, R.; Ventura, T.; Elizur, A. Alternative Feed Raw Materials Modulate Intestinal Microbiota and Its Relationship with Digestibility in Yellowtail Kingfish Seriola lalandi. Fishes 2020, 5, 14. [Google Scholar] [CrossRef]
- Larios-Soriano, E.; Re-Araujo, A.D.; Gómez-Gil, B.; Tovar-Ramírez, D.; Trejo-Escamilla, I.; Galaviz, M.A. Reciprocal Effect of Temperature and Dietary Lipids on Metabolic Performance and Gut Microbiota of Yellowtail Kingfish (Seriola lalandi) Juveniles. Aquac. Res. 2021, 52, 6189–6204. [Google Scholar] [CrossRef]
- Hartviksen, M.; Vecino, J.L.G.; Ringø, E.; Bakke, A.M.; Wadsworth, S.; Krogdahl, Å.; Ruohonen, K.; Kettunen, A. Alternative Dietary Protein Sources for Atlantic Salmon (Salmo salar L.) Effect on Intestinal Microbiota, Intestinal and Liver Histology and Growth. Aquac. Nutr. 2014, 20, 381–398. [Google Scholar] [CrossRef]
- Ingerslev, H.C.; von Gersdorff Jørgensen, L.; Lenz Strube, M.; Larsen, N.; Dalsgaard, I.; Boye, M.; Madsen, L. The Development of the Gut Microbiota in Rainbow Trout (Oncorhynchus mykiss) Is Affected by First Feeding and Diet Type. Aquaculture 2014, 424–425, 24–34. [Google Scholar] [CrossRef]
- Arias-Jayo, N.; Abecia, L.; Alonso-Sáez, L.; Ramirez-Garcia, A.; Rodriguez, A.; Pardo, M.A. High-Fat Diet Consumption Induces Microbiota Dysbiosis and Intestinal Inflammation in Zebrafish. Microb. Ecol. 2018, 76, 1089–1101. [Google Scholar] [CrossRef]
- Sumithra, T.G.; Sharma, S.R.K.; Gayathri, S.; Ebeneezar, S.; Reshma, K.J.; Anikuttan, K.K.; Narasimapallavan, G.I.; Rameshkumar, P.; Sakthivel, M.; Prabu, D.L.; et al. Comparative Evaluation of Fish Larval Preservation Methods on Microbiome Profiles to Aid in Metagenomics Research. Appl. Microbiol. Biotechnol. 2022, 106, 4719–4735. [Google Scholar] [CrossRef] [PubMed]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The Gut Microbiota of Marine Fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef] [PubMed]
- Sparagon, W.J.; Gentry, E.C.; Minich, J.J.; Vollbrecht, L.; Laurens, L.M.L.; Allen, E.E.; Sims, N.A.; Dorrestein, P.C.; Kelly, L.W.; Nelson, C.E. Fine Scale Transitions of the Microbiota and Metabolome along the Gastrointestinal Tract of Herbivorous Fishes. Anim. Microbiome 2022, 4, 33. [Google Scholar] [CrossRef]
- Legrand, T.P.R.A.; Catalano, S.R.; Wos-Oxley, M.L.; Wynne, J.W.; Weyrich, L.S.; Oxley, A.P.A. Antibiotic-Induced Alterations and Repopulation Dynamics of Yellowtail Kingfish Microbiota. Anim. Microbiome 2020, 2, 26. [Google Scholar] [CrossRef]
- Bledsoe, J.W.; Pietrak, M.R.; Burr, G.S.; Peterson, B.C.; Small, B.C. Functional Feeds Marginally Alter Immune Expression and Microbiota of Atlantic Salmon (Salmo salar) Gut, Gill, and Skin Mucosa Though Evidence of Tissue-Specific Signatures and Host–Microbe Coadaptation Remain. Anim. Microbiome 2022, 4, 20. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, Y.; Chen, K.; Yu, N.; Zhou, Z.; Chen, L.; Du, Z.; Li, E. Characterization of the Intestinal Microbiota in Pacific White Shrimp, Litopenaeus vannamei, Fed Diets with Different Lipid Sources. Aquaculture 2014, 434, 449–455. [Google Scholar] [CrossRef]
- Ramírez, C.; Romero, J. Fine Flounder (Paralichthys adspersus) Microbiome Showed Important Differences between Wild and Reared Specimens. Front. Microbiol. 2017, 8, 271. [Google Scholar] [CrossRef]
- Ramírez, C.; Romero, J. The Microbiome of Seriola lalandi of Wild and Aquaculture Origin Reveals Differences in Composition and Potential Function. Front. Microbiol. 2017, 8, 1844. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial Effects on Host Energy Metabolism of Short-Chain Fatty Acids and Vitamins Produced by Commensal and Probiotic Bacteria. Microb. Cell Fact. 2017, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- USDA FoodData Central. Available online: https://fdc.nal.usda.gov/fdc-app.html#/food-details/173711/nutrients (accessed on 1 March 2023).
- Hernandez de-Dios, M.A.; Tovar-Ramírez, D.; Maldonado García, D.; Galaviz-Espinoza, M.A.; Spanopoulos Zarco, M.; Maldonado-García, M.C. Functional Additives as a Boost to Reproductive Performance in Marine Fish: A Review. Fishes 2022, 7, 262. [Google Scholar] [CrossRef]
- Hennersdorf, P.; Mrotzek, G.; Abdul-Aziz, M.A.; Saluz, H.P. Metagenomic Analysis between Free-Living and Cultured Epinephelus Fuscoguttatus under Different Environmental Conditions in Indonesian Waters. Mar. Pollut. Bull. 2016, 110, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Nazemroaya, S.; Sahari, M.A.; Rezaei, M. Effect of Frozen Storage on Fatty Acid Composition and Changes in Lipid Content of Scomberomorus commersoni and Carcharhinus dussumieri. J. Appl. Ichthyol. 2009, 25, 91–95. [Google Scholar] [CrossRef]
- Nakazawa, N.; Okazaki, E. Recent Research on Factors Influencing the Quality of Frozen Seafood. Fish. Sci. 2020, 86, 231–244. [Google Scholar] [CrossRef]
- Veyrand-Quirós, B.; Guzmán-Villanueva, L.T.; Reyes, A.G.; Rodríguez-Jaramillo, C.; Salas-Leiva, J.; Tovar-Ramírez, D.; Balcázar, J.L.; Quiroz-Guzman, E. Assessment of Bacteriophage VB_Pd_PDCC-1 on Bacterial Dynamics during Ontogenetic Development of the Longfin Yellowtail (Seriola rivoliana). Appl. Microbiol. Biotechnol. 2021, 105, 2877–2887. [Google Scholar] [CrossRef]
- González-Félix, M.L.; Gatlin, D.M.; Urquidez-Bejarano, P.; de la Reé-Rodríguez, C.; Duarte-Rodríguez, L.; Sánchez, F.; Casas-Reyes, A.; Yamamoto, F.Y.; Ochoa-Leyva, A.; Perez-Velazquez, M. Effects of Commercial Dietary Prebiotic and Probiotic Supplements on Growth, Innate Immune Responses, and Intestinal Microbiota and Histology of Totoaba Macdonaldi. Aquaculture 2018, 491, 239–251. [Google Scholar] [CrossRef]
- Leyva-López, N.; Osuna-García, E.; Hernández, C.; Gómez-Gil, B.; Soto-Rodríguez, S.; Guerrero, A. A Preliminary Study of the Effect of Total Fishmeal Replacement with Different Dietary Sources on the Gut Microbiota of Spotted Rose Snapper Juvenile (Lutjanus guttatus Steindachner, 1869). Aquac. Res. 2020, 51, 4771–4784. [Google Scholar] [CrossRef]
- Reyes, G.; Betancourt, I.; Andrade, B.; Panchana, F.; Román, R.; Sorroza, L.; Trujillo, L.E.; Bayot, B. Microbiome of Penaeus vannamei Larvae and Potential Biomarkers Associated with High and Low Survival in Shrimp Hatchery Tanks Affected by Acute Hepatopancreatic Necrosis Disease. Front. Microbiol. 2022, 13, 1227. [Google Scholar] [CrossRef]
- Guibert, I.; Lecellier, G.; Torda, G.; Pochon, X.; Berteaux-Lecellier, V. Metabarcoding Reveals Distinct Microbiotypes in the Giant Clam Tridacna maxima. Microbiome 2020, 8, 57. [Google Scholar] [CrossRef]
- Shimada, M.; Yunis-Aguinaga, J.; Cueva-Quiroz, V.; Filho, J.; Mouriño, J.; Claudiano, G.; Moraes, F.; Moraes, J. Isolation and Characterization of Pathology in Case of Massive Mortality by Photobacterium damselae subsp. piscicida in Rachycentron canadum. Biosci. J. 2019, 36, 1732–1741. [Google Scholar] [CrossRef]
- Actis, L.A.; Tolmasky, M.E.; Crosa, J.H. Vibriosis. In Fish Diseases and Disorders: Viral, Bacterial and Fungal Infections; Woo, P.T.K., Bruno, D.W., Eds.; CABI: Wallingford, UK, 2011; Volume 3, pp. 570–605. [Google Scholar]
- Ma, S.; Shu, X.; Wang, W.X. Responses of Two Marine Fish to Organically Complexed Zn: Insights from Microbial Community and Liver Transcriptomics. Sci. Total Environ. 2022, 835, 155457. [Google Scholar] [CrossRef] [PubMed]
- Větrovský, T.; Baldrian, P. The Variability of the 16S RRNA Gene in Bacterial Genomes and Its Consequences for Bacterial Community Analyses. PLoS ONE 2013, 8, e57923. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Chen, C.; Jia, L.; He, X.; Zhang, B. Comparison of the Intestinal Microbiota Composition and Function in Healthy and Diseased Yunlong Grouper. AMB Express 2019, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, S.J.R.; Lee, K.C.; Handley, K.M.; Angert, E.R.; White, W.L.; Clements, K.D. Substrate Degradation Pathways, Conserved Functions and Community Composition of the Hindgut Microbiota in the Herbivorous Marine Fish Kyphosus sydneyanus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2022, 272, 111283. [Google Scholar] [CrossRef] [PubMed]
- Parra, M.; Espinoza, D.; Valdes, N.; Vargas, R.; Gonzalez, A.; Modak, B.; Tello, M. Microbiota Modulates the Immunomodulatory Effects of Filifolinone on Atlantic Salmon. Microorganisms 2020, 8, 1320. [Google Scholar] [CrossRef] [PubMed]
- Andlid, T.; Vázquez-Juárez, R.; Gustafsson, L. Yeasts Isolated from the Intestine of Rainbow Trout Adhere to and Grow in Intestinal Mucus. Mol. Mar. Biol. Biotechnol. 1998, 7, 115–126. [Google Scholar]
- Li, Y.; Bruni, L.; Jaramillo-Torres, A.; Gajardo, K.; Kortner, T.M.; Krogdahl, Å. Differential Response of Digesta- and Mucosa-Associated Intestinal Microbiota to Dietary Insect Meal during the Seawater Phase of Atlantic Salmon. Anim. Microbiome 2021, 3, 8. [Google Scholar] [CrossRef]
- Nagahama, T. Yeast Biodiversity in Freshwater, Marine and Deep-Sea Environments. In Biodiversity and Ecophysiology of Yeasts; Péter, G., Rosa, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 241–262. ISBN 2626-8868. [Google Scholar]
- Zhou, A.; Xie, S.; Junaid, M.; Sun, D.; Tang, H.; Chuan, J.; Li, X.; Xu, G.; Zou, J. First Insight into the Environmental Microbial Communities Associated with Potentially Pathogenic Strains in Pond Cultured Tilapia (Oreochromis niloticus) at Various Growth Stages Based on 16S, 18S, and ITS2 RRNA Gene Amplicons Sequencing. Aquaculture 2021, 532, 736007. [Google Scholar] [CrossRef]
- Yanong, R.P.E. Fungal Diseases of Fish. Vet. Clin. N. Am. Exot. Anim. Pract. 2003, 6, 377–400. [Google Scholar] [CrossRef]
- Navarrete, P.; Tovar-Ramírez, D. Use of Yeasts as Probiotics in Fish Aquaculture. In Sustainable Aquaculture Techniques; Hernandez-Vergara, M., Perez-Rostro, C., Eds.; IntechOpen: London, UK, 2014; pp. 135–172. ISBN 978-953-51-1224-2. [Google Scholar]
- Harikrishnan, R.; Kim, M.C.; Kim, J.S.; Balasundaram, C.; Heo, M.S. Immunomodulatory Effect of Probiotics Enriched Diets on Uronema Marinum Infected Olive Flounder. Fish Shellfish Immunol. 2011, 30, 964–971. [Google Scholar] [CrossRef] [PubMed]
- Abu-Elala, N.; Marzouk, M.; Moustafa, M. Use of Different Saccharomyces cerevisiae Biotic Forms as Immune-Modulator and Growth Promoter for Oreochromis niloticus Challenged with Some Fish Pathogens. Int. J. Vet. Sci. Med. 2013, 1, 21–29. [Google Scholar] [CrossRef]
- Chiu, C.H.; Cheng, C.H.; Gua, W.R.; Guu, Y.K.; Cheng, W. Dietary Administration of the Probiotic, Saccharomyces cerevisiae P13, Enhanced the Growth, Innate Immune Responses, and Disease Resistance of the Grouper, Epinephelus coioides. Fish Shellfish Immunol. 2010, 29, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Lissouba, P.; Mousseau, J.; Rizet, G.; Rossignol, J.L. Fine Structure of Genes in the Ascomycete Ascobolus Immersus. Adv. Genet. 1963, 11, 343–380. [Google Scholar] [CrossRef]
- Lamb, B.C. Ascobolus. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: London, UK, 2013; pp. 204–205. [Google Scholar] [CrossRef]
- van Brummelen, J. A World Monograph of the Genera Ascobolus and Saccobolus (Ascomycetes, Pezizales). Persoonia Suppl. 1967, 1, 1–260. [Google Scholar]
- Santus, W.; Devlin, J.R.; Behnsen, J. Crossing Kingdoms: How the Mycobiota and Fungal-Bacterial Interactions Impact Host Health and Disease. Infect. Immun. 2021, 89, e00648-20. [Google Scholar] [CrossRef]



| Index | Diet | p-Value | |
|---|---|---|---|
| FF | FFP | ||
| Chao1 | 645.22 ± 126.38 | 668.37 ± 156.33 | 0.760 |
| Richness | 634.00 ± 125.32 | 659.38 ± 152.42 | 0.733 |
| Simpson | 0.99 ± 0.00 | 0.99 ± 0.00 | 0.127 |
| Inv. Simpson | 124.96 ± 36.71 | 167.12 ± 60.44 | 0.133 |
| Shannon | 5.18 ± 0.31 | 5.45 ± 0.30 | 0.112 |
| ACE | 642.51 ± 125.57 | 667.17 ± 155.20 | 0.743 |
| Index | Diet | p-Value | |
|---|---|---|---|
| FF | FFP | ||
| Chao1 | 68.29 ± 30.50 | 68.14 ± 13.39 | 0.148 |
| Richness | 62.33 ± 25.70 | 64.83 ± 11.69 | 0.109 |
| Simpson | 0.89 ± 0.11 | 0.92 ± 0.03 | 0.872 |
| Inv. Simpson | 16.55 ± 10.20 | 15.42 ± 8.22 | 0.336 |
| Shannon | 3.07 ± 0.75 | 3.22 ± 0.31 | 1.000 |
| ACE | 67.12 ± 28.92 | 68.09 ± 13.51 | 0.109 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reinoso, S.; Gutiérrez, M.S.; Reyes-Jara, A.; Toro, M.; García, K.; Reyes, G.; Argüello-Guevara, W.; Bohórquez-Cruz, M.; Sonnenholzner, S.; Navarrete, P. Feed Regime Slightly Modifies the Bacterial but Not the Fungal Communities in the Intestinal Mucosal Microbiota of Cobia Fish (Rachycentron canadum). Microorganisms 2023, 11, 2315. https://doi.org/10.3390/microorganisms11092315
Reinoso S, Gutiérrez MS, Reyes-Jara A, Toro M, García K, Reyes G, Argüello-Guevara W, Bohórquez-Cruz M, Sonnenholzner S, Navarrete P. Feed Regime Slightly Modifies the Bacterial but Not the Fungal Communities in the Intestinal Mucosal Microbiota of Cobia Fish (Rachycentron canadum). Microorganisms. 2023; 11(9):2315. https://doi.org/10.3390/microorganisms11092315
Chicago/Turabian StyleReinoso, Samira, María Soledad Gutiérrez, Angélica Reyes-Jara, Magaly Toro, Katherine García, Guillermo Reyes, Wilfrido Argüello-Guevara, Milton Bohórquez-Cruz, Stanislaus Sonnenholzner, and Paola Navarrete. 2023. "Feed Regime Slightly Modifies the Bacterial but Not the Fungal Communities in the Intestinal Mucosal Microbiota of Cobia Fish (Rachycentron canadum)" Microorganisms 11, no. 9: 2315. https://doi.org/10.3390/microorganisms11092315
APA StyleReinoso, S., Gutiérrez, M. S., Reyes-Jara, A., Toro, M., García, K., Reyes, G., Argüello-Guevara, W., Bohórquez-Cruz, M., Sonnenholzner, S., & Navarrete, P. (2023). Feed Regime Slightly Modifies the Bacterial but Not the Fungal Communities in the Intestinal Mucosal Microbiota of Cobia Fish (Rachycentron canadum). Microorganisms, 11(9), 2315. https://doi.org/10.3390/microorganisms11092315











