Recent Advances in Assessing the Clinical Implications of Epstein-Barr Virus Infection and Their Application to the Diagnosis and Treatment of Nasopharyngeal Carcinoma
Abstract
:1. Introduction
1.1. Histopathology of NPC and EBV Infection
1.2. EBV Gene Expression in NPC
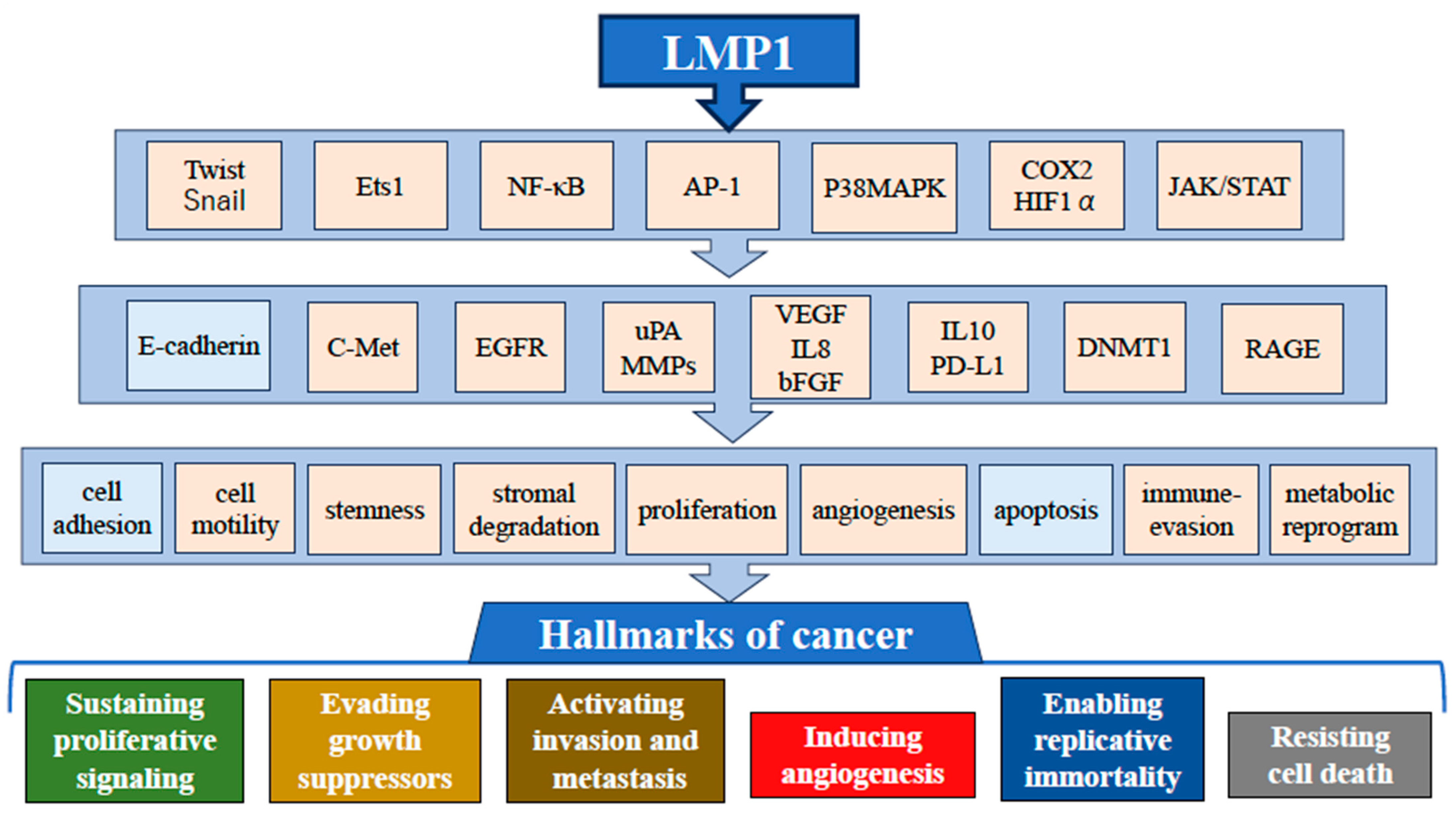
1.3. Effect of LMP1 Expression in NPC
2. Effect of BZLF1 Expression in NPC
2.1. Genetic and Epigenetic Alteration in NPC Host Genome
2.2. Potential Risk Factors Other Than EBV
2.3. Prognostic Relevance of Cell-Free EBV DNA
3. Immune Microenvironment of NPC and EBV
3.1. Immune Evasion Mechanism in NPC
3.2. Superantigen Induction by EBV
3.3. Modulation of Immune Checkpoint in NPC
3.4. Challenge for EBV Vaccine Development
4. Early Diagnosis of NPC
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wen, K.W.; Wang, L.; Menke, J.R.; Damania, B. Cancers associated with human gammaherpesviruses. FEBS J. 2022, 289, 7631–7669. [Google Scholar] [CrossRef] [PubMed]
- Epstein, M.A.; Achong, B.G.; Barr, Y.M. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 1964, 7335, 702–703. [Google Scholar] [CrossRef] [PubMed]
- Old, L.J.; Boyse, E.A.; Oettgen, H.F.; Harven, E.D.; Geering, G.; Williamson, B.; Clifford, P. Precipitating antibody in human serum to an antigen present in cultured Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. USA 1966, 56, 1699–1704. [Google Scholar] [CrossRef]
- Zur Hausen, H.; Schulte-Holthausen, I.; Klein, G.; Henle, W.; Henle, G.; Clifford, P.; Santesson, L. EBV-DNA in biopsies of Burkitt’s tumors and anaplastic carcinomas of the nasopharynx. Nature 1970, 228, 1056–1058. [Google Scholar] [CrossRef] [PubMed]
- Wolf, H.; zur Hausen, H.; Becker, V. EB viral genomes in epithelial nasopharyngeal carcinoma cells. Nat. New Biol. 1973, 244, 245–247. [Google Scholar] [CrossRef]
- Shanmugaratnam, K. Histological typing of nasopharyngeal carcinoma. IARC Sci. Publ. 1978, 20, 3–12. [Google Scholar]
- Yoshizaki, T. EBV-related serological biomarkers for nasopharyngeal cancer remain a hot topic. Ann. Nasopharynx Cancer 2018, 2, 5–7. [Google Scholar] [CrossRef]
- Neel, H.B., 3rd; Pearson, G.R.; Weiland, L.H.; Taylor, W.F.; Goepfert, H.H.; Pilch, B.Z.; Goodman, M.; Lanier, A.P.; Huang, A.T.; Hyams, V.J.; et al. Application of Epstein-Barr virus serology to the diagnosis and staging of North American patients with nasopharyngeal carcinoma. Otolaryngol. Head Neck Surg. 1983, 91, 255–262. [Google Scholar] [CrossRef]
- Chen, Y.P.; Chan, A.T.C.; Le, Q.T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef]
- Sam, C.K.; Prasad, U.; Pathmanathan, R. Serological markers in the diagnosis of histopathological types of nasopharyngeal carcinoma. Eur. J. Surg. Oncol. 1989, 15, 357–360. [Google Scholar]
- Schieber, J.; Pring, M.; Ness, A.; Liu, Z.; Hsu, W.L.; Brenner, N.; Butt, J.; Waterboer, T.; Simon, J. Development of a Duplex Serological Multiplex Assay for the Simultaneous Detection of Epstein-Barr Virus IgA and IgG Antibodies in Nasopharyngeal Carcinoma Patients. Cancers 2023, 15, 2578. [Google Scholar] [CrossRef] [PubMed]
- Lam, W.K.J.; King, A.D.; Miller, J.A.; Liu, Z.; Yu, K.J.; Chua, M.L.K.; Ma, B.B.Y.; Chen, M.Y.; Pinsky, B.A.; Lou, P.J.; et al. Recommendations for Epstein-Barr virus-based screening for nasopharyngeal cancer in high- and intermediate-risk regions. J. Natl. Cancer Inst. 2023, 115, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Fuwa, N.; Kodaira, T.; Daimon, T.; Yoshizaki, T. The long-term outcomes of alternating chemoradiotherapy for locoregionally advanced nasopharyngeal carcinoma: A multiinstitutional phase II study. Cancer Med. 2016, 4, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- Kano, M.; Kondo, S.; Wakisaka, N.; Moriyama-Kita, M.; Nakanishi, Y.; Endo, K.; Murono, S.; Nakamura, H.; Yoshizaki, T. The influence of human papillomavirus on nasopharyngeal carcinoma in Japan. Auris Nasus Larynx 2017, 44, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Khong, B.; Kwok, S.; Cao, H.; West, R.B.; Le, Q.T.; Kong, C.S. Human papillomavirus 16 detected in nasopharyngeal carcinomas in white Americans but not in endemic Southern Chinese patients. Head Neck 2014, 36, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Dogan, S.; Hedberg, M.L.; Ferris, R.L.; Rath, T.J.; Assaad, A.M.; Chiosea, S.I. Human papillomavirus and Epstein-Barr virus in nasopharyngeal carcinoma in a low-incidence population. Head Neck 2014, 36, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.; Suh, Y.E.; Paleri, V.; Devlin, D.; Ayaz, B.; Pertl, L.; Thavaraj, S. Oncogenic human papillomavirus-associated nasopharyngeal carcinoma: An observational study of correlation with ethnicity, histological subtype and outcome in a UK population. Infect. Agents Cancer 2013, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Laantri, N.; Attaleb, M.; Kandil, M.; Naji, F.; Mouttaki, T.; Dardari, R.; Belghmi, K.; Benchakroun, N.; El Mzibri, M.; Khyatti, M. Human papillomavirus detection in moroccan patients with nasopharyngeal carcinoma. Infect. Agents Cancer 2011, 6, 3. [Google Scholar] [CrossRef]
- Mirzamani, N.; Salehian, S.; Farhadi, M.; Amin Tehran, E. Detection of EBV and HPV in nasopharyngeal carcinoma by in situ hybridization Exp. Mol. Pathol. 2006, 81, 231–234. [Google Scholar] [CrossRef]
- Rassekh, C.H.; Rady, P.L.; Arany, I.; Tyring, S.K.; Knudsen, S.; Calhoun, K.H.; Seikaly, H.; Bailey, B.J. Combined Epstein-Barr virus and human papillomavirus infection in nasopharyngeal carcinoma. Laryngoscope 1998, 108, 362–367. [Google Scholar] [CrossRef]
- Robbins, H.A.; Ferreiro-Iglesias, A.; Waterboer, T.; Brenner, N.; Nygard, M.; Bender, N.; Schroeder, L.; Hildesheim, A.; Pawlita, M.; D’Souza, G.; et al. Absolute Risk of Oropharyngeal Cancer after an HPV16-E6 Serology Test and Potential Implications for Screening: Results from the Human Papillomavirus Cancer Cohort Consortium. J. Clin. Oncol. 2022, 40, 3613–3622. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.W.; Tsang, C.M.; Lo, K.W. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160270. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein-Barr virus: More than 50 years old and still providing surprises. Nat. Rev. Cancer 2016, 16, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Jha, H.C.; Pei, Y.; Robertson, E.S. Epstein-Barr virus: Diseases linked to infection and transformation. Front. Microbiol. 2016, 7, 1602. [Google Scholar] [CrossRef] [PubMed]
- Bechtel, D.; Kurth, J.; Unkel, C.; Kuppers, R. Transformation of BCR-deficient germinal-center B cells by EBV supports a major role of the virus in the pathogenesis of Hodgkin and posttransplantation lymphomas. Blood 2005, 106, 4345–4350. [Google Scholar] [CrossRef] [PubMed]
- Rowe, M.; Rowe, D.T.; Gregory, C.D.; Young, L.S.; Farrell, P.J.; Rupani, H.; Rickinson, A.B. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt’s lymphoma cells. EMBO J. 1987, 6, 2743–2751. [Google Scholar] [CrossRef]
- Kang, D.; Skalsky, R.L.; Cullen, B.R. EBV BART microRNAs target multiple pro-apoptotic cellular genes to promote epithelial cell survival. PLoS Pathog. 2015, 11, e1004979. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Pathmanathan, R.; Prasad, U.; Sadler, R.; Flynn, K.; Raab-Traub, N. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N. Engl. J. Med. 1995, 333, 693–698. [Google Scholar] [CrossRef]
- Mosialos, G.; Birkenbach, M.; Yalamanchili, R.; VanArsdale, T.; Ware, C.; Kieff, E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 1995, 80, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.T.; Chang, Y.T.; Chen, S.C.; Chuang, Y.C.; Chen, Y.R.; Lin, C.S.; Chen, J.Y. Epstein-Barr virus latent membrane protein 1 represses p53-mediated DNA repair and transcriptional activity. Oncogene 2005, 24, 2635–2646. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.; Rowe, M.; Gregory, C.; Croom-Carter, D.; Wang, F.; Longnecker, R.; Kieff, E.; Rickinson, A. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 1991, 65, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Hong, L.; Cheng, C.; Li, N.; Zhao, X.; Shi, F.; Liu, J.; Fan, J.; Zhou, J.; Bode, A.M.; et al. DNMT1 mediates metabolic reprogramming induced by Epstein-Barr virus latent membrane protein 1 and reversed by grifolin in nasopharyngeal carcinoma. Cell Death Dis. 2018, 9, 619. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, A.; Wakisaka, N.; Kondo, S.; Murono, S.; Furukawa, M.; Yoshizaki, T. Induction of receptor for advanced glycation end products by EBV latent membrane protein 1 and its correlation with angiogenesis and cervical lymph node metastasis in nasopharyngeal carcinoma. Clin. Cancer Res. 2008, 14, 5368–5375. [Google Scholar] [CrossRef]
- Kondo, S.; Wakisaka, N.; Muramatsu, M.; Zen, Y.; Endo, K.; Murono, S.; Sugimoto, H.; Yamaoka, S.; Pagano, J.S.; Yoshizaki, T. Epstein-Barr virus latent membrane protein 1 induces cancer stem/progenitor-like cells in nasopharyngeal epithelial cell lines. J. Virol. 2011, 85, 11255–11264. [Google Scholar] [CrossRef]
- Horikawa, T.; Yang, J.; Kondo, S.; Yoshizaki, T.; Joab, I.; Furukawa, M.; Pagano, J.S. Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res. 2007, 67, 1970–1978. [Google Scholar] [CrossRef]
- Ressing, M.E.; van Gent, M.; Gram, A.M.; Hooykaas, M.J.; Piersma, S.J.; Wiertz, E.J. Immune evasion by Epstein-Barr virus. Curr. Top. Microbiol. Immunol. 2015, 391, 355–381. [Google Scholar] [CrossRef]
- Kase, K.; Kondo, S.; Wakisaka, N.; Dochi, H.; Mizokami, H.; Kobayashi, E.; Kano, M.; Komori, T.; Hirai, N.; Ueno, T.; et al. Epstein-Barr Virus LMP1 Induces Soluble PD-L1 in Nasopharyngeal Carcinoma. Microorganisms 2021, 9, 603. [Google Scholar] [CrossRef]
- Fåhraeus, R.; Chen, W.; Trivedi, P.; Klein, G.; Obrink, B. Decreased expression of E-cadherin and increased invasive capacity in EBV-LMP-transfected human epithelial and murine adenocarcinoma cells. Int. J. Cancer 1992, 52, 834–838. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Sato, H.; Furukawa, M.; Pagano, J. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc. Natl. Acad. Sci. USA 1998, 95, 3621–3626. [Google Scholar] [CrossRef] [PubMed]
- Yoshizaki, T.; Horikawa, T.; Ren, Q.; Wakisaka, N.; Takeshita, H.; Sheen, T.S.; Lee, S.Y.; Sato, H.; Furukawa, M. Induction of interleukin-8 by Epstein-Barr virus latent membrane pretein-1 and its correlation with angiogenesis in nasopharyngeal carcinoma. Clin. Cancer Res. 2001, 7, 1946–1951. [Google Scholar] [PubMed]
- Murono, S.; Inoue, H.; Tanabe, T.; Joab, I.; Yoshizaki, T.; Furukawa, M.; Pagano, S. Induction of cyclooxygenase-2 by Epstein-Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc. Natl. Acad. Sci. USA 2001, 98, 6905–6910. [Google Scholar] [CrossRef] [PubMed]
- Yoshizaki, T.; Kondo, S.; Endo, K.; Nakanishi, Y.; Aga, M.; Kobayashi, E.; Hirai, N.; Sugimoto, H.; Hatano, M.; Ueno, T.; et al. Modulation of the tumor microenvironment by Epstein-Barr virus latent membrane protein 1 in nasopharyngeal carcinoma. Cancer Sci. 2018, 109, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Dai, W.; Cheung, A.K.; Ko, J.M.; Kan, R.; Wong, B.W.; Leong, M.M.; Deng, M.; Kwok, T.C.; Chan, J.Y.; et al. Whole-exome sequencing identifies multiple loss-of-function mutations of NF-kappaB pathway regulators in nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA 2016, 113, 11283–11288. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chung, G.T.; Lui, V.W.; To, K.F.; Ma, B.B.; Chow, C.; Woo, J.K.; Yip, K.Y.; Seo, J.; Hui, E.P.; et al. Exome and genome sequencing of nasopharynx cancer identifies NF-kappaB pathway activating mutations. Nat. Commun. 2017, 8, 14121. [Google Scholar] [CrossRef] [PubMed]
- Countryman, J.; Miller, G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 1985, 82, 4085–4089. [Google Scholar] [CrossRef]
- Ramayanti, O.; Juwana, H.; Verkuijlen, S.A.; Adham, M.; Pegtel, M.D.; Greijer, A.E.; Middeldorp, J.M. Epstein-Barr virus mRNA profiles and viral DNA methylation status in nasopharyngeal brushings from nasopharyngeal carcinoma patients reflect tumor origin. Int. J. Cancer 2017, 140, 149–162. [Google Scholar] [CrossRef]
- Germini, D.; Sall, F.B.; Shmakova, A.; Wiels, J.; Dokudovskaya, S.; Drouet, E.; Vassetzky, Y. Oncogenic Properties of the EBV ZEBRA Protein. Cancers 2020, 12, 1479. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Sato, H.; Murono, S.; Pagano, J.S.; Furukawa, M. Matrix metalloproteinase 9 is induced by the Epstein-Barr virus BZLF1 transactivator. Clin. Exp. Metastasis 1999, 17, 431–436. [Google Scholar] [CrossRef]
- Morrison, T.E.; Mauser, A.; Wong, A.; Ting, J.P.; Kenney, S.C. Inhibition of IFN-gamma signaling by an Epstein-Barr virus immediate-early protein. Immunity 2001, 15, 787–799. [Google Scholar] [CrossRef]
- Rosemarie, Q.; Sugden, B. Epstein-Barr Virus: How Its Lytic Phase Contributes to Oncogenesis. Microorganisms 2020, 8, 1824. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Miwa, H.; Takeshita, H.; Sato, H.; Furukawa, M. Elevation of antibody against Epstein-Barr virus genes BRLF1 and BZLF1 in nasopharyngeal carcinoma. J. Cancer Res. Clin. Oncol. 2000, 126, 69–73. [Google Scholar]
- Zhang, G.; Li, Z.; Zhou, Q. Utility of Serum EB Virus Zta Antibody in the Diagnostic of Nasopharyngeal Carcinoma: Evidences from 2126 Cases and 15,644 Controls. Front. Oncol. 2019, 9, 1391. [Google Scholar] [CrossRef]
- Dochi, H.; Kondo, S.; Murata, T.; Fukuyo, M.; Nanbo, A.; Wakae, K.; Jiang, W.P.; Hamabe-Horiike, T.; Tanaka, M.; Nishiuchi, T.; et al. Estrogen induces the expression of EBV lytic protein ZEBRA, a marker of poor prognosis in nasopharyngeal carcinoma. Cancer Sci. 2022, 113, 2862–2877. [Google Scholar] [CrossRef]
- Chien, Y.C.; Chen, J.Y.; Liu, M.Y.; Yang, H.I.; Hsu, M.M.; Chen, C.J.; Yang, C.S. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N. Engl. J. Med. 2001, 345, 1877–1882. [Google Scholar] [CrossRef]
- Tsai, M.H.; Lin, X.; Shumilov, A.; Bernhardt, K.; Feederle, R.; Poirey, R.; Kopp-Schneider, A.; Pereira, B.; Almeida, R.; Delecluse, H.J. The biological properties of different Epstein-Barr virus strains explain their association with various types of cancers. Oncotarget 2017, 8, 10238–10254. [Google Scholar] [CrossRef]
- Okuno, Y.; Murata, T.; Sato, Y.; Muramatsu, H.; Ito, Y.; Watanabe, T.; Okuno, T.; Murakami, N.; Yoshida, K.; Sawada, A.; et al. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat. Microbiol. 2019, 4, 404–413. [Google Scholar] [CrossRef]
- Kondo, S.; Okuno, Y.; Murata, T.; Dochi, H.; Wakisaka, N.; Mizokami, H.; Moriyama-Kita, M.; Kobayashi, E.; Kano, M.; Komori, T.; et al. EBV genome variations enhance clinicopathological features of nasopharyngeal carcinoma in a non-endemic region. Cancer Sci. 2022, 113, 2446–2456. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yao, Y.; Chen, H.; Zhang, S.; Cao, S.M.; Zhang, Z.; Luo, B.; Liu, Z.; Li, Z.; Xiang, T.; et al. Genome sequencing analysis identifies Epstein-Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat. Genet. 2019, 51, 1131–1136. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Yajima, M.; Ikuta, K. Epstein-Barr virus strain variation and cancer. Cancer Sci. 2019, 110, 1132–1139. [Google Scholar] [CrossRef]
- Chan, A.S.; To, K.F.; Lo, K.W.; Ding, M.; Li, X.; Johnson, P.; Huang, D.P. Frequent chromosome 9p losses in histologically normal nasopharyngeal epithelia from southern Chinese. Int. J. Cancer 2002, 102, 300–303. [Google Scholar] [CrossRef]
- Chan, A.S.; To, K.F.; Lo, K.W.; Mak, K.F.; Pak, W.; Chiu, B.; Tse, G.M.; Ding, M.; Li, X.; Lee, J.C.; et al. High frequency of chromosome 3p deletion in histologically normal nasopharyngeal epithelia from southern Chinese. Cancer Res. 2000, 60, 5365–5370. [Google Scholar]
- Hui, A.B.; Or, Y.Y.; Takano, H.; Tsang, R.K.; To, K.F.; Guan, X.Y.; Sham, J.S.; Hung, K.W.; Lam, C.N.; van Hasselt, C.A.; et al. Array-based comparative genomic hybridization analysis identified cyclin D1 as a target oncogene at 11q13.3 in nasopharyngeal carcinoma. Cancer Res. 2005, 65, 8125–8133. [Google Scholar] [CrossRef]
- Lo, K.W.; Teo, P.M.; Hui, A.B.; To, K.F.; Tsang, Y.S.; Chan, S.Y.; Mak, K.F.; Lee, J.C.; Huang, D.P. High resolution allelotype of microdissected primary nasopharyngeal carcinoma. Cancer Res. 2000, 60, 3348–3353. [Google Scholar]
- Lin, D.C.; Meng, X.; Hazawa, M.; Nagata, Y.; Varela, A.M.; Xu, L.; Sato, Y.; Liu, L.Z.; Ding, L.W.; Sharma, A.; et al. The genomic landscape of nasopharyngeal carcinoma. Nat. Genet. 2014, 46, 866–871. [Google Scholar] [CrossRef]
- Lo, K.W.; Huang, D.P.; Lau, K.M. p16 gene alterations in nasopharyngeal carcinoma. Cancer Res. 1995, 55, 2039–2043. [Google Scholar]
- Kaneda, A.; Matsusaka, K.; Aburatani, H.; Fukayama, M. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012, 72, 3445–3450. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Y.; Guo, B.B.; Chan, F.K.; Tao, Q. Oncogenic induction of cellular high CpG methylation by Epstein-Barr virus in malignant epithelial cells. Chin. J. Cancer 2014, 33, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.W.; Kwong, J.; Hui, A.B.; Chan, S.Y.; To, K.F.; Chan, A.S.; Chow, L.S.; Teo, P.M.; Johnson, P.J.; Huang, D.P. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001, 61, 3877–3881. [Google Scholar] [PubMed]
- Dai, W.; Cheung, A.K.; Ko, J.M.; Cheng, Y.; Zheng, H.; Ngan, R.K.; Ng, W.T.; Lee, A.W.; Yau, C.C.; Lee, V.H.; et al. Comparative methylome analysis in solid tumors reveals aberrant methylation at chromosome 6p in nasopharyngeal carcinoma. Cancer Med. 2015, 4, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.W.; Cheung, S.T.; Leung, S.F.; van Hasselt, A.; Tsang, Y.S.; Mak, K.F.; Chung, Y.F.; Woo, J.K.; Lee, J.C.; Huang, D.P. Hypermethylation of the p16 gene in nasopharyngeal carcinoma. Cancer Res. 1996, 56, 2721–2725. [Google Scholar] [PubMed]
- Kwong, J.; Lo, W.; To, K.F.; Teo, P.M.; Johnson, P.J.; Huang, D.P. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin. Cancer Res. 2002, 8, 131–137. [Google Scholar]
- Guo, X.; Johnson, R.C.; Deng, H.; Liao, J.; Guan, L.; Nelson, G.W.; Tang, M.; Zheng, Y.; de The, G.; O’Brien, S.J.; et al. Evaluation of nonviral risk factors for nasopharyngeal carcinoma in a high-risk population of Southern China. Int. J. Cancer 2009, 124, 2942–2947. [Google Scholar] [CrossRef]
- Tsao, S.W.; Yip, Y.L.; Tsang, C.M.; Pang, P.S.; Lau, V.M.; Zhang, G.; Lo, K.W. Etiological factors of nasopharyngeal carcinoma. Oral Oncol. 2014, 50, 330–338. [Google Scholar] [CrossRef]
- Liu, Z.; Chang, E.T.; Liu, Q.; Cai, Y.; Zhang, Z.; Chen, G.; Xie, S.H.; Cao, S.M.; Shao, J.Y.; Jia, W.H.; et al. Oral hygiene and risk of nasopharyngeal carcinoma—A population-based case-control study in China. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1201–1207. [Google Scholar] [CrossRef]
- Liu, Z.; Chang, E.T.; Liu, Q.; Cai, Y.; Zhang, Z.; Chen, G.; Huang, Q.H.; Xie, S.H.; Cao, S.M.; Shao, J.Y.; et al. Quantification of familial risk of nasopharyngeal carcinoma in a high-incidence area. Cancer 2017, 123, 2716–2725. [Google Scholar] [CrossRef]
- Chang, E.T.; Liu, Z.; Hildesheim, A.; Liu, Q.; Cai, Y.; Zhang, Z.; Chen, G.; Xie, S.H.; Cao, S.M.; Shao, J.Y.; et al. Active and passive smoking and risk of nasopharyngeal carcinoma: A population-based case-control study in southern China. Am. J. Epidemiol. 2017, 185, 1272–1280. [Google Scholar] [CrossRef]
- Park, E.M.; Chelvanambi, M.; Bhutiani, N.; Kroemer, G.; Zitvogel, L.; Wargo, J.A. Targeting the gut and tumor microbiota in cancer. Nat. Med. 2022, 28, 690–703. [Google Scholar] [CrossRef]
- Qiao, H.; Tan, X.R.; Li, H.; Li, J.Y.; Chen, X.Z.; Li, Y.Q.; Li, W.F.; Tang, L.L.; Zhou, G.Q.; Zhang, Y.; et al. Association of Intratumoral Microbiota with Prognosis in Patients with Nasopharyngeal Carcinoma from 2 Hospitals in China. JAMA Oncol. 2022, 8, 1301–1309. [Google Scholar] [CrossRef]
- Imai, K.; Inoue, H.; Tamura, M.; Cueno, M.E.; Inoue, H.; Takeichi, O.; Kusama, K.; Saito, I.; Ochiai, K. The periodontal pathogen Porphyromonas gingivalis induces the Epstein-Barr virus lytic switch transactivator ZEBRA by histone modification. Biochimie 2012, 94, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.L.; Chen, Y.P.; Mao, Y.P.; Wang, Z.X.; Guo, R.; Chen, L.; Tian, L.; Lin, A.H.; Li, L.; Sun, Y.; et al. Validation of the 8th edition of the uicc/ajcc staging system for nasopharyngeal carcinoma from endemic areas in the intensity-modulated radiotherapy era. J. Natl. Compr. Cancer Netw. 2017, 15, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.Q.; Li, C.F.; Li, J.; Chen, W.H.; Chen, Q.Y.; Yuan, L.X.; Lai, X.P.; He, Y.; Xu, Y.X.; Hu, D.P.; et al. Establishment and validation of prognostic nomograms for endemic nasopharyngeal carcinoma. J. Natl. Cancer Inst. 2015, 108, djv291. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Liu, N.; Chen, X.Z.; Sun, Y.; Li, B.; Ren, X.Y.; Qin, W.F.; Jiang, N.; Xu, Y.F.; Li, Y.Q.; et al. Genome-wide identification of a methylation gene panel as a prognostic biomarker in nasopharyngeal carcinoma. Mol. Cancer Ther. 2015, 14, 2864–2873. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Chen, N.Y.; Cui, R.X.; Li, W.; Li, Y.; Wei, R.; Zhang, M.; Sun, Y.; Huang, B.; Chen, M.; et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: A microRNA expression analysis. Lancet Oncol. 2012, 13, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.R.; Li, Y.Q.; Liang, S.B.; Jiang, W.; Liu, F.; Ge, W.; Tang, L.; Mao, Y.; He, M.; Yang, X.; et al. Development and validation of a gene expression-based signature to predict distant metastasis in locoregionally advanced nasopharyngeal carcinoma: A retrospective, multicentre, cohort study. Lancet Oncol. 2018, 19, 382–393. [Google Scholar] [CrossRef]
- Xu, C.; Chen, Y.P.; Liu, X.; Li, W.F.; Chen, L.; Mao, Y.P.; Zhang, Y.; Guo, R.; Zhou, G.Q.; Tang, L.L.; et al. Establishing and applying nomograms based on the 8th edition of the UICC/AJCC staging system to select patients with nasopharyngeal carcinoma who benefit from induction chemotherapy plus concurrent chemoradiotherapy. Oral Oncol. 2017, 69, 99–107. [Google Scholar] [CrossRef]
- Guo, R.; Tang, L.L.; Mao, Y.P.; Du, X.J.; Chen, L.; Zhang, Z.C.; Liu, L.Z.; Tian, L.; Luo, X.T.; Xie, Y.B.; et al. Proposed modifications and incorporation of plasma Epstein-Barr virus DNA improve the TNM staging system for Epstein-Barr virus-related nasopharyngeal carcinoma. Cancer 2019, 125, 79–89. [Google Scholar] [CrossRef]
- Liu, L.T.; Tang, L.Q.; Chen, Q.Y.; Zhang, L.; Guo, S.S.; Guo, L.; Mo, H.Y.; Zhao, C.; Guo, X.; Cao, K.J.; et al. The prognostic value of plasma Epstein-barr viral DNA and tumor response to neoadjuvant chemotherapy in advanced-stage nasopharyngeal carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2015, 93, 862–869. [Google Scholar] [CrossRef]
- Chan, A.T.C.; Hui, E.P.; Ngan, R.K.C.; Tung, S.Y.; Cheng, A.C.K.; Ng, W.T.; Lee, V.H.F.; Ma, B.B.Y.; Cheng, H.C.; Wong, F.C.S.; et al. Analysis of plasma Epstein-Barr virus DNA in nasopharyngeal cancer after chemoradiation to identify high-risk patients for adjuvant chemotherapy: A randomized controlled trial. J. Clin. Oncol. 2018, 36, 3091–3100. [Google Scholar] [CrossRef] [PubMed]
- Le, Q.T.; Zhang, Q.; Cao, H.; Cheng, A.J.; Pinsky, B.A.; Hong, R.L.; Chang, J.T.; Wang, C.W.; Tsao, K.C.; Lo, Y.M.D.; et al. An international collaboration to harmonize the quantitative plasma Epstein-Barr virus DNA assay for future biomarker-guided trials in nasopharyngeal carcinoma. Clin. Cancer Res. 2013, 19, 2208–2215. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Le, Q.T.; Yom, S.S.; Pinsky, B.A.; Bratman, S.V.; Ng, R.H.; El Mubarak, H.S.; Chan, K.C.; Sander, M.; Conley, B.A. Current state of PCR-based Epstein-Barr virus DNA testing for nasopharyngeal cancer. J. Natl. Cancer Inst. 2017, 109, djx007. [Google Scholar] [CrossRef] [PubMed]
- Kiyono, H.; Fukuyama, S. NALT- versus PEYER’S-patch-mediated mucosal immunity. Nat. Rev. Immunol. 2004, 4, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zou, C.; Zhang, S.; Chu, T.S.M.; Zhang, Y.; Chen, W.; Zhao, C.; Yang, L.; Xu, Z.; Dong, S.; et al. Reshaping the systemic tumor immune environment (STIE) and tumor immune microenvironment (TIME) to enhance immunotherapy efficacy in solid tumors. J. Hematol. Oncol. 2022, 15, 87. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Chen, Y.P.; Zhang, Y.; Jiang, W.; Liu, N.; Yun, J.P.; Sun, Y.; He, Q.M.; Tang, X.R.; Wen, X.; et al. Prognostic significance of tumor-infiltrating lymphocytes in nondisseminated nasopharyngeal carcinoma: A large-scale cohort study. Int. J. Cancer 2018, 142, 2558–2566. [Google Scholar] [CrossRef] [PubMed]
- Crotzer, V.L.; Christian, R.E.; Brooks, J.M.; Shabanowitz, J.; Settlage, R.E.; Marto, J.A.; White, F.M.; Rickinson, A.B.; Hunt, D.F. Engelhard VH. Immunodominance among EBV-derived epitopes restricted by HLA-B27 does not correlate with epitope abundance in EBV-transformed B-lymphoblastoid cell lines. J. Immunol. 2000, 164, 6120–6129. [Google Scholar] [CrossRef]
- Hill, A.B.; Lee, S.P.; Haurum, J.S.; Murray, N.; Yao, Q.Y.; Rowe, M.; Signoret, N.; Rickinson, A.B.; McMichael, A.J. Class I major histocompatibility complex-restricted cytotoxic T lymphocytes specific for Epstein-Barr virus (EBV)-transformed B lymphoblastoid cell lines against which they were raised. J. Exp. Med. 1995, 181, 2221–2228. [Google Scholar] [CrossRef]
- Münz, C. Epstein-barr virus nuclear antigen 1: From immunologically invisible to a promising T cell target. J. Exp. Med. 2004, 199, 1301–1304. [Google Scholar] [CrossRef]
- Clausse, B.; Fizazi, K.; Walczak, V.; Tetaud, C.; Wiels, J.; Tursz, T.; Busson, P. High concentration of the EBV Latent Membrane Protein 1 in glycosphingolipid-rich complexes from both epithelial and lymphoid cells. Virolgy 1997, 228, 285–293. [Google Scholar] [CrossRef]
- Smith, C.; Wakisaka, N.; Crough, T.; Peet, J.; Yoshizaki, T.; Beagley, L.; Khanna, R. Discerning regulation of cis- and trans-presentation of CD8+ T-cell epitopes by EBV-encoded oncogene LMP-1 through self-aggregation. Blood 2009, 113, 6148–6152. [Google Scholar] [CrossRef] [PubMed]
- Kieff, E.; Ricknson, A. Epstein-Barr virus and its replication. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 2001; pp. 2511–2573. [Google Scholar]
- Li, Y.; Long, X.; Huang, L.; Yang, M.; Yuan, Y.; Wang, Y.; Delecluse, H.J.; Kuang, E. Epstein-Barr Virus BZLF1-Mediated Downregulation of Proinflammatory Factors Is Essential for Optimal Lytic Viral Replication. J. Virol. 2015, 90, 887–903. [Google Scholar] [CrossRef] [PubMed]
- Conrad, B.; Weissmahr, R.N.; Böni, J.; Arcari, R.; Schüpbach, J.; Mach, B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell 1997, 90, 303–313. [Google Scholar] [CrossRef]
- Sutkowski, N.; Conrad, B.; Thorley-Lawson, D.A.; Huber, B.T. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 2001, 15, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Sutkowski, N.; Chen, G.; Calderon, G.; Huber, B.T. Epstein-Barr virus latent membrane protein LMP-2A is sufficient for transactivation of the human endogenous retrovirus HERV-K18 superantigen. J. Virol. 2004, 78, 7852–7860. [Google Scholar] [CrossRef] [PubMed]
- Agathanggelou, A.; Niedobitek, G.; Chen, R.; Nicholls, J.; Yin, W.; Young, L.S. Expression of immune regulatory molecules in Epstein-Barr virus-associated nasopharyngeal carcinomas with prominent lymphoid stroma. Evidence for a functional interaction between epithelial tumor cells and infiltrating lymphoid cells. Am. J. Pathol. 1995, 147, 1152–1160. [Google Scholar] [PubMed]
- Niedobitek, G.; Agathanggelou, A.; Nicholls, J.M. Epstein-Barr virus infection and the pathogenesis of nasopharyngeal carcinoma: Viral gene expression, tumour cell phenotype, and the role of the lymphoid stroma. Semin. Cancer Biol. 1996, 7, 165–174. [Google Scholar] [CrossRef]
- Fang, W.; Zhang, J.; Hong, S.; Zhan, J.; Chen, N.; Qin, T.; Tang, Y.; Zhang, Y.; Kang, S.; Zhou, T.; et al. EBV-driven LMP1 and IFN-γ up-regulate PD-L1 in nasopharyngeal carcinoma: Implications for oncotargeted therapy. Oncotarget 2014, 5, 12189–12202. [Google Scholar] [CrossRef]
- Zhang, J.; Fang, W.; Qin, T.; Yang, Y.; Hong, S.; Liang, W.; Ma, Y.; Zhao, H.; Huang, Y.; Xue, C. Co-expression of PD-1 and PD-L1 predicts poor outcome in nasopharyngeal carcinoma. Med. Oncol. 2015, 32, 86. [Google Scholar] [CrossRef]
- Smith, C.; McGrath, M.; Neller, M.A.; Matthews, K.K.; Crooks, P.; Le Texier, L.; Panizza, B.; Porceddu, S.; Khanna, R. Complete response to PD-1 blockade following EBV-specific T-cell therapy in metastatic nasopharyngeal carcinoma. NPJ Precis. Oncol. 2021, 5, 24. [Google Scholar] [CrossRef]
- Lee, V.H.; Lo, A.W.; Leung, C.Y.; Shek, W.H.; Kwong, D.L.; Lam, C.O.; Tong, C.C.; Sze, C.L.; Leung, T.W. Correlation of PD-L1 expression of tumor cells with survival outcomes after radical intensity-modulated radiation therapy for non-metastatic nasopharyngeal carcinoma. PLoS ONE 2016, 11, e0157969. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Cai, M.Y.; Chen, C.L.; Hu, H.; Lin, H.X.; Li, M.; Weng, D.S.; Zhao, J.J.; Guo, L.; Xia, J.C. Tumor cells PD-L1 expression as a favorable prognosis factor in nasopharyngeal carcinoma patients with pre-existing intratumor-infiltrating lymphocytes. Oncoimmunology 2017, 6, e1312240. [Google Scholar] [CrossRef] [PubMed]
- Larbcharoensub, N.; Mahaprom, K.; Jiarpinitnun, C.; Trachu, N.; Tubthong, N.; Pattaranutaporn, P.; Sirachainan, E.; Ngamphaiboon, N. Characterization of PD-L1 and PD-1 expression and CD8+tumor-infiltrating lymphocyte in Epstein-Barr virus-associated nasopharyngeal carcinoma. Am. J. Clin. Oncol. 2018, 41, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Azuma, K.; Kawahara, A.; Sasada, T.; Matsuo, N.; Kakuma, T.; Kamimura, H.; Maeda, R.; Hattori, C.; On, K.; et al. Prognostic stratification of patients with nasopharyngeal carcinoma based on tumor immune microenvironment. Head Neck 2018, 40, 2007–2019. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.T.; Ye, S.B.; Liu, Y.N.; He, J.; Chen, Q.Y.; Mai, H.Q.; Zhang, C.X.; Cui, J.; Zhang, X.S.; Busson, P.; et al. LMP1-mediated glycolysis induces myeloid-derived suppressor cell expansion in nasopharyngeal carcinoma. PLoS Pathog. 2017, 13, e1006503. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.W.; Ng, W.T.; Chan, L.; Hung, W.M.; Chan, C.; Sze, H.C.; Chan, O.S.; Chang, A.T.; Yeung, R.M. Evolution of treatment for nasopharyngeal cancer--success and setback in the intensity-modulated radiotherapy era. Radiother. Oncol. 2014, 110, 377–384. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, W.; Lei, F.; Yu, X.; Li, Z.; Liu, X.; Ni, Y.; Deng, L.; Ji, M. Long-term survival after nasopharyngeal carcinoma treatment in a local prefecture-level hospital in southern China. Cancer Manag. Res. 2020, 12, 1329–1338. [Google Scholar] [CrossRef]
- Ji, M.F.; Sheng, W.; Cheng, W.M.; Ng, M.H.; Wu, B.H.; Yu, X.; Wei, K.R.; Li, F.G.; Lian, S.F.; Wang, P.P.; et al. Incidence and mortality of nasopharyngeal carcinoma: Interim analysis of a cluster randomized controlled screening trial (PRO-NPC-001) in southern China. Ann. Oncol. 2019, 30, 1630–1637. [Google Scholar] [CrossRef]
- Tay, J.K.; Lim, M.Y.; Kanagalingam, J. Screening in nasopharyngeal carcinoma: Current strategies and future directions. Curr. Otorhinolaryngol. Rep. 2014, 2, 1–7. [Google Scholar] [CrossRef]
- He, Z.; Zhang, K.; Zhao, N.; Wang, Y.; Hou, W.; Meng, Q.; Li, C.; Chen, J.; Li, J. Deep learning for real-time detection of nasopharyngeal carcinoma during nasopharyngeal endoscopy. iScience 2023, 26, 07463. [Google Scholar] [CrossRef]
- Coghill, A.E.; Hsu, W.L.; Pfeiffer, R.M.; Juwana, H.; Yu, K.J.; Lou, P.J.; Wang, C.P.; Chen, J.Y.; Chen, C.J.; Middeldorp, J.M.; et al. Epstein–Barr virus serology as a potential screening marker for nasopharyngeal carcinoma among high-risk individuals from multiplex families in Taiwan. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, H.; Ji, M. Epstein–Barr virus-based nasopharyngeal carcinoma population screening. Ann. Nasopharynx Cancer 2022, 6, 3. [Google Scholar] [CrossRef]
- Yuan, Y.; Ye, F.; Wu, J.H.; Fu, X.Y.; Huang, Z.X.; Zhang, T. Early screening of nasopharyngeal carcinoma. Head Neck 2023, 45, 2700–2709. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Dai, W.; Kwong, D.L.; Szeto, C.Y.; Wong, E.H.; Ng, W.T.; Lee, A.W.; Ngan, R.K.; Yau, C.C.; Tung, S.Y.; et al. Epigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution melting. Int. J. Cancer 2015, 136, E127–E135. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, D.; Hutajulu, S.H.; Nawaz, I.; Nguyen Van, D.; Huang, G.; Haryana, S.M.; Middeldorp, J.M.; Ernberg, I.; Hu, L.F. Development of a noninvasive method, multiplex methylation specific PCR (MMSP), for early diagnosis of nasopharyngeal carcinoma. PLoS ONE 2012, 7, e45908. [Google Scholar] [CrossRef]
- Li, T.; Li, F.; Guo, X.; Hong, C.; Yu, X.; Wu, B.; Lian, S.; Song, L.; Tang, J.; Wen, S.; et al. Anti-Epstein-Barr Virus BNLF2b for Mass Screening for Nasopharyngeal Cancer. N. Engl. J. Med. 2023, 389, 808–819. [Google Scholar] [CrossRef]
| WHO I | WHO II | WHO III | |
|---|---|---|---|
| Differentiation status | well differentiated | moderately to poorly differentiated | undifferentiated |
| Histological category in WHO classification | keratinizing | nonkeratinizing-differentiated | nonkeratinizing-undifferentiated |
| TIL infiltration | fair to moderate | heavy | |
| EBERs in tumor | (−) or faint | (+) | |
| EBV antibodies | not elevated | elevated | |
| Chemoradiosensitivity | moderate | good | |
| Metastatic property | low to moderate | high | |
| Epidemiology | 20% in non-endemic area; <5% in endemic areas | 80% in non-endemic areas; >95% in endemic areas | |
| Screening Procedure | Swab Cytology | Serological Test | Circulating EBV-DNA | Cell Free DNA Methylation | Nasopharyngeal Endoscopy |
|---|---|---|---|---|---|
| Benefits | Easy sampling | Most popular methods | Higher sensitivity and specificity | Higher sensitivity and specificity | Higher positive predictive value of approximately 90% |
| Quick detection | Possible combination of multiple antibody types | Higher positive predictive value but still <20% | Higher positive predictive value of approximately 90% | Combination of deep learning model improve accuracy | |
| Applicable for EBV negative NPC | Applicable for EBV negative NPC | ||||
| Drawbacks | Influenced by sample quality, physician’s skill, cytologist’s skill | Low level of positive predictive value (10%) | Influenced by laboratory skill | More advanced technique required | Large scale screening is not possible |
| Not globally standardized | Lack of large-scale study | Higher cost | |||
| Higher cost |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshizaki, T.; Kondo, S.; Dochi, H.; Kobayashi, E.; Mizokami, H.; Komura, S.; Endo, K. Recent Advances in Assessing the Clinical Implications of Epstein-Barr Virus Infection and Their Application to the Diagnosis and Treatment of Nasopharyngeal Carcinoma. Microorganisms 2024, 12, 14. https://doi.org/10.3390/microorganisms12010014
Yoshizaki T, Kondo S, Dochi H, Kobayashi E, Mizokami H, Komura S, Endo K. Recent Advances in Assessing the Clinical Implications of Epstein-Barr Virus Infection and Their Application to the Diagnosis and Treatment of Nasopharyngeal Carcinoma. Microorganisms. 2024; 12(1):14. https://doi.org/10.3390/microorganisms12010014
Chicago/Turabian StyleYoshizaki, Tomokazu, Satoru Kondo, Hirotomo Dochi, Eiji Kobayashi, Harue Mizokami, Shigetaka Komura, and Kazuhira Endo. 2024. "Recent Advances in Assessing the Clinical Implications of Epstein-Barr Virus Infection and Their Application to the Diagnosis and Treatment of Nasopharyngeal Carcinoma" Microorganisms 12, no. 1: 14. https://doi.org/10.3390/microorganisms12010014





