An Indirect Fluorescence Microscopy Method to Assess Vaginal Lactobacillus Concentrations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Conditions and Strains
2.2. Colony-Forming Units Counting Method
2.3. Flow Cytometry Quantification
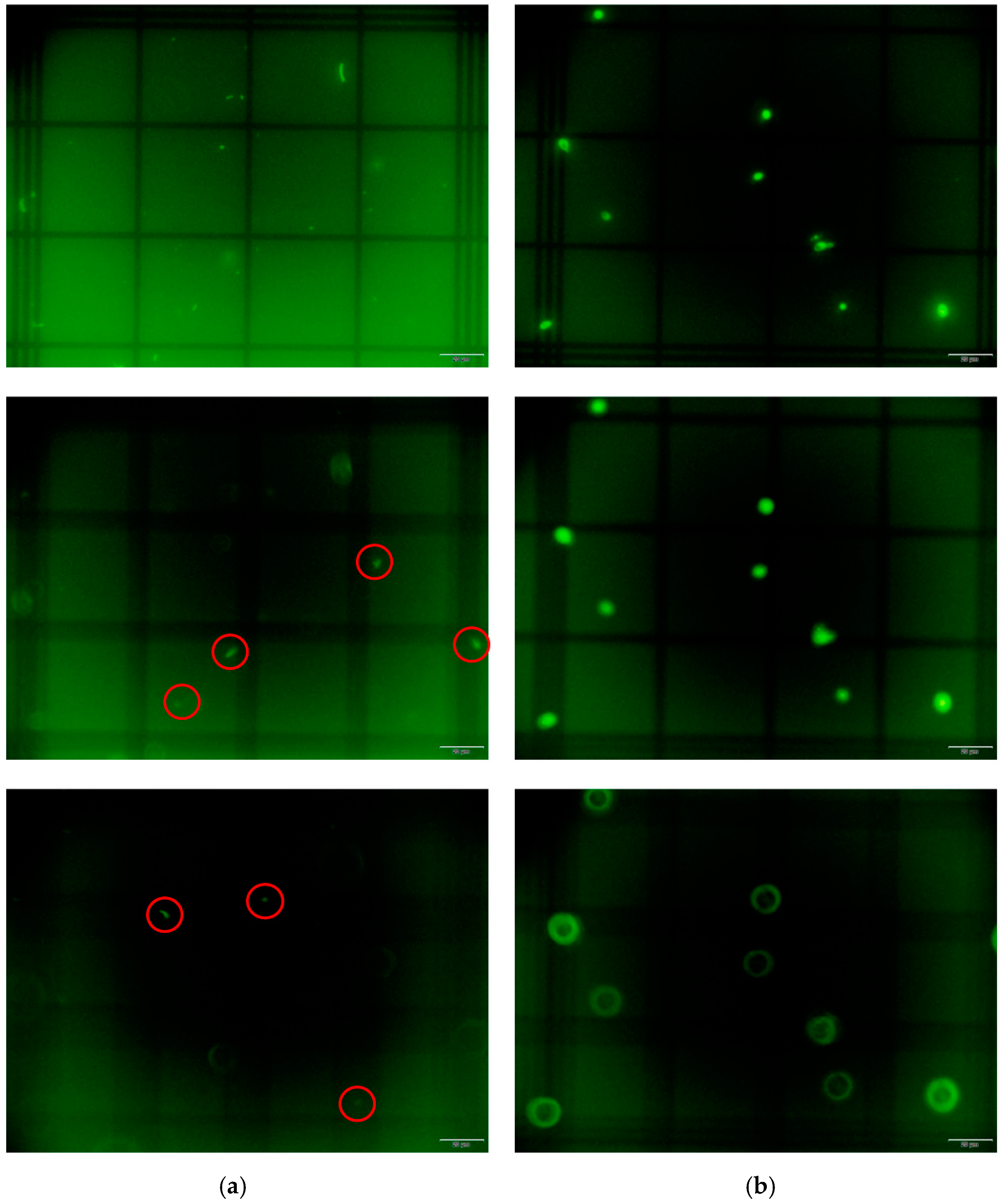
2.4. Visualization of Bacteria and Yeast in the Neubauer Chamber
2.5. Fluorescence Microscopy Quantification of Total Counts
2.6. Indirect Assessment of Bacterial Concentration by Measurement of Bacterial Surface Area
2.7. Statistical Analysis
3. Results
3.1. Comparison of CFU and FC in the Quantification of Vaginal Lactobacilli
3.2. Neubauer Chamber to Quantify Bacteria vs. Yeast
3.3. Correlation of Surface Area Bacterial Measurement with Initial Bacterial Density
3.4. Quantification of Lactobacillus spp. Concentration by an Indirect Microscopy Method
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chee, W.; Chew, S.; Than, L. Vaginal Microbiota and the Potential of Lactobacillus Derivatives in Maintaining Vaginal Health. Microb. Cell Fact. 2020, 19, 203. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Forney, L.J. Vaginal Microbiome of Reproductive-Age Women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef] [PubMed]
- Witkin, S.; Linhares, I. Why Do Lactobacilli Dominate the Human Vaginal Microbiota? BJOG Int. J. Obstet. Gynaecol. 2016, 124, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Petrova, M.I.; Reid, G.; Vaneechoutte, M.; Lebeer, S. Lactobacillus Iners: Friend or Foe? Trends Microbiol. 2017, 25, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Guo, R.; Wang, J.; Zhou, W.; Ling, Z. Contribution of Lactobacillus Iners to Vaginal Health and Diseases: A Systematic Review. Front. Cell. Infect. Microbiol. 2021, 11, 1177. [Google Scholar] [CrossRef] [PubMed]
- Pramanick, R.; Mayadeo, N.; Warke, H.; Begum, S.; Aich, P.; Aranha, C. Vaginal Microbiota of Asymptomatic Bacterial Vaginosis and Vulvovaginal Candidiasis: Are They Different from Normal Microbiota? Microb. Pathog. 2019, 134, 103599. [Google Scholar] [CrossRef]
- Bagnall, P.; Rizzolo, D. Bacterial Vaginosis: A Practical Review. J. Am. Acad. Physician Assist. 2017, 30, 15–21. [Google Scholar] [CrossRef]
- Reid, G. Is Bacterial Vaginosis a Disease? Appl. Microbiol. Biotechnol. 2018, 102, 553–558. [Google Scholar] [CrossRef]
- van de Wijgert, J.H.; Jespers, V. The Global Health Impact of Vaginal Dysbiosis. Res. Microbiol. 2017, 168, 859–864. [Google Scholar] [CrossRef]
- Turpin, R.; Tuddenham, S.; He, X.; Klebanoff, M.A.; Ghanem, K.G.; Brotman, R.M. Bacterial Vaginosis and Behavioral Factors Associated with Incident Pelvic Inflammatory Disease in the Longitudinal Study of Vaginal Flora. J. Infect. Dis. 2021, 224, S137–S144. [Google Scholar] [CrossRef]
- Shimaoka, M.; Yo, Y.; Doh, K.; Kotani, Y.; Suzuki, A.; Tsuji, I.; Mandai, M.; Matsumura, N. Association between Preterm Delivery and Bacterial Vaginosis with or without Treatment. Sci. Rep. 2019, 9, 509. [Google Scholar] [CrossRef] [PubMed]
- Hay, P. Bacterial Vaginosis. Medicine 2014, 42, 359–363. [Google Scholar] [CrossRef]
- Ravel, J.; Moreno, I.; Simón, C. Bacterial Vaginosis and Its Association with Infertility, Endometritis, and Pelvic Inflammatory Disease. Am. J. Obstet. Gynecol. 2021, 224, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.R.; Lingappa, J.R.; Baeten, J.M.; Ngayo, M.O.; Spiegel, C.A.; Hong, T.; Donnell, D.; Celum, C.; Kapiga, S.; Delany, S.; et al. Bacterial Vaginosis Associated with Increased Risk of Female-to-Male HIV-1 Transmission: A Prospective Cohort Analysis among African Couples. PLoS Med. 2012, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Bautista, C.T.; Wurapa, E.; Sateren, W.B.; Morris, S.; Hollingsworth, B.; Sanchez, J.L. Bacterial Vaginosis: A Synthesis of the Literature on Etiology, Prevalence, Risk Factors, and Relationship with Chlamydia and Gonorrhea Infections. Mil. Med. Res. 2016, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Abbe, C.; Mitchell, C.M. Bacterial Vaginosis: A Review of Approaches to Treatment and Prevention. Front. Reprod. Health 2023, 5, 1100029. [Google Scholar] [CrossRef] [PubMed]
- Onderdonk, A.B.; Delaney, M.L.; Fichorova, R.N. The Human Microbiome during Bacterial Vaginosis. Clin. Microbiol. Rev. 2016, 29, 223–238. [Google Scholar] [CrossRef]
- Muzny, C.; Taylor, C.; Swords, W.; Tamhane, A.; Chattopadhyay, D.; Cerca, N.; Schwebke, J. An Updated Conceptual Model on the Pathogenesis of Bacterial Vaginosis. J. Infect. Dis. 2019, 220, 1399–1405. [Google Scholar] [CrossRef]
- Castro, J.; Rosca, A.; Muzny, C.; Cerca, N. Atopobium vaginae and Prevotella bivia Are Able to Incorporate and Influence Gene Expression in a Pre-Formed Gardnerella vaginalis Biofilm. Pathogens 2021, 10, 247. [Google Scholar] [CrossRef]
- Castro, J.; Machado, D.; Cerca, N. Unveiling the Role of Gardnerella vaginalis in Polymicrobial Bacterial Vaginosis Biofilms: The Impact of Other Vaginal Pathogens Living as Neighbors. ISME J. 2019, 13, 1306–1317. [Google Scholar] [CrossRef]
- Duar, R.M.; Lin, X.B.; Zheng, J.; Elisa, P.; Martino, E.; Michael, G.; Bernard, C.; Sciences, B.; Tg, A.B. Lifestyles in Transition: Evolution and Natural History of the Genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, 27–48. [Google Scholar] [CrossRef] [PubMed]
- Tachedjian, G.; Aldunate, M.; Bradshaw, C.S.; Cone, R.A. The Role of Lactic Acid Production by Probiotic Lactobacillus Species in Vaginal Health. Res. Microbiol. 2017, 168, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Parente, E.; Ricciardi, A. Aerobic Metabolism in the Genus Lactobacillus: Impact on Stress Response and Potential Applications in the Food Industry. J. Appl. Microbiol. 2017, 122, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Säde, E.; Johansson, P.; Heinonen, T.; Hultman, J.; Bjorkroth, J. Growth and Metabolic Characteristics of Fastidious Meat-Derived Lactobacillus algidus Strains. Int. J. Food Microbiol. 2020, 313, 108379. [Google Scholar] [CrossRef] [PubMed]
- Davis, C. Enumeration of Probiotic Strains: Review of Culture-Dependent and Alternative Techniques to Quantify Viable Bacteria. J. Microbiol. Methods 2014, 103, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.; Candice, L.; Bazana, G.; Fl, M.; Fuentefria, A.M. Curve fitting and linearization of UV–Vis spectrophotometric measurements to estimate yeast in inoculum preparation. Anal. Biochem. 2021, 625, 114216. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.J.; Lee, J.E.; Lee, C.H. Importance of Lactic Acid Bacteria in Asian Fermented Foods. Microb. Cell Fact. 2011, 10, S5. [Google Scholar] [CrossRef]
- Yu, J.; Wang, H.M.; Zha, M.S.; Qing, Y.T.; Bai, N.; Ren, Y.; Xi, X.X.; Liu, W.J.; Menghe, B.L.G.; Zhang, H.P. Molecular Identification and Quantification of Lactic Acid Bacteria in Traditional Fermented Dairy Foods of Russia. J. Dairy Sci. 2015, 98, 5143–5154. [Google Scholar] [CrossRef]
- Lhomme, E.; Orain, S.; Courcoux, P.; Onno, B.; Dousset, X. The Predominance of Lactobacillus sanfranciscensis in French Organic Sourdoughs and Its Impact on Related Bread Characteristics. Int. J. Food Microbiol. 2015, 213, 40–48. [Google Scholar] [CrossRef]
- Kusters, J.G.; Reuland, E.A.; Bouter, S.; Koenig, P.; Dorigo-Zetsma, J.W. A Multiplex Real-Time PCR Assay for Routine Diagnosis of Bacterial Vaginosis. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1779–1785. [Google Scholar] [CrossRef]
- Gobert, G.; Cotillard, A.; Fourmestraux, C.; Pruvost, L.; Miguet, J.; Boyer, M. Droplet Digital PCR Improves Absolute Quantification of Viable Lactic Acid Bacteria in Faecal Samples. J. Microbiol. Methods 2018, 148, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Jahromi, M.F.; Altaher, Y.W.; Shokryazdan, P.; Ebrahimi, R.; Ebrahimi, M.; Idrus, Z.; Tufarelli, V.; Liang, J.B. Dietary Supplementation of a Mixture of Lactobacillus Strains Enhances Performance of Broiler Chickens Raised under Heat Stress Conditions. Int. J. Biometeorol. 2016, 60, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Bogachev, M.I.; Volkov, V.Y.; Markelov, O.A.; Trizna, E.Y.; Baydamshina, R.; Melnikov, V.; Murtazina, R.R.; Zelenikhin, P.V.; Sharafutdinov, S.; Kayumov, A.R. Fast and Simple Tool for the Quantification of Biofilm-Embedded Cells Sub-Populations from Fluorescent Microscopic Images. PLoS ONE 2018, 13, e0193267. [Google Scholar] [CrossRef] [PubMed]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.J.; Osborn, A.M. Advantages and Limitations of Quantitative PCR (Q-PCR)-Based Approaches in Microbial Ecology. FEMS Microbiol. Ecol. 2009, 67, 6–20. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.; França, A.; Muzny, C.A.; Taylor, C.M.; Cerca, N. DNA Extraction Leads to Bias in Bacterial Quantification by QPCR. Appl. Microbiol. Biotechnol. 2022, 106, 7993–8006. [Google Scholar] [CrossRef] [PubMed]
- Hammes, F.; Egli, T. Cytometric Methods for Measuring Bacteria in Water: Advantages, Pitfalls and Applications. Anal. Bioanal. Chem. 2010, 397, 1083–1095. [Google Scholar] [CrossRef]
- Van Nevel, S.; Koetzsch, S.; Proctor, C.R.; Besmer, M.D.; Prest, E.I.; Vrouwenvelder, J.S.; Knezev, A.; Boon, N.; Hammes, F. Flow Cytometric Bacterial Cell Counts Challenge Conventional Heterotrophic Plate Counts for Routine Microbiological Drinking Water Monitoring. Water Res. 2017, 113, 191–206. [Google Scholar] [CrossRef]
- Davey, H.M. Life, Death, and in-between: Meanings and Methods in Microbiology. Appl. Environ. Microbiol. 2011, 77, 5571–5576. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.; Olsen, G.; Davis, J.; Disz, T.; Edwards, R.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, 206–214. [Google Scholar] [CrossRef]
- Stewart, C.; Giannini, J. Inexpensive, Open Source Epifluorescence Microscopes. J. Chem. Educ. 2016, 93, 1310–1315. [Google Scholar] [CrossRef]
- Bui, S.; Dalvin, S.; Vågseth, T.; Oppedal, F.; Fossøy, F.; Brandsegg, H.; Skern-Mauritzen, R. Finding the Needle in the Haystack: Comparison of Methods for Salmon Louse Enumeration in Plankton Samples. Aquac. Res. 2021, 52, 3591–3604. [Google Scholar] [CrossRef]
- Seo, E.; Ahn, T.; Zo, Y. Agreement, Precision, and Accuracy of Epifluorescence Microscopy Methods for Enumeration of Total Bacterial Numbers†. Appl. Environ. Microbiol. 2010, 76, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- Gordeev, A.A.; Chetverin, A.B. Methods for Screening Live Cells. Biochemistry 2018, 83, 81–102. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.; Salgueiro, D.; Harwich, M.; Jefferson, K.K.; Cerca, N. Quantitative Analysis of Initial Adhesion of Bacterial Vaginosis-Associated Anaerobes to ME-180 Cells. Anaerobe 2013, 23, 1–4. [Google Scholar] [CrossRef]
- Castro, J.; Henriques, A.; Henriques, M.; Jefferson, K.K. Reciprocal Interference between Lactobacillus spp. and Gardnerella Vaginalis on Initial Adherence to Epithelial Cells. Int. J. Med. Microbiol. 2013, 10, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, L.; Costa, R.; Henriques, M.; Rodrigues, M.E. Simulated Vaginal Fluid: Candida Resistant Strains’ Biofilm Characterization and Vapor Phase of Essential Oil Effect. J. Med. Mycol. 2023, 33, 101329. [Google Scholar] [CrossRef]
- Castro, J.; Lima, A.; Sousa, L.; Rosca, A.S.; Muzny, C.A.; Cerca, N. Crystal Violet Staining Alone Is Not Adequate to Assess Synergism or Antagonism in Multi-Species Bio Fi Lms of Bacteria Associated with Bacterial Vaginosis. Front. Cell. Infect. Microbiol. 2022, 11, 1375. [Google Scholar] [CrossRef]
- Wohlsen, T.; Bates, J.; Vesey, G.; Robinson, W.A.; Katouli, M. Evaluation of the Methods for Enumerating Coliform Bacteria from Water Samples Using Precise Reference Standards. Lett. Appl. Microbiol. 2006, 42, 350–356. [Google Scholar] [CrossRef]
- Sakamoto, M.; Takeuchi, Y.; Umeda, M.; Ishikawa, I.; Benno, Y. Rapid Detection and Quantification of Five Periodontopathic Bacteria by Real-Time PCR. Microbiol. Immunol. 2001, 45, 39–44. [Google Scholar] [CrossRef]
- Clavel, M.; Barraud, O.; Moucadel, V.; Meynier, F.; Karam, E.; Ploy, M.C.; François, B.; Pichon, N.; Vignon, P.; Droual, R.; et al. Molecular Quantification of Bacteria from Respiratory Samples in Patients with Suspected Ventilator-Associated Pneumonia. Clin. Microbiol. Infect. 2016, 22, 812.e1–812.e7. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, N.; Xu, Y.; Goodacre, R. Detection and Quantification of Bacterial Spoilage in Milk and Pork Meat Using MALDI-TOF-MS and Multivariate Analysis. Anal. Chem. 2012, 84, 5951–5958. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.Q.; Yu, Y.N.; Gao, R.W.; Wang, H.; Zhang, J.; Li, R.; Long, X.H.; Shen, Q.R.; Chen, W.; Cai, F. High-Throughput Absolute Quantification Sequencing Reveals the Effect of Different Fertilizer Applications on Bacterial Community in a Tomato Cultivated Coastal Saline Soil. Sci. Total Environ. 2019, 687, 601–609. [Google Scholar] [CrossRef] [PubMed]
- López-Gutiérrez, J.C.; Henry, S.; Hallet, S.; Martin-Laurent, F.; Catroux, G.; Philippot, L. Quantification of a Novel Group of Nitrate-Reducing Bacteria in the Environment by Real-Time PCR. J. Microbiol. Methods 2004, 57, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.J.; Feng, Y.S.; Sung, W.P.; Surampalli, R.Y. Quantification and Analysis of Airborne Bacterial Characteristics in a Nursing Care Institution. J. Air Waste Manag. Assoc. 2011, 61, 732–739. [Google Scholar] [CrossRef] [PubMed]
- García, M.R.; Cabo, M.L. Optimization of E. coli Inactivation by Benzalkonium Chloride Reveals the Importance of Quantifying the Inoculum Effect on Chemical Disinfection. Front. Microbiol. 2018, 9, 1259. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, V.; Dey, A.; Ghosh, S.; Bajpai, S.; Jha, M.K. Quantification of Bacterial Adherence on Different Textile Fabrics. Int. Biodeterior. Biodegrad. 2011, 65, 1169–1174. [Google Scholar] [CrossRef]
- Zhang, L. Machine Learning for Enumeration of Cell Colony Forming Units. Vis. Comput. Ind. Biomed. Art 2022, 5, 26. [Google Scholar] [CrossRef]
- Stingl, K.; Heise, J.; Thieck, M.; Wulsten, I.F.; Pacholewicz, E.; Iwobi, A.N.; Govindaswamy, J.; Zeller-Péronnet, V.; Scheuring, S.; Luu, H.Q.; et al. Challenging the “Gold Standard” of Colony-Forming Units-Validation of a Multiplex Real-Time PCR for Quantification of Viable Campylobacter spp. in Meat Rinses. Int. J. Food Microbiol. 2021, 359, 109417. [Google Scholar] [CrossRef]
- Zandri, G.; Pasquaroli, S.; Vignaroli, C.; Talevi, S.; Manso, E.; Donelli, G.; Biavasco, F. Detection of Viable but Non-Culturable Staphylococci in Biofilms from Central Venous Catheters Negative on Standard Microbiological Assays. Clin. Microbiol. Infect. 2012, 18, E259–E261. [Google Scholar] [CrossRef]
- Knipper, A.D.; Plaza-Rodríguez, C.; Filter, M.; Wulsten, I.F.; Stingl, K.; Crease, T. Modeling the Survival of Campylobacter jejuni in Raw Milk Considering the Viable but Non-Culturable Cells (VBNC). J. Food Saf. 2023, 43, e13077. [Google Scholar] [CrossRef]
- Lindbäck, T.; Rottenberg, M.E.; Roche, S.M.; Rørvik, L.M. The Ability to Enter into an Avirulent Viable but Non-Culturable (VBNC) Form Is Widespread among Listeria monocytogenes Isolates from Salmon, Patients and Environment. Vet. Res. 2010, 41, 8. [Google Scholar] [CrossRef] [PubMed]
- Afari, G.K.; Hung, Y.C. Detection and Verification of the Viable but Nonculturable (VBNC) State of Escherichia coli O157:H7 and Listeria monocytogenes Using Flow Cytometry and Standard Plating. J. Food Sci. 2018, 83, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Saravanan, V.; Karunasagar, I.; Karunasagar, I. Detection of Vibrio parahaemolyticus in Tropical Shellfish by SYBR Green Real-Time PCR and Evaluation of Three Enrichment Media. Int. J. Food Microbiol. 2009, 129, 124–130. [Google Scholar] [CrossRef]
- Lopes, S.P.; Azevedo, N.F.; Pereira, M.O. Quantitative Assessment of Individual Populations within Polymicrobial Biofilms. Sci. Rep. 2018, 8, 9494. [Google Scholar] [CrossRef] [PubMed]
- Clais, S.; Boulet, G.; Van Kerckhoven, M.; Lanckacker, E.; Delputte, P.; Maes, L.; Cos, P. Comparison of Viable Plate Count, Turbidity Measurement and Real-Time PCR for Quantification of Porphyromonas gingivalis. Lett. Appl. Microbiol. 2015, 60, 79–84. [Google Scholar] [CrossRef]
- Ricchi, M.; Bertasio, C.; Boniotti, M.B.; Vicari, N.; Russo, S.; Tilola, M.; Bellotti, M.A.; Bertasi, B. Comparison among the Quantification of Bacterial Pathogens by QPCR, DPCR, and Cultural Methods. Front. Microbiol. 2017, 8, 1174. [Google Scholar] [CrossRef]
- Lahtinen, S.J.; Gueimonde, M.; Ouwehand, A.C.; Reinikainen, J.P.; Salminen, S.J. Comparison of Four Methods to Enumerate Probiotic Bifidobacteria in a Fermented Food Product. Food Microbiol. 2006, 23, 571–577. [Google Scholar] [CrossRef]
- Castro, J.; Alves, P.; Sousa, C.; Cereija, T.; França, A.; Jefferson, K.; Cerca, N. Using an In-Vitro Biofilm Model to Assess the Virulence Potential of Bacterial Vaginosis or Non-Bacterial Vaginosis Gardnerella vaginalis Isolates. Sci. Rep. 2015, 5, 11640. [Google Scholar] [CrossRef]
- Castro, J.; Paula, A.; Elisa, M.; Cerca, N. Lactobacillus Crispatus Represses Vaginolysin Expression by BV Associated Gardnerella vaginalis and Reduces Cell Cytotoxicity. Anaerobe 2018, 50, 60–63. [Google Scholar] [CrossRef]
- Ping, Y.; Jie, H.; Shi, Y.; Chen, S.; Shao, L.; Xu, Y.; Zhang, J.D.; Molnár, I. Antimicrobial Substances and Mechanisms of Lactobacillus rhamnosus against Gardnerella vaginalis. Probiotics Antimicrob. Proteins 2023, 15, 400–410. [Google Scholar] [CrossRef]
- Atassi, F.; Brassart, D.; Grob, P.; Graf, F.; Servin, A.L. Lactobacillus Strains Isolated from the Vaginal Microbiota of Healthy Women Inhibit Prevotella bivia and Gardnerella vaginalis in Coculture and Cell Culture. Pathog. Dis. 2006, 48, 424–432. [Google Scholar] [CrossRef]
- Atassi, F.; Brassart, D.; Grob, P.; Graf, F.; Servin, A.L. In Vitro Antibacterial Activity of Lactobacillus helveticus Strain KS300 against Diarrhoeagenic, Uropathogenic and Vaginosis-Associated Bacteria. J. Appl. Microbiol. 2006, 101, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Forney, L.J.; Ravel, J. Vaginal Microbiome: Rethinking Health and Disease. Annu. Rev. Microbiol. 2012, 66, 371–389. [Google Scholar] [CrossRef] [PubMed]
- Gamalero, E.; Lingua, G.; Caprì, F.G.; Fusconi, A.; Berta, G.; Lemanceau, P. Colonization Pattern of Primary Tomato Roots by Pseudomonas fluorescens A6RI Characterized by Dilution Plating, Flow Cytometry, Fluorescence, Confocal and Scanning Electron Microscopy. FEMS Microbiol. Ecol. 2004, 48, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Bressan, M.; Trinsoutrot Gattin, I.; Desaire, S.; Castel, L.; Gangneux, C.; Laval, K. A Rapid Flow Cytometry Method to Assess Bacterial Abundance in Agricultural Soil. Appl. Soil Ecol. 2015, 88, 60–68. [Google Scholar] [CrossRef]
- Rajab, S.; Tabandeh, F.; Khodabandeh, M. Anaerobe The Effect of Lactobacillus Cell Size on Its Probiotic Characteristics. Anaerobe 2020, 62, 102103. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Margarida, A.; Ana, C.; Sampaio, C. The Effect of Benzyl Isothiocyanate on Candida albicans Growth, Cell Size, Morphogenesis, and Ultrastructure. World J. Microbiol. Biotechnol. 2020, 36, 153. [Google Scholar] [CrossRef]
- Hasan, M.M.; Alam, M.W.; Wahid, K.A.; Miah, S.; Lukong, K.E. A Low-Cost Digital Microscope with Real-Time Fluorescent Imaging Capability. PLoS ONE 2016, 11, e0167863. [Google Scholar] [CrossRef]
- Maecker, H.T.; Rinfret, A.; Souza, P.D.; Darden, J.; Roig, E.; Landry, C.; Hayes, P.; Birungi, J.; Anzala, O.; Garcia, M.; et al. Standardization of Cytokine Flow Cytometry Assays. BMC Immunol. 2005, 18, 13. [Google Scholar] [CrossRef]
- Marie, D.; Lopes, A.; Brandini, F.P.; Vaulot, D. Estimating Microbial Populations by Flow Cytometry: Comparison between Instruments. Limnol. Oceanogr. Methods 2016, 14, 750–758. [Google Scholar] [CrossRef]
- Qiu, P. Toward Exhaustive Gating of Flow Cytometry Data. In Proceedings of the Proceedings-IEEE International Workshop on Genomic Signal Processing and Statistics, Washington, DC, USA, 2–4 December 2012; pp. 183–186. [Google Scholar]
- Christaki, U.; Courties, C.; Massana, R.; Catala, P.; Lebaron, P.; Gasol, J.M.; Zubkov, M.V. Optimized Routine Flow Cytometric Enumeration of Heterotrophic Flagellates Using SYBR Green I. Limnol. Oceanogr. Methods 2011, 9, 329–339. [Google Scholar] [CrossRef]
- Vaahtovuo, J.; Korkeama, M.; Munukka, E.; Viljanen, M.K.; Toivanen, P. Quantification of Bacteria in Human Feces Using 16S RRNA-Hybridization, DNA-Staining and Flow Cytometry. J. Microbiol. Methods 2005, 63, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.C. Accuracy and Precision in Quantitative Fluorescence Microscopy. J. Cell Biol. 2009, 185, 1135–1148. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Pfaffl, M.W.; Zhao, S.; Spiess, A.N.; Boggy, G.; Blom, J.; Rutledge, R.G.; Sisti, D.; Lievens, A.; De Preter, K.; et al. Evaluation of QPCR Curve Analysis Methods for Reliable Biomarker Discovery: Bias, Resolution, Precision, and Implications. Methods 2013, 59, 32–46. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lima, Â.; Muzny, C.A.; Cerca, N. An Indirect Fluorescence Microscopy Method to Assess Vaginal Lactobacillus Concentrations. Microorganisms 2024, 12, 114. https://doi.org/10.3390/microorganisms12010114
Lima Â, Muzny CA, Cerca N. An Indirect Fluorescence Microscopy Method to Assess Vaginal Lactobacillus Concentrations. Microorganisms. 2024; 12(1):114. https://doi.org/10.3390/microorganisms12010114
Chicago/Turabian StyleLima, Ângela, Christina A. Muzny, and Nuno Cerca. 2024. "An Indirect Fluorescence Microscopy Method to Assess Vaginal Lactobacillus Concentrations" Microorganisms 12, no. 1: 114. https://doi.org/10.3390/microorganisms12010114


_Di_Marco.png)


