Methotrexate and Non-Surgical Periodontal Treatment Change the Oral–Gut Microbiota in Rheumatoid Arthritis: A Prospective Cohort Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design, Setting, and Ethical Clearance
2.2. Participants and RA Diagnostic Criteria
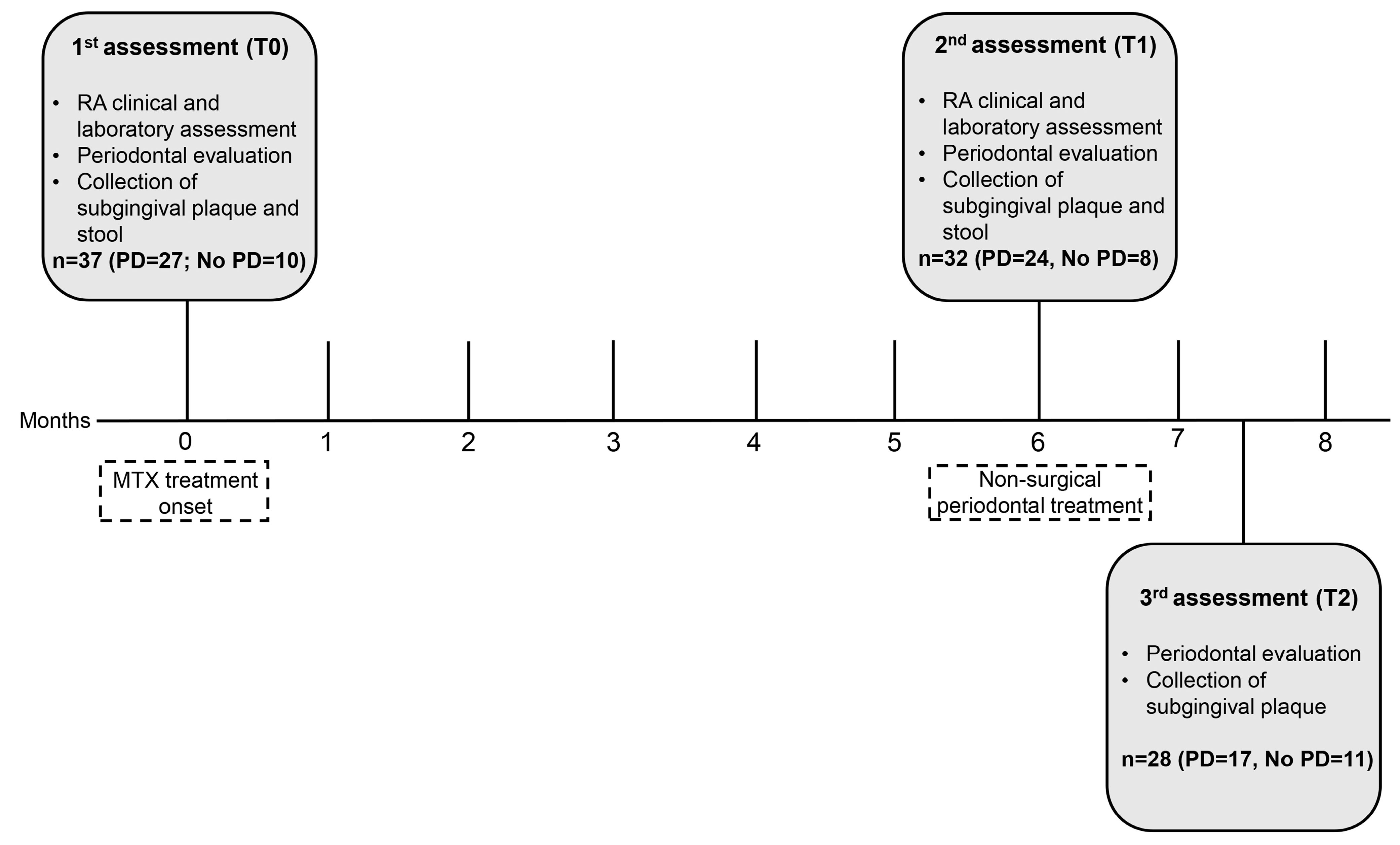
2.3. Data Assessment and Collection
2.4. Clinical and Laboratory RA Assessment
2.5. Periodontal Evaluation and Treatment
2.6. Subgingival Plaque and Stool Samples for Microbiome Analysis
2.6.1. DNA Extraction and 16S rRNA Amplicon Library Preparation and Sequencing
2.6.2. Read Processing and Taxonomic Classification
2.6.3. Data Analysis
2.7. Statistical Analysis
3. Results
3.1. Clinicodemographic Data
3.2. NSPT Modified Periodontal Parameters
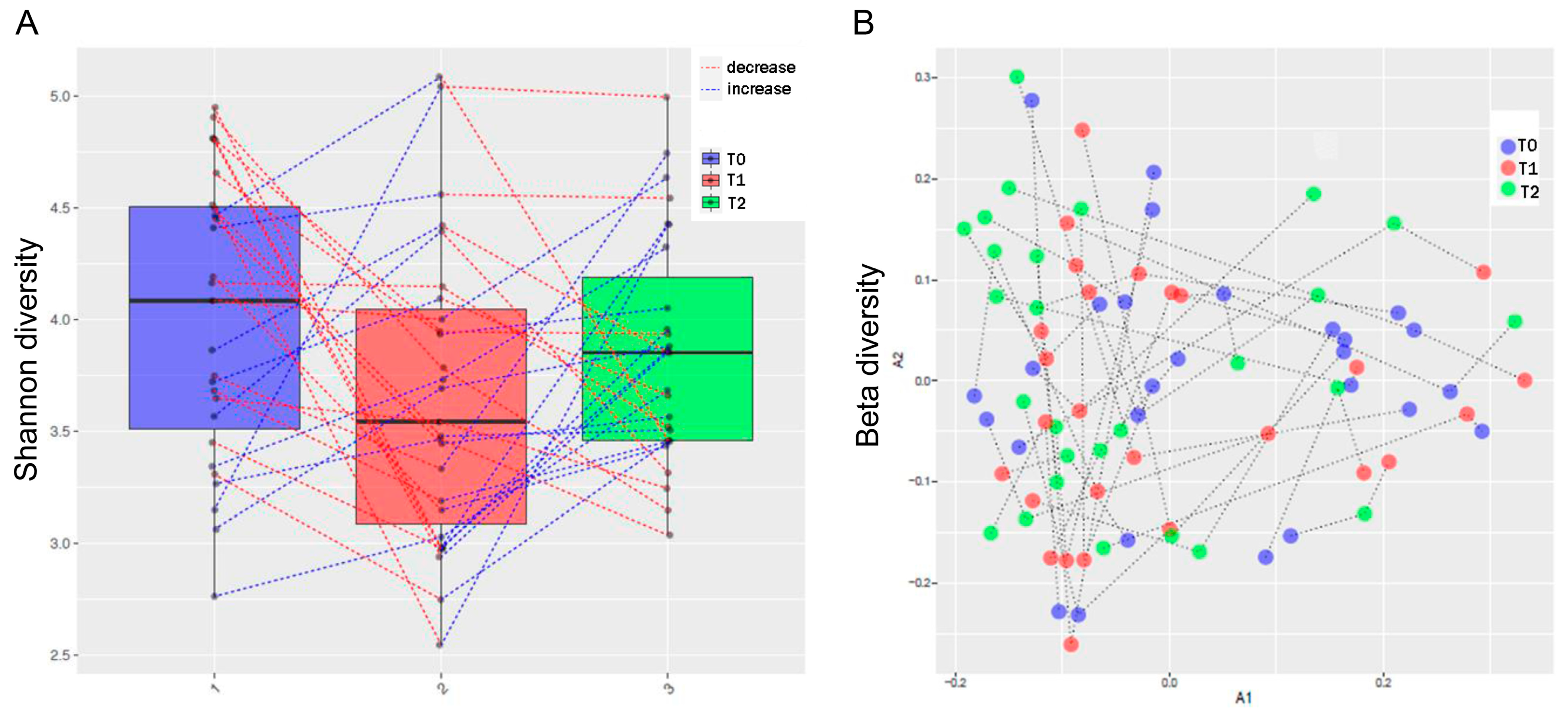
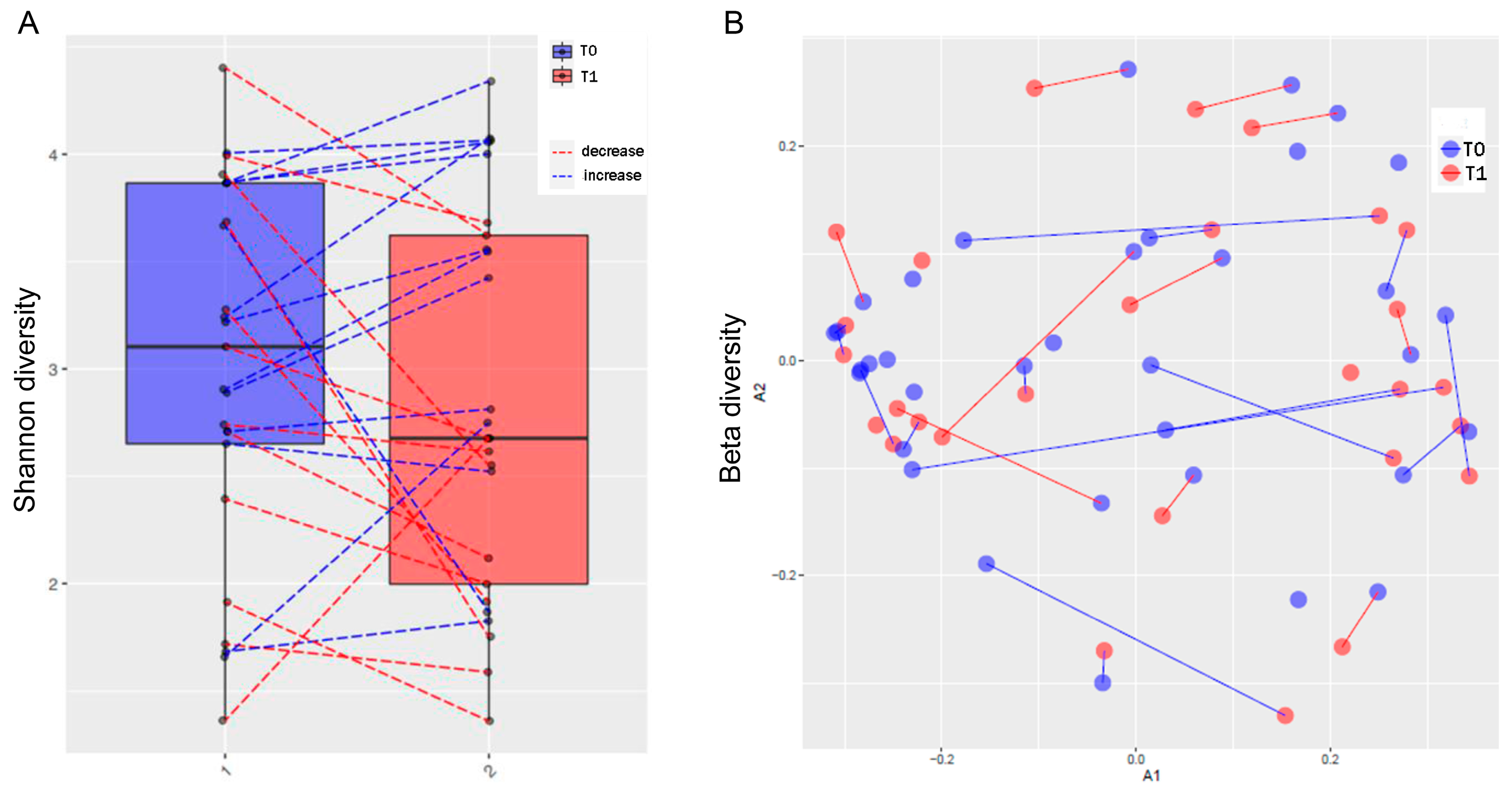
3.3. MTX and NSPT Affected Subgingival Microbial Richness and Diversity
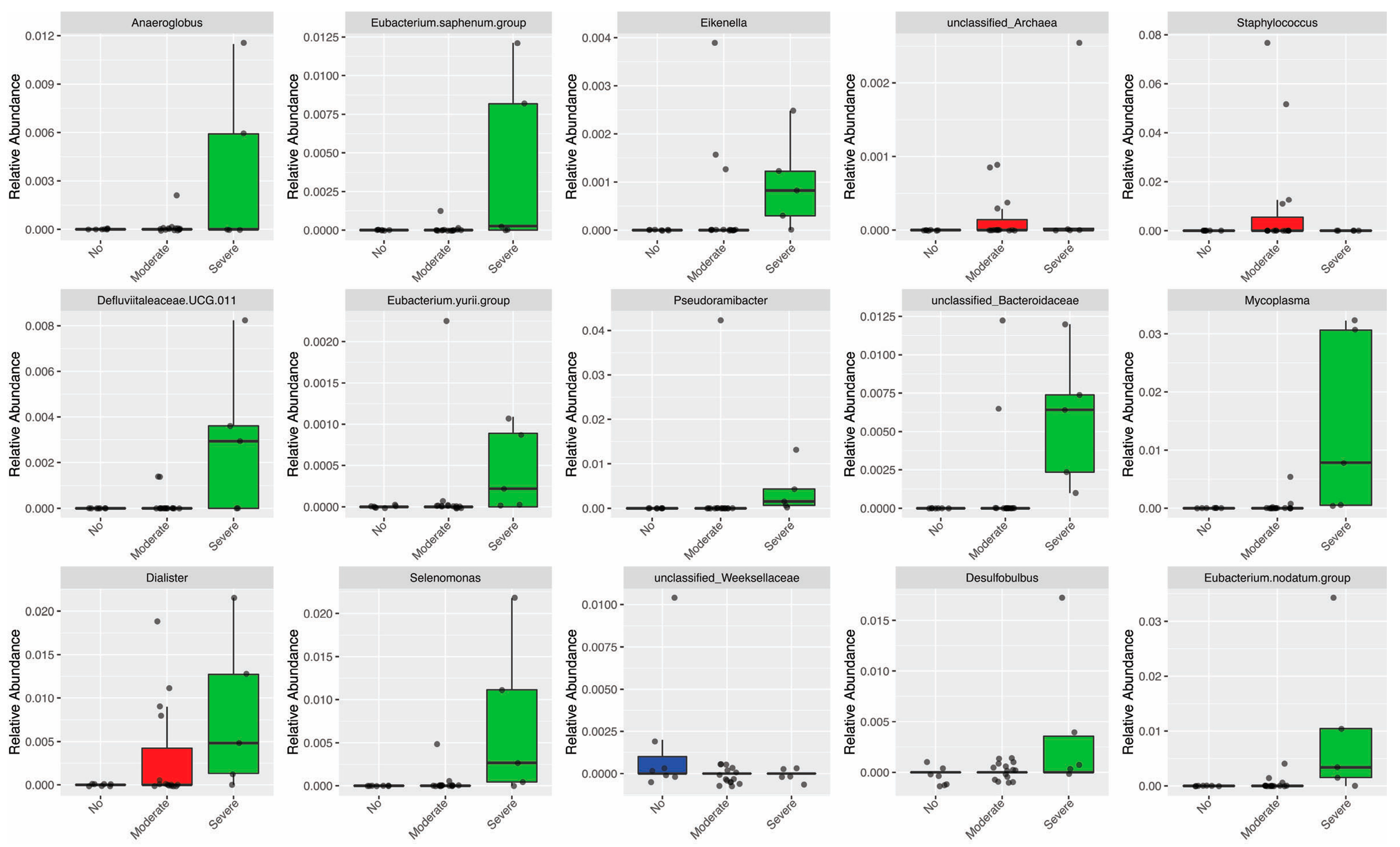
3.4. Periodontitis Influenced the Relative Abundance of Different Genera in the Oral Microbiota
3.5. MTX Treatment and Gut Microbiome Diversity
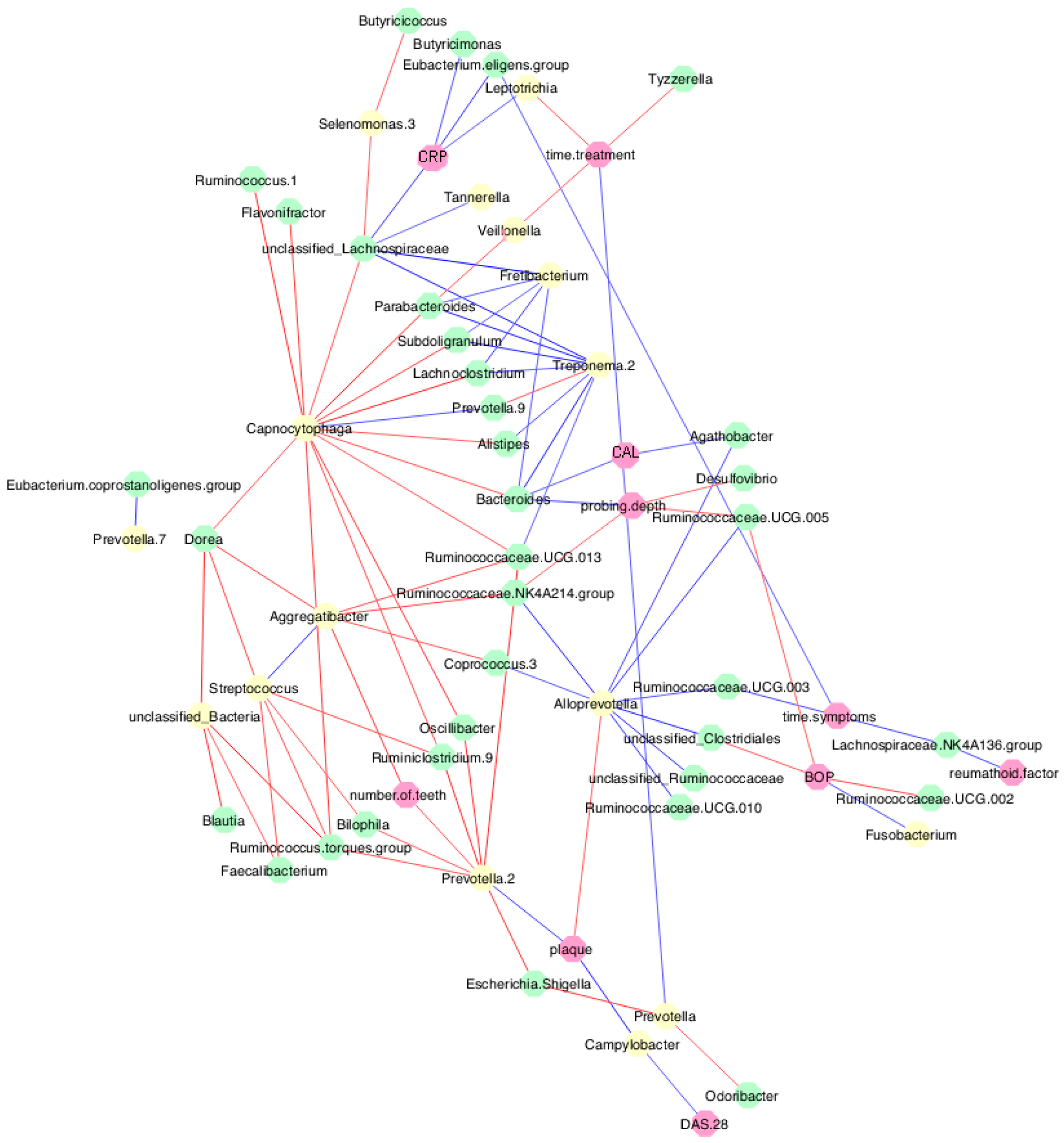
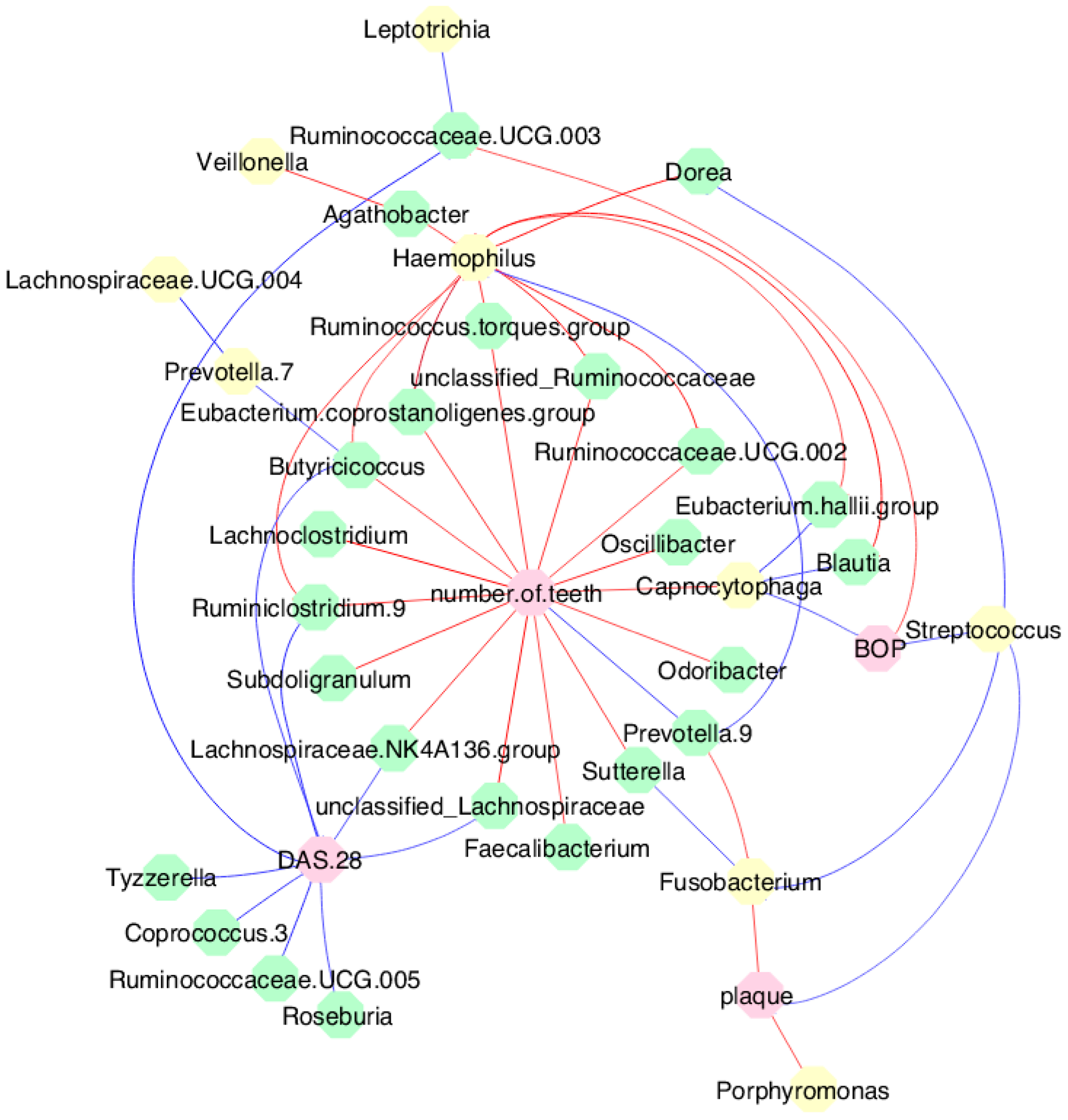
3.6. Periodontal Parameters and RA Activity Influenced the Oral and Gut Microbiota
3.7. MTX Treatment Defined New Associations of Periodontal and RA Parameters with Oral and Gut Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Potempa, J.; Mydel, P.; Koziel, J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat. Rev. Rheumatol. 2017, 13, 606–620. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Aletaha, D.; Barton, A.; Burmester, G.R.; Emery, P.; Firestein, G.S.; Kavanaugh, A.; McInnes, I.B.; Solomon, D.H.; Strand, V.; et al. Rheumatoid arthritis. Nat. Rev. Dis Primers. 2018, 4, 18001. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Takeda, K. Role of Gut microbiota in rheumatoid arthritis. J. Clin. Med. 2017, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Bolstad, A.I.; Fevang, B.S.; Lie, S.A. Increased risk of periodontitis in patients with rheumatoid arthritis: A nationwide register study in Norway. J. Clin. Periodontol. 2023, 50, 1022–1032. [Google Scholar] [CrossRef]
- Krutyhołowa, A.; Strzelec, K.; Dziedzic, A.; Bereta, G.P.; Łazarz-Bartyzel, K.; Potempa, J.; Gawron, K. Host and bacterial factors linking periodontitis and rheumatoid arthritis. Front. Immunol. 2022, 13, 980805. [Google Scholar] [CrossRef]
- Farquharson, D.; Butcher, J.P.; Culshaw, S. Periodontitis, Porphyromonas, and the pathogenesis of rheumatoid arthritis. Mucosal Immunol. 2012, 5, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Ebbers, M.; Lübcke, P.M.; Volzke, J.; Kriebel, K.; Hieke, C.; Engelmann, R.; Lang, H.; Kreikemeyer, B.; Müller-Hilke, B. Interplay between P. gingivalis, F. nucleatum and A. actinomycetemcomitans in murine alveolar bone loss, arthritis onset and progression. Sci. Rep. 2018, 8, 15129. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef]
- Corrêa, J.D.; Fernandes, G.R.; Calderaro, D.C.; Mendonça, S.M.S.; Silva, J.M.; Albiero, M.L.; Cunha, F.Q.; Xiao, E.; Ferreira, G.A.; Teixeira, A.L.; et al. Oral microbial dysbiosis linked to worsened periodontal condition in rheumatoid arthritis patients. Sci. Rep. 2019, 9, 8379. [Google Scholar] [CrossRef]
- Beyer, K.; Zaura, E.; Brandt, B.W.; Buijs, M.J.; Brun, J.G.; Crielaard, W.; Bolstad, A.I. Subgingival microbiome of rheumatoid arthritis patients in relation to their disease status and periodontal health. PLoS ONE 2018, 13, e0202278. [Google Scholar] [CrossRef]
- Eriksson, K.; Fei, G.; Lundmark, A.; Benchimol, D.; Lee, L.; Hu, Y.O.O.; Kats, A.; Saevarsdottir, S.; Catrina, A.I.; Klinge, B.; et al. Periodontal health and oral microbiota in patients with rheumatoid arthritis. J. Clin. Med. 2019, 8, 630. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chen, Q.; Lin, P.; Xu, R.; He, D.; Ji, W.; Bian, Y.; Shen, Y.; Li, Q.; Liu, C.; et al. Characteristics of gut microbiota in patients with rheumatoid arthritis in Shanghai, China. Front. Cell. Infect. Microbiol. 2019, 9, 369. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.J.; Cao, N.W.; Zhou, H.Y.; Meng, X.; Guo, B.; Zhang, H.Y.; Li, B.Z. The oral and gut microbiome in rheumatoid arthritis patients: A systematic review. Rheumatology 2021, 60, 1054–1066. [Google Scholar] [CrossRef] [PubMed]
- du Teil Espina, M.; Gabarrini, G.; Harmsen, H.J.M.; Westra, J.; van Winkelhoff, A.J.; van Dijl, J.M. Talk to your gut: The oral-gut microbiome axis and its immunomodulatory role in the etiology of rheumatoid arthritis. FEMS Microbiol. Rev. 2019, 43, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Mikuls, T.R.; Thiele, G.M.; Deane, K.D.; Payne, J.B.; O’Dell, J.R.; Yu, F.; Sayles, H.; Weisman, M.H.; Gregersen, P.K.; Buckner, J.H.; et al. Porphyromonas gingivalis and disease-related autoantibodies in individuals at increased risk of rheumatoid arthritis. Arthritis Rheum. 2012, 64, 3522–3530. [Google Scholar] [CrossRef]
- Konig, M.F.; Abusleme, L.; Reinholdt, J.; Palmer, R.J.; Teles, R.P.; Sampson, K.; Rosen, A.; Nigrovic, P.A.; Sokolove, J.; Giles, J.T.; et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016, 8, 369ra176. [Google Scholar] [CrossRef]
- Schwenzer, A.; Quirke, A.M.; Marzeda, A.M.; Wong, A.; Montgomery, A.B.; Sayles, H.R.; Eick, S.; Gawron, K.; Chomyszyn-Gajewska, M.; Łazarz-Bartyzel, K.; et al. Association of distinct fine specificities of anti-citrullinated peptide antibodies with elevated immune responses to Prevotella intermedia in a subgroup of patients with rheumatoid arthritis and periodontitis. Arthritis Rheumatol. 2017, 69, 2303–2313. [Google Scholar] [CrossRef]
- Martu, M.A.; Solomon, S.M.; Sufaru, I.G.; Jelihovschi, I.; Martu, S.; Rezus, E.; Surdu, A.E.; Onea, R.M.; Grecu, G.P.; Foia, L. Study on the prevalence of periodontopathogenic bacteria in serum and subgingival bacterial plaque in patients with rheumatoid arthritis. Rev. Chim. 2017, 68, 1946–1950. [Google Scholar] [CrossRef]
- Möller, B.; Kollert, F.; Sculean, A.; Villiger, P.M. Infectious triggers in periodontitis and the gut in rheumatoid arthritis (RA): A complex story about association and causality. Front. Immunol. 2020, 11, 1108. [Google Scholar] [CrossRef]
- Giollo, A.; Fuzzi, E.; Doria, A. Methotrexate in early rheumatoid arthritis: Is the anchor drug still holding? Autoimmun. Rev. 2022, 21, 103031. [Google Scholar] [CrossRef]
- Bodkhe, R.; Balakrishnan, B.; Taneja, V. The role of microbiome in rheumatoid arthritis treatment. Ther. Adv. Musculoskelet. Dis. 2019, 11, 1759720X19844632. [Google Scholar] [CrossRef] [PubMed]
- Lübcke, P.M.; Ebbers, M.N.B.; Volzke, J.; Bull, J.; Kneitz, S.; Engelmann, R.; Lang, H.; Kreikemeyer, B.; Müller-Hilke, B. Periodontal treatment prevents arthritis in mice and methotrexate ameliorates periodontal bone loss. Sci. Rep. 2019, 9, 8128. [Google Scholar] [CrossRef] [PubMed]
- de Arruda, J.A.A.; Corrêa, J.D.; Singh, Y.; Oliveira, S.R.; Machado, C.C.; Schneider, A.H.; Medeiros, J.D.; Fernandes, G.R.; Macari, S.; Barrioni, B.R.; et al. Methotrexate promotes recovery of arthritis-induced alveolar bone loss and modifies the composition of the oral-gut microbiota. Anaerobe 2022, 75, 102577. [Google Scholar] [CrossRef] [PubMed]
- Knottnerus, A.; Tugwell, P. STROBE—A checklist to Strengthen the Reporting of Observational Studies in Epidemiology. J. Clin. Epidemiol. 2008, 61, 323. [Google Scholar] [CrossRef] [PubMed]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., 3rd; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010, 62, 2569–2581. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.L.; Pisetsky, D.S. Early rheumatoid arthritis. Curr. Opin. Rheumatol. 2007, 19, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.R.; de Arruda, J.A.A.; Schneider, A.H.; Carvalho, V.F.; Machado, C.C.; Corrêa, J.D.; Moura, M.F.; Duffles, L.F.; de Souza, F.F.L.; Ferreira, G.A.; et al. Are neutrophil extracellular traps the link for the cross-talk between periodontitis and rheumatoid arthritis physiopathology? Rheumatology 2021, 61, 174–184. [Google Scholar] [CrossRef]
- Aletaha, D.; Smolen, J.S. Diagnosis and management of rheumatoid arthritis: A review. JAMA 2018, 320, 1360–1372. [Google Scholar] [CrossRef]
- Eke, P.I.; Page, R.C.; Wei, L.; Thornton-Evans, G.; Genco, R.J. Update of the case definitions for population-based surveillance of periodontitis. J. Periodontol. 2012, 83, 1449–1454. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- de Paula, A.C.L.; Medeiros, J.D.; Fernandes, G.R.; da Silva, V.L.; Diniz, C.G. Microbiome of industrialized Minas frescal cheese reveals high prevalence of putative bacteria: A concern in the one health context. LWT 2021, 139, 110791. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Oliveira, F.S.; Correa, F.B.; Morais, D.K.; Fernandes, G.B. TAG.ME: Taxonomic assignment of genetic markers for ecology. bioRxiv 2018. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Soft. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Drost, H.G. Philentropy: Information theory and distance quantification with R. J. Open Source Softw. 2018, 3, 765. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Maciejewski, M.; Sands, C.; Nair, N.; Ling, S.; Verstappen, S.; Hyrich, K.; Barton, A.; Ziemek, D.; Lewis, M.R.; Plant, D. Prediction of response of methotrexate in patients with rheumatoid arthritis using serum lipidomics. Sci. Rep. 2021, 11, 7266. [Google Scholar] [CrossRef]
- Zaragoza-García, O.; Castro-Alarcón, N.; Pérez-Rubio, G.; Guzmán-Guzmán, I.P. DMARDs-Gut microbiota feedback: Implications in the response to therapy. Biomolecules 2020, 10, 1479. [Google Scholar] [CrossRef]
- Han, M.; Zhang, N.; Mao, Y.; Huang, B.; Ren, M.; Peng, Z.; Bai, Z.; Chen, L.; Liu, Y.; Wang, S.; et al. The potential of gut microbiota metabolic capability to detect drug response in rheumatoid arthritis patients. Front. Microbiol. 2022, 13, 839015. [Google Scholar] [CrossRef] [PubMed]
- Nayak, R.R.; Alexander, M.; Deshpande, I.; Stapleton-Gray, K.; Rimal, B.; Patterson, A.D.; Ubeda, C.; Scher, J.U.; Turnbaugh, P.J. Methotrexate impacts conserved pathways in diverse human gut bacteria leading to decreased host immune activation. Cell Host Microbe 2021, 29, 362–377.e11. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, D.; Borges, R.; Ribeiro, R.A.; de Souza, R.F.; Amado, P.P.P.; Saraiva, L.; Horliana, A.C.R.T.; Faveri, M.; Mayer, M.P.A. Oral dysbiosis in severe forms of periodontitis is associated with gut dysbiosis and correlated with salivary inflammatory mediators: A preliminary study. Front. Oral Health 2021, 2, 722495. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, J.D.; Calderaro, D.C.; Ferreira, G.A.; Mendonça, S.M.; Fernandes, G.R.; Xiao, E.; Teixeira, A.L.; Leys, E.J.; Graves, D.T.; Silva, T.A. Subgingival microbiota dysbiosis in systemic lupus erythematosus: Association with periodontal status. Microbiome 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Ubeda, C.; Equinda, M.; Khanin, R.; Buischi, Y.; Viale, A.; Lipuma, L.; Attur, M.; Pillinger, M.H.; Weissmann, G.; et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012, 64, 3083–3094. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zheng, Y.; Bian, X.; Ge, H.; Wang, J.; Zhang, Z. Non-surgical periodontal treatment improves rheumatoid arthritis disease activity: A meta-analysis. Clin. Oral Investig. 2021, 25, 4975–4985. [Google Scholar] [CrossRef]
- Äyräväinen, L.; Leirisalo-Repo, M.; Kuuliala, A.; Ahola, K.; Koivuniemi, R.; Meurman, J.H.; Heikkinen, A.M. Periodontitis in early and chronic rheumatoid arthritis: A prospective follow-up study in Finnish population. BMJ Open 2017, 7, e011916. [Google Scholar] [CrossRef]
- Kordtabar, S.; Aghaie, M.; Fakhari, E.; Vakili, M.A. Periodontal condition in patients with rheumatoid arthritis: Effect of anti-rheumatic Drugs. J. Dent. 2019, 20, 190–194. [Google Scholar] [CrossRef]
- Jung, G.U.; Han, J.Y.; Hwang, K.G.; Park, C.J.; Stathopoulou, P.G.; Fiorellini, J.P. Effects of conventional synthetic disease-modifying antirheumatic drugs on response to periodontal treatment in patients with rheumatoid arthritis. Biomed. Res. Int. 2018, 2018, 1465402. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, C.; Gao, L.; Zhang, D.; Li, C.; Liu, J. Influence of anti-rheumatic agents on the periodontal condition of patients with rheumatoid arthritis and periodontitis: A systematic review and meta-analysis. J. Periodontal. Res. 2021, 56, 1099–1115. [Google Scholar] [CrossRef]
- Balta, M.G.; Papathanasiou, E.; Blix, I.J.; Van Dyke, T.E. Host modulation and treatment of periodontal disease. J. Dent. Res. 2021, 100, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Belstrøm, D.; Grande, M.A.; Sembler-Møller, M.L.; Kirkby, N.; Cotton, S.L.; Paster, B.J.; Holmstrup, P. Influence of periodontal treatment on subgingival and salivary microbiotas. J. Periodontol. 2018, 89, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Luan, Q.; Chen, F.; Chen, Z.; Zhang, Q.; Yu, X. Shift in the subgingival microbiome following scaling and root planing in generalized aggressive periodontitis. J. Clin. Periodontol. 2018, 45, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Porsch, M.; Grosse, I.; Hoffmann, K.; Schaller, H.G.; Reichert, S. Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch. Oral Biol. 2019, 99, 169–176. [Google Scholar] [CrossRef]
- Greenwood, D.; Afacan, B.; Emingil, G.; Bostanci, N.; Belibasakis, G.N. Salivary microbiome shifts in response to periodontal treatment outcome. Proteom. Clin. Appl. 2020, 14, e2000011. [Google Scholar] [CrossRef] [PubMed]
- Martu, M.A.; Luchian, I.; Mares, M.; Solomon, S.; Ciurcanu, O.; Danila, V.; Rezus, E.; Foia, L. The effectiveness of laser applications and photodynamic therapy on relevant periodontal pathogens (Aggregatibacter actinomycetemcomitans) associated with immunomodulating anti-rheumatic drugs. Bioengineering 2023, 10, 61. [Google Scholar] [CrossRef]
- Schwarzberg, K.; Le, R.; Bharti, B.; Lindsay, S.; Casaburi, G.; Salvatore, F.; Saber, M.H.; Alonaizan, F.; Slots, J.; Gottlieb, R.A.; et al. The personal human oral microbiome obscures the effects of treatment on periodontal disease. PLoS ONE 2014, 9, e86708. [Google Scholar] [CrossRef]
- Ziebolz, D.; Pabel, S.O.; Lange, K.; Krohn-Grimberghe, B.; Hornecker, E.; Mausberg, R.F. Clinical periodontal and microbiologic parameters in patients with rheumatoid arthritis. J. Periodontol. 2011, 82, 1424–1432. [Google Scholar] [CrossRef]
| Variables | n (%) |
|---|---|
| Age, median (min–max) | 51 (22–70) |
| Sex | |
| Female | 33 (89.2) |
| Male | 4 (10.8) |
| RA | |
| Early | 22 (59.5) |
| Established | 15 (40.5) |
| Tobacco smoking | |
| Never | 22 (59.5) |
| Still | 8 (21.6) |
| Stopped | 7 (18.9) |
| Alcohol consumption | |
| Never | 31 (83.8) |
| Still | 2 (5.4) |
| Stopped | 4 (10.8) |
| RF (IU/mL), median (min–max) | 15.00 (0–1890) |
| ACPA (IU/mL), median (min–max) | 69.75 (0–340) |
| CRP (mg/dL), median (min–max) | 0.77 (0–14) |
| VAS (mm), median (min–max) | 60 (0–100) |
| ESR (mm/h), median (min–max) | 18.00 (2–85) |
| DAS 28, median (min–max) | 4.99 (0.63–8.06) |
| Disease activity | |
| Remission | 7 (18.9) |
| Low | 6 (16.2) |
| Moderate | 11 (29.7) |
| High | 13 (35.1) |
| MTX treatment * (n, %) | |
| Failure | 16 (43.2) |
| Responsive | 17 (45.9) |
| Adverse events | 2 (5.4) |
| NA | 2 (5.4) |
| Time of treatment α, median (min–max) | 3 (0–408) |
| Periodontitis ∞ (n, %) | |
| None | 10 (27.0) |
| Mild | - |
| Moderate | 21 (56.8) |
| Severe | 6 (16.2) |
| Variables | T0, n = 37 | T1, n = 32 | T2, n = 28 | p-Value |
|---|---|---|---|---|
| Probing depth (mm), median (min–max) | 1.92 (0.86–3.16) | 1.70 (0.64–3.70) | 1.55 a,b (0.52–2.98) | <0.001 § |
| CAL (mm), median (min–max) | 2.05 (1.06–5.15) | 1.89 (0.83–7.14) | 1.83 a (0.78–6.92) | 0.026 § |
| BOP (%), median (min–max) | 9.00 (0–53) | 24.00 a (6–85) | 10.00 b (0–35) | <0.001 § |
| Plaque index (%), median (min–max) | 42.00 (0–100) | 37.50 (6–92) | 18.00 a,b (3–50) | <0.001 § |
| Periodontitis (n, %) | ||||
| None | 10 (27.0) | 8 (21.6) | 11 (29.7) | <0.05 ¶ |
| Mild | 0 (0.0) | 1 (2.7) | 0 (0.0) | |
| Moderate | 21 (56.8) | 17 (45.9) | 13 a,b (35.1) | |
| Severe | 6 (16.2) | 6 (16.2) | 4 (10.8) | |
| NA | - | 5 (13.6) | 9 (24.4) | |
| DAS 28, median (min–max) | 4.99 (0.63–8.06) | 4.49 (0.14–7.76) | - | 0.082 § |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, S.R.; de Arruda, J.A.A.; Corrêa, J.D.; Carvalho, V.F.; Medeiros, J.D.; Schneider, A.H.; Machado, C.C.; Duffles, L.F.; Fernandes, G.d.R.; Calderaro, D.C.; et al. Methotrexate and Non-Surgical Periodontal Treatment Change the Oral–Gut Microbiota in Rheumatoid Arthritis: A Prospective Cohort Study. Microorganisms 2024, 12, 68. https://doi.org/10.3390/microorganisms12010068
Oliveira SR, de Arruda JAA, Corrêa JD, Carvalho VF, Medeiros JD, Schneider AH, Machado CC, Duffles LF, Fernandes GdR, Calderaro DC, et al. Methotrexate and Non-Surgical Periodontal Treatment Change the Oral–Gut Microbiota in Rheumatoid Arthritis: A Prospective Cohort Study. Microorganisms. 2024; 12(1):68. https://doi.org/10.3390/microorganisms12010068
Chicago/Turabian StyleOliveira, Sicília Rezende, José Alcides Almeida de Arruda, Jôice Dias Corrêa, Valessa Florindo Carvalho, Julliane Dutra Medeiros, Ayda Henriques Schneider, Caio Cavalcante Machado, Letícia Fernanda Duffles, Gabriel da Rocha Fernandes, Débora Cerqueira Calderaro, and et al. 2024. "Methotrexate and Non-Surgical Periodontal Treatment Change the Oral–Gut Microbiota in Rheumatoid Arthritis: A Prospective Cohort Study" Microorganisms 12, no. 1: 68. https://doi.org/10.3390/microorganisms12010068
APA StyleOliveira, S. R., de Arruda, J. A. A., Corrêa, J. D., Carvalho, V. F., Medeiros, J. D., Schneider, A. H., Machado, C. C., Duffles, L. F., Fernandes, G. d. R., Calderaro, D. C., Júnior, M. T., Abreu, L. G., Fukada, S. Y., Oliveira, R. D. R., Louzada-Júnior, P., Cunha, F. Q., & Silva, T. A. (2024). Methotrexate and Non-Surgical Periodontal Treatment Change the Oral–Gut Microbiota in Rheumatoid Arthritis: A Prospective Cohort Study. Microorganisms, 12(1), 68. https://doi.org/10.3390/microorganisms12010068








