Molecular Characterization of Carbapenem and Colistin Resistance in Klebsiella pneumoniae Isolates Obtained from Clinical Samples at a University Hospital Center in Algeria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Clinical Sample Collection
2.2. Antimicrobial Susceptibility Testing
2.3. Molecular Investigation of Antibiotic Resistance Genes
2.4. Primer and TaqMan Probes
2.5. Multiplex Real-Time PCR
2.6. Standard PCR
2.7. Primers and Probes for PCR and qPCR
2.8. Statistical Analysis
3. Results
3.1. Distribution of Strains by Sample Source, Hospital Department, and Patient Demographics
3.2. Antimicrobial Susceptibility
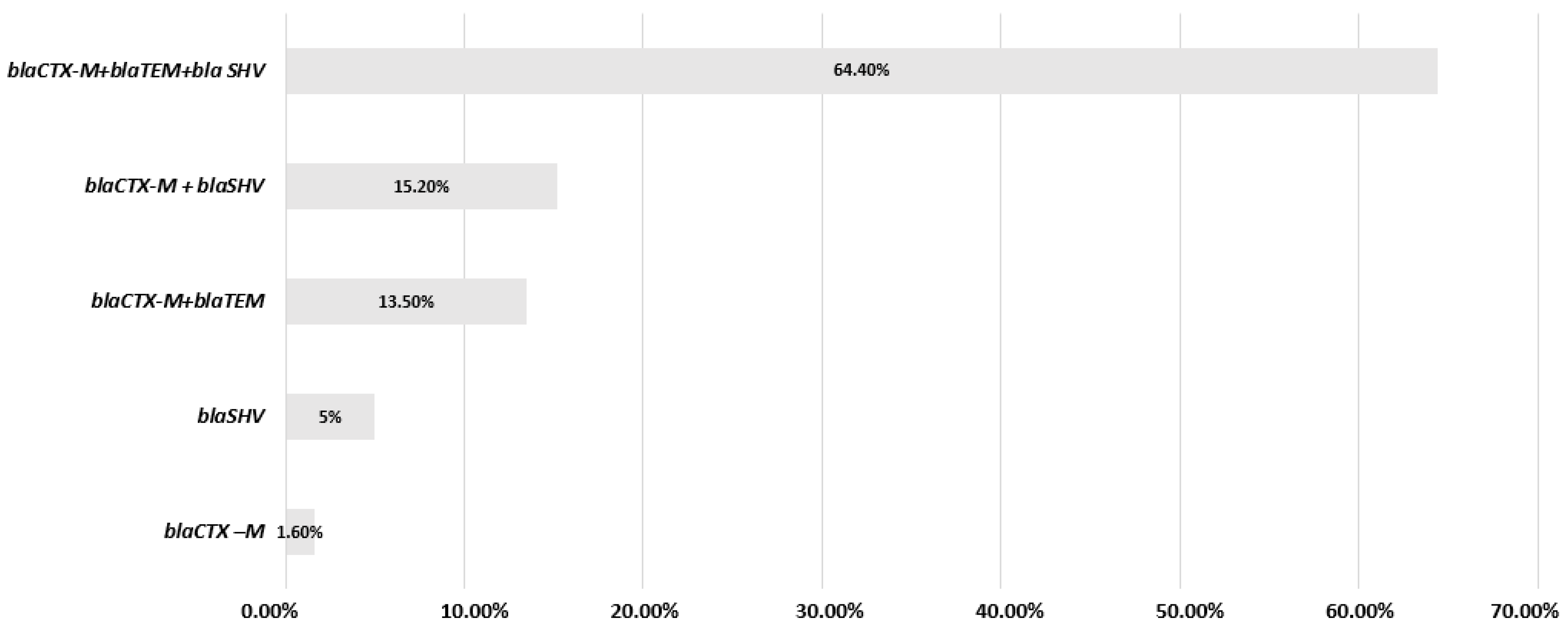
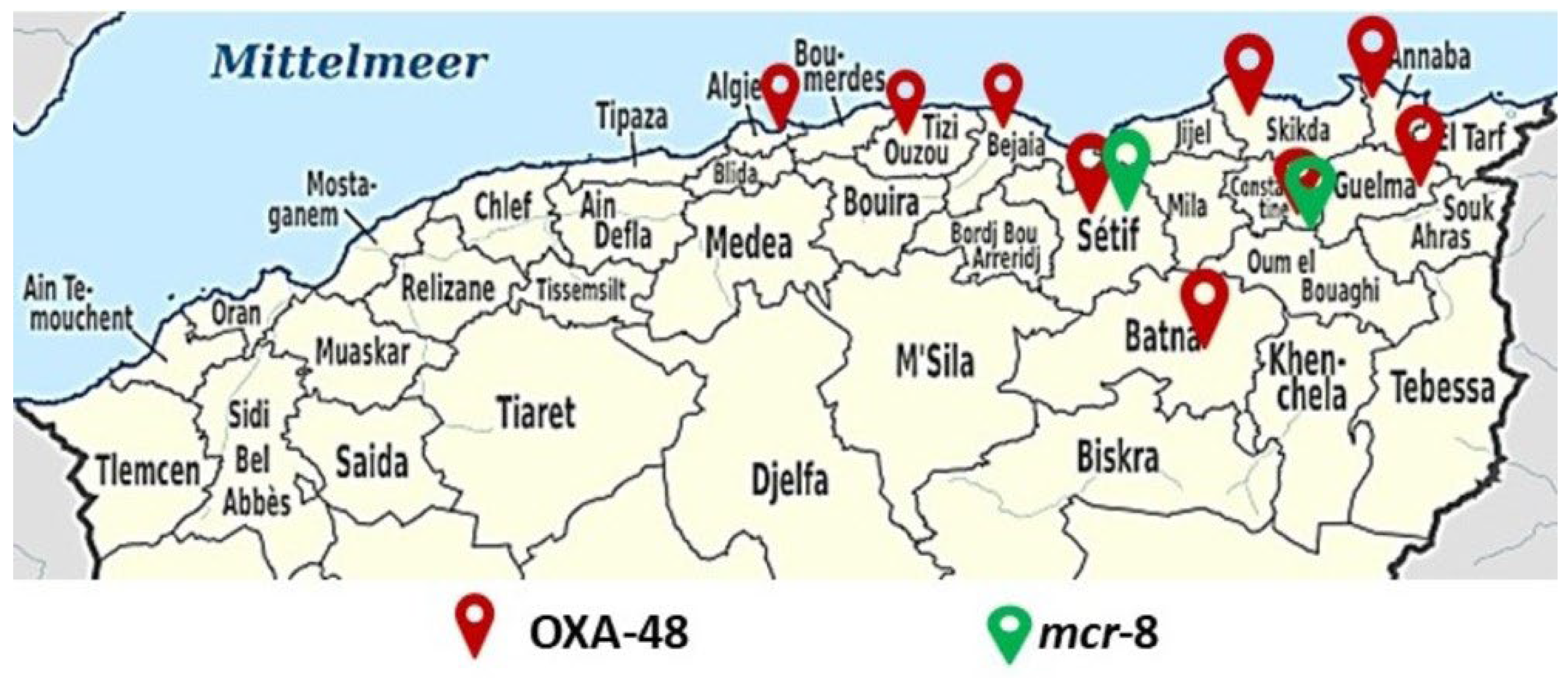
3.3. Molecular Characterization of β-Lactamases, Carbapenemases, mcr Genes and mgrB Modifications
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Committee Approval
References
- Coque, T.M.; Baquero, F.; Canton, R. Increasing Prevalence of ESBL-Producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 1–11. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global Spread of Carbapenemase Producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Xu, T.; Feng, W.; Sun, F.; Qian, Y. Clinical and Resistance Characterization of Carbapenem-Resistant Klebsiella pneumoniae Isolated from Intensive Care Units in China. Ann. Transl. Med. 2022, 10, 1109. [Google Scholar] [CrossRef] [PubMed]
- Manenzhe, R.I.; Zar, H.J.; Nicol, M.P.; Kaba, M. The Spread of Carbapenemase-Producing Bacteria in Africa: A Systematic Review. J. Antimicrob. Chemother. 2015, 70, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Nabti, L.Z.; Sahli, F.; Olowo-Okere, A.; Benslama, A.; Harrar, A.; Lupande-Mwenebitu, D.; Diene, S.M.; Rolain, J.-M. Molecular Characterization of Clinical Carbapenem-Resistant Enterobacteriaceae Isolates from Sétif, Algeria. Microb. Drug Resist. 2022, 28, 274–279. [Google Scholar] [CrossRef]
- Al Bshabshe, A.; Al-Hakami, A.; Alshehri, B.; Al-Shahrani, K.A.; Alshehri, A.A.; Al Shahrani, M.B.; Assiry, I.; Joseph, M.R.; Alkahtani, A.M.; Hamid, M.E. Rising Klebsiella pneumoniae Infections and Its Expanding Drug Resistance in the Intensive Care Unit of a Tertiary Healthcare Hospital, Saudi Arabia. Cureus 2020, 12, e10060. [Google Scholar] [CrossRef]
- Reyes, J.; Aguilar, A.C.; Caicedo, A. Carbapenem-Resistant Klebsiella pneumoniae: Microbiology Key Points for Clinical Practice. Int. J. Gen. Med. 2019, 12, 437–446. [Google Scholar] [CrossRef]
- Tseng, C.-H.; Huang, Y.-T.; Mao, Y.-C.; Lai, C.-H.; Yeh, T.-K.; Ho, C.-M.; Liu, P.-Y. Insight into the Mechanisms of Carbapenem Resistance in Klebsiella pneumoniae: A Study on IS26 Integrons, Beta-Lactamases, Porin Modifications, and Plasmidome Analysis. Antibiotics 2023, 12, 749. [Google Scholar] [CrossRef]
- Armin, S.; Fallah, F.; Karimi, A.; Azimi, T.; Kafil, H.S.; Zahedani, S.S.; Ghanaiee, R.M.; Azimi, L. Multicentre Study of the Main Carbapenem Resistance Mechanisms in Important Members of the Enterobacteriaceae Family in Iran. New Microbes New Infect. 2021, 41, 100860. [Google Scholar] [CrossRef]
- Janssen, A.B.; Doorduijn, D.J.; Mills, G.; Rogers, M.R.C.; Bonten, M.J.M.; Rooijakkers, S.H.M.; Willems, R.J.L.; Bengoechea, J.A.; van Schaik, W. Evolution of Colistin Resistance in the Klebsiella pneumoniae Complex Follows Multiple Evolutionary Trajectories with Variable Effects on Fitness and Virulence Characteristics. Antimicrob. Agents Chemother. 2020, 65, 10–1128. [Google Scholar] [CrossRef]
- Higashino, H.R.; Marchi, A.P.; Martins, R.C.R.; Batista, M.V.; Perdigão Neto, L.V.; Lima, V.A.C.d.C.; Rossi, F.; Guimarães, T.; Levin, A.S.; Rocha, V.; et al. Colistin-Resistant Klebsiella pneumoniae Co-Harboring KPC and MCR-1 in a Hematopoietic Stem Cell Transplantation Unit. Bone Marrow Transpl. 2019, 54, 1118–1120. [Google Scholar] [CrossRef]
- Nation, R.L.; Li, J. Colistin in the 21st Century. Curr. Opin. Infect. Dis. 2009, 22, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Giani, T.; Arena, F.; Vaggelli, G.; Conte, V.; Chiarelli, A.; De Angelis, L.H.; Fornaini, R.; Grazzini, M.; Niccolini, F.; Pecile, P.; et al. Large Nosocomial Outbreak of Colistin-Resistant, Carbapenemase-Producing Klebsiella pneumoniae Traced to Clonal Expansion of an Mgrb Deletion Mutant. J. Clin. Microbiol. 2015, 53, 3341–3344. [Google Scholar] [CrossRef] [PubMed]
- Cannatelli, A.; D’Andrea, M.M.; Giani, T.; Di Pilato, V.; Arena, F.; Ambretti, S.; Gaibani, P.; Rossolini, G.M. In Vivo Emergence of Colistin Resistance in Klebsiella pneumoniae Producing KPC-Type Carbapenemases Mediated by Insertional Inactivation of the PhoQ/PhoP MgrB Regulator. Antimicrob. Agents Chemother. 2013, 57, 5521–5526. [Google Scholar] [CrossRef] [PubMed]
- Cannatelli, A.; Giani, T.; D’Andrea, M.M.; Di Pilato, V.; Arena, F.; Conte, V.; Tryfinopoulou, K.; Vatopoulos, A.; Rossolini, G.M. MgrB Inactivation Is a Common Mechanism of Colistin Resistance in KPC-Producing Klebsiella pneumoniae of Clinical Origin. Antimicrob. Agents Chemother. 2014, 58, 5696–5703. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Boyce, J.D. Mechanisms of Polymyxin Resistance. In Polymyxin Antibiotics: From Laboratory Bench to Bedside; Springer: Cham, Switzerland, 2019; pp. 55–71. [Google Scholar]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of Novel Mobile Colistin Resistance Gene Mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, Y.; Wang, Z.; Hu, N.; Liu, Q.; Zhou, W.; Li, X.; Hu, L.; Guo, J.; Huang, X.; et al. Molecular Mechanisms of Colistin Resistance in Klebsiella pneumoniae in a Tertiary Care Teaching Hospital. Front. Cell Infect. Microbiol. 2021, 11, 673503. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, H.; Liu, Y.-H.; Feng, Y. Towards Understanding MCR-like Colistin Resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef]
- Narimisa, N.; Goodarzi, F.; Bavari, S. Prevalence of Colistin Resistance of Klebsiella pneumoniae Isolates in Iran: A Systematic Review and Meta-Analysis. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 29. [Google Scholar] [CrossRef]
- Llobet, E.; Tomás, J.M.; Bengoechea, J.A. Capsule Polysaccharide Is a Bacterial Decoy for Antimicrobial Peptides. Microbiology 2008, 154, 3877–3886. [Google Scholar] [CrossRef] [PubMed]
- Berrazeg, M.; Hadjadj, L.; Ayad, A.; Drissi, M.; Rolain, J.M. First Detected Human Case in Algeria of Mcr-1 Plasmid-Mediated Colistin Resistance in a 2011 Escherichia Coli Isolate. Antimicrob. Agents Chemother. 2016, 60, 6996–6997. [Google Scholar] [CrossRef] [PubMed]
- Diene, S.M.; Bruder, N.; Raoult, D.; Rolain, J.-M. Real-Time PCR Assay Allows Detection of the New Delhi Metallo-β-Lactamase (NDM-1)-Encoding Gene in France. Int. J. Antimicrob. Agents 2011, 37, 544–546. [Google Scholar] [CrossRef] [PubMed]
- Rolain, J.M.; Canton, R.; Cornaglia, G. Emergence of Antibiotic Resistance: Need for a New Paradigm. Clin. Microbiol. Infect. 2012, 18, 615–616. [Google Scholar] [CrossRef] [PubMed]
- Roschanski, N.; Fischer, J.; Guerra, B.; Roesler, U. Development of a Multiplex Real-Time PCR for the Rapid Detection of the Predominant Beta-Lactamase Genes CTX-M, SHV, TEM and CIT-Type AmpCs in Enterobacteriaceae. PLoS ONE 2014, 9, e100956. [Google Scholar] [CrossRef]
- Kruger, T.; Szabo, D.; Keddy, K.H.; Deeley, K.; Marsh, J.W.; Hujer, A.M.; Bonomo, R.A.; Paterson, D.L. Infections with Nontyphoidal Salmonella Species Producing TEM-63 or a Novel TEM Enzyme, TEM-131, in South Africa. Antimicrob. Agents Chemother. 2004, 48, 4263–4270. [Google Scholar] [CrossRef]
- Oka, K.; Tetsuka, N.; Morioka, H.; Iguchi, M.; Kawamura, K.; Hayashi, K.; Yanagiya, T.; Morokuma, Y.; Watari, T.; Kiyosuke, M.; et al. Genetic and Epidemiological Analysis of ESBL-Producing Klebsiella pneumoniae in Three Japanese University Hospitals. J. Infect. Chemother. 2022, 28, 1286–1294. [Google Scholar] [CrossRef]
- Poirel, L.; Naas, T.; Nordmann, P. Diversity, Epidemiology, and Genetics of Class D β-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 24–38. [Google Scholar] [CrossRef]
- Jena, J.; Sahoo, R.K.; Debata, N.K.; Subudhi, E. Prevalence of TEM, SHV, and CTX-M Genes of Extended-Spectrum β-Lactamase-Producing Escherichia Coli Strains Isolated from Urinary Tract Infections in Adults. 3 Biotech 2017, 7, 244. [Google Scholar] [CrossRef]
- Cantón, R.; Coque, T.M. The CTX-M β-Lactamase Pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef]
- Podschun, R.; Ullmann, U. Klebsiella Spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Keynan, Y.; Rubinstein, E. The Changing Face of Klebsiella pneumoniae Infections in the Community. Int. J. Antimicrob. Agents 2007, 30, 385–389. [Google Scholar] [CrossRef] [PubMed]
- McNulty, C.A.M.; Lecky, D.M.; Xu-McCrae, L.; Nakiboneka-Ssenabulya, D.; Chung, K.T.; Nichols, T.; Thomas, H.L.; Thomas, M.; Alvarez-Buylla, A.; Turner, K.; et al. CTX-M ESBL-Producing Enterobacteriaceae: Estimated Prevalence in Adults in England in 2014. J. Antimicrob. Chemother. 2018, 73, 1368–1388. [Google Scholar] [CrossRef] [PubMed]
- Müller-Schulte, E.; Tuo, M.N.; Akoua-Koffi, C.; Schaumburg, F.; Becker, S.L. High Prevalence of ESBL-Producing Klebsiella pneumoniae in Clinical Samples from Central Côte d’Ivoire. Int. J. Infect. Dis. 2020, 91, 207–209. [Google Scholar] [CrossRef]
- Founou, R.C.; Founou, L.L.; Allam, M.; Ismail, A.; Essack, S.Y. Whole Genome Sequencing of Extended Spectrum β-Lactamase (ESBL)-Producing Klebsiella pneumoniae Isolated from Hospitalized Patients in KwaZulu-Natal, South Africa. Sci. Rep. 2019, 9, 6266. [Google Scholar] [CrossRef]
- Natoubi, S.; Barguigua, A.; Zriouil, S.B.; Baghdad, N.; Timinouni, M.; Hilali, A.; Amghar, S.; Zerouali, K. Incidence of Extended-Spectrum Be-Ta-Lactamase-Producing Klebsiella pneumoniae among Patients and in the Environment of Hassan II Hospital, Settat, Morocco. Adv. Microbiol. 2016, 6, 152–161. [Google Scholar] [CrossRef]
- Bariz, K.; De Mendonça, R.; Denis, O.; Nonhoff, C.; Azzam, A.; Houali, K. Multidrug Resistance of the Extended-Spectrum Beta-Lactamase-Producing Klebsiella pneumoniae Isolated in Tizi-Ouzou (Algeria). Cell. Mol. Biol. 2019, 65, 11–17. [Google Scholar] [CrossRef]
- Aysert-Yildiz, P.; Özgen-Top, Ö.; Habibi, H.; Dizbay, M. Efficacy and Safety of Intravenous Fosfomycin for the Treatment of Carbapenem-Resistant Klebsiella pneumoniae. J. Chemother. 2023, 35, 471–476. [Google Scholar] [CrossRef]
- Ghayaz, F.; Kelishomi, F.Z.; Amereh, S.; Aali, E.; Javadi, A.; Peymani, A.; Nikkhahi, F. In Vitro Activity of Fosfomycin on Multidrug-Resistant Strains of Klebsiella pneumoniae and Klebsiella Oxytoca Causing Urinary Tract Infection. Curr. Microbiol. 2023, 80, 115. [Google Scholar] [CrossRef]
- Ahmadi, Z.; Noormohammadi, Z.; Ranjbar, R.; Behzadi, P. Prevalence of Tetracycline Resistance Genes Tet (A, B, C, 39) in Klebsiella pneumoniae Isolated from Tehran, Iran. Iran. J. Med. Microbiol. 2022, 16, 141–147. [Google Scholar] [CrossRef]
- Caubey, M.; Suchitra, M.S. Occurrence of TEM, SHV and CTX-M β Lactamases in Clinical Isolates of Proteus Species in a Tertiary Care Center. Infect. Disord. Drug Targets 2017, 18, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Mairi, A.; Meyer, S.; Tilloy, V.; Barraud, O.; Touati, A. Whole Genome Sequencing of Extended-Spectrum Beta-Lactamase-Producing Klebsiella pneumoniae Isolated from Neonatal Bloodstream Infections at a Neonatal Care Unit, Algeria. Microb. Drug Resist. 2022, 28, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. The Difficult-to-Control Spread of Carbapenemase Producers among Enterobacteriaceae Worldwide. Clin. Microbiol. Infect. 2014, 20, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Giakkoupi, P.; Papagiannitsis, C.C.; Miriagou, V.; Pappa, O.; Polemis, M.; Tryfinopoulou, K.; Tzouvelekis, L.S.; Vatopoulos, A.C. An Update of the Evolving Epidemic of BlaKPC-2-Carrying Klebsiella pneumoniae in Greece (2009–2010). J. Antimicrob. Chemother. 2011, 66, 1510–1513. [Google Scholar] [CrossRef]
- Patel, G.; Huprikar, S.; Factor, S.H.; Jenkins, S.G.; Calfee, D.P. Outcomes of Carbapenem-Resistant Klebsiella pneumoniae Infection and the Impact of Antimicrobial and Adjunctive Therapies. Infect. Control Hosp. Epidemiol. 2008, 29, 1099–1106. [Google Scholar] [CrossRef]
- Olowo-Okere, A.; Ibrahim, Y.K.E.; Olayinka, B.O. Molecular Characterisation of Extended-Spectrum β-Lactamase-Producing Gram-Negative Bacterial Isolates from Surgical Wounds of Patients at a Hospital in North Central Nigeria. J. Glob. Antimicrob. Resist. 2018, 14, 85–89. [Google Scholar] [CrossRef]
- Saseedharan, S. Act Fast as Time Is Less: High Faecal Carriage of CarbapenemResistant Enterobacteriaceae in Critical Care Patients. J. Clin. Diagn. Res. 2016, 10, DC01. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Surveillance Surveillance Atlas of Infectious Diseases; European Centre for Disease Prevention and Control: Solna, Sweden, 2019. [Google Scholar]
- Maseda, E.; Salgado, P.; Anillo, V.; Ruiz-Carrascoso, G.; Gómez-Gil, R.; Martín-Funke, C.; Gimenez, M.-J.; Granizo, J.-J.; Aguilar, L.; Gilsanz, F. Risk Factors for Colonization by Carbapenemase-Producing Enterobacteria at Admission to a Surgical ICU: A Retrospective Study. Enferm. Infecc. Microbiol. Clin. 2017, 35, 333–337. [Google Scholar] [CrossRef]
- David, S.; Reuter, S.; Harris, S.R.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of Carbapenem-Resistant Klebsiella pneumoniae in Europe Is Driven by Nosocomial Spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef]
- Martínez-Martínez, L.; Conejo, M.C.; Pascual, A.; Hernández-Allés, S.; Ballesta, S.; Ramírez De Arellano-Ramos, E.; Benedí, V.J.; Perea, E.J. Activities of Imipenem and Cephalosporins against Clonally Related Strains of Escherichia Coli Hyperproducing Chromosomal Beta-Lactamase and Showing Altered Porin Profiles. Antimicrob. Agents Chemother. 2000, 44, 2534–2536. [Google Scholar] [CrossRef]
- Nordmann, P.; Carrer, A. Les Carbapénèmases des Entérobactéries. Arch. Pediatr. 2010, 17, S154–S162. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Hindler, J.A.; Humphries, R.M. Colistin MIC Variability by Method for Contemporary Clinical Isolates of Multidrug-Resistant Gram-Negative Bacilli. J. Clin. Microbiol. 2013, 51, 1678–1684. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P. Résistance Aux Carbapénèmes Chez Les Bacilles à Gram Négatif. Med. Sci. 2010, 26, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, E.; Kojima, S.; Nikaido, H. Klebsiella pneumoniae Major Porins OmpK35 and OmpK36 Allow More Efficient Diffusion of β-Lactams than Their Escherichia Coli Homologs OmpF and OmpC. J. Bacteriol. 2016, 198, 3200–3208. [Google Scholar] [CrossRef]
- Aggoune, N.; Tali-Maamar, H.; Assaous, F.; Guettou, B.; Laliam, R.; Benamrouche, N.; Zerouki, A.; Naim, M.; Rahal, K. Wide Spread of Oxa-48-Producing Enterobacteriaceae in Algerian Hospitals: A Four Years’ Study. J. Infect. Dev. Ctries. 2018, 12, 1039–1044. [Google Scholar] [CrossRef]
- Memariani, M.; Najar-Peerayeh, S.; Salehi, T.Z.; Mostafavi, S.K.S. Occurrence of SHV, TEM and CTX-M β-Lactamase Genes among Enteropathogenic Escherichia Coli Strains Isolated from Children with Diarrhea. Jundishapur J. Microbiol. 2015, 8, e15620. [Google Scholar] [CrossRef]
- Blot, K.; Hammami, N.; Blot, S.; Vogelaers, D.; Lambert, M.L. Gram-Negative Central Line-Associated Bloodstream Infection Incidence Peak during the Summer: A National Seasonality Cohort Study. Sci. Rep. 2022, 12, 5202. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, F.; Hu, Y.; Zhang, G.; Zhu, B.; Gao, G.F. Detection of Mobile Colistin Resistance Gene Mcr-9 in Carbapenem-Resistant Klebsiella pneumoniae Strains of Human Origin in Europe. J. Infect. 2020, 80, 578–606. [Google Scholar] [CrossRef]
- Abdelhamid, S.M.; Abd-Elaal, H.M.; Matareed, M.O.; Baraka, K. Genotyping and Virulence Analysis of Drug Resistant Clinical Klebsiella pneumoniae Isolates in Egypt. J. Pure Appl. Microbiol. 2020, 14, 1967–1975. [Google Scholar] [CrossRef]
- Kulkarni, A.A.; Ebadi, M.; Zhang, S.; Meybodi, M.A.; Ali, A.M.; Defor, T.; Shanley, R.; Weisdorf, D.; Ryan, C.; Vasu, S.; et al. Comparative Analysis of Antibiotic Exposure Association with Clinical Outcomes of Chemotherapy versus Immunotherapy across Three Tumour Types. ESMO Open 2020, 5, e000803. [Google Scholar] [CrossRef] [PubMed]
- Fuster, B.; Salvador, C.; Tormo, N.; García-González, N.; Gimeno, C.; González-Candelas, F. Molecular Epidemiology and Drug-Resistance Mechanisms in Carbapenem-Resistant Klebsiella pneumoniae Isolated in Patients from a Tertiary Hospital in Valencia, Spain. J. Glob. Antimicrob. Resist. 2020, 22, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Poudyal, A.; Howden, B.P.; Bell, J.M.; Gao, W.; Owen, R.J.; Turnidge, J.D.; Nation, R.L.; Li, J. In Vitro Pharmacodynamics of Colistin against Multidrug-Resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2008, 62, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Zahedi Bialvaei, A.; Eslami, P.; Ganji, L.; Dolatyar Dehkharghani, A.; Asgari, F.; Koupahi, H.; Barzegarian Pashacolaei, H.R.; Rahbar, M. Prevalence and Epidemiological Investigation of MgrB-Dependent Colistin Resistance in Extensively Drug Resistant Klebsiella pneumoniae in Iran. Sci. Rep. 2023, 13, 10680. [Google Scholar] [CrossRef] [PubMed]
- Mansour, W.; Haenni, M.; Saras, E.; Grami, R.; Mani, Y.; Ben Haj Khalifa, A.; el Atrouss, S.; Kheder, M.; Fekih Hassen, M.; Boujâafar, N.; et al. Outbreak of Colistin-Resistant Carbapenemase-Producing Klebsiella pneumoniae in Tunisia. J. Glob. Antimicrob. Resist. 2017, 10, 88–94. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a Novel Mobile Colistin Resistance Gene, Mcr-8, in NDM-Producing Klebsiella pneumoniae Article. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene Mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella Enterica Serotype Typhimurium Isolate. MBio 2019, 10, 10–1128. [Google Scholar] [CrossRef]
- Olowo-Okere, A.; Yacouba, A. Molecular Mechanisms of Colistin Resistance in Africa: A Systematic Review of Literature. Germs 2020, 10, 367. [Google Scholar] [CrossRef]
- Belbel, Z.; Lalaoui, R.; Bakour, S.; Nedjai, S.; Djahmi, N.; Rolain, J.M. First Report of Colistin Resistance in an OXA-48- and a CTX-M-15 Producing Klebsiella pneumoniae Clinical Isolate in Algeria Due to PmrB Protein Modification and MgrB Inactivation. J. Glob. Antimicrob. Resist. 2018, 14, 158–160. [Google Scholar] [CrossRef]
- Nabti, L.Z.; Sahli, F.; Ngaiganam, E.P.; Radji, N.; Mezaghcha, W.; Lupande-Mwenebitu, D.; Baron, S.A.; Rolain, J.M.; Diene, S.M. Development of Real-Time PCR Assay Allowed Describing the First Clinical Klebsiella pneumoniae Isolate Harboring Plasmid-Mediated Colistin Resistance Mcr-8 Gene in Algeria. J. Glob. Antimicrob. Resist. 2020, 20, 266–271. [Google Scholar] [CrossRef]


| Gene | Primer/Probe | Sequence (5′-3′) | Size (bp) | Reference |
|---|---|---|---|---|
| blaOXA-51 | OXA-51-F1 OXA-51-R1 OXA-51- Probe | GCTCGTGCTTCGACCGAGTA TTTTTGCCCGTCCCACTTAAA FAM-TCGGCCTTGAGCACCATAAGGCA-TAMRA | 117 | [24] |
| blaOXA-23 | OXA-23-F1 OXA-23-R1 OXA-23- Probe | TGCTCTAAGCCGCGCAAATA TGACCTTTTCTCGCCCTTCC FAM-GCCCTGATCGGATTGGAGAACCA-TAMRA | 130 | [24] |
| blaOXA-24 | OXA-24-F OXA-24-R OXA-24- Probe | CAAATGAGATTTTCAAATGGGATGG TCCGTCTTGCAAGCTCTTGAT FAM-GGTGAGGCAATGGCATTGTCAGCA-TAMRA | 123 | [24] |
| blaOXA-58 | OXA-58-F OXA-58-R OXA-58- Probe | CGCAGAGGGGAGAATCGTCT TTGCCCATCTGCCTTTTCAA FAM-GGGGAATGGCTGTAGACCCGC-TAMRA | 102 | [24] |
| blaNDM-1 | NDM-1-F NDM-1-R NDM-1-probe | GCGCAACACAGCCTGACTTT CAGCCACCAAAAGCGATGTC FAM-CAACCGCGCCCAACTTTGGC-TAMRA | 155 | [24] |
| blaOXA-48 | OXA-48-F OXA-48-R OXA-48- Probe | TCTTAAACGGGCGAACCAAG GCGTCTGTCCATCCCACTTA FAM-AGCTTGATCGCCCTCGATTTGC-TAMRA | 125 | [25] |
| blaKPC | KPC-F KPC-R KPC-probe | GATACCACGTTCCGTCTGGA GGTCGTGTTTCCCTTTAGCC FAM-CGCGCGCCGTGACGGAAAGC-TAMRA | 180 | [25] |
| bla VIM | VIM-F VIM-R VIM-probe | CACAGYGGCMCTTCTCGCGGAGA GCGTACGTYGCCACYCCAGCC FAMAGTCTCCACGCACTTTCATGACCGCGTCGGC G-TAMRA | 132 | Jean.M. ROLAIN Laboratory [23] |
| blaCTX-M | CTX-M F CTX-M R CTX-M Probe | CGGGCRATGGCGCARAC TGCRCCGGTSGTATTGCC CCARCGGGCGCAGYTGGTGAC | 105 | [26] |
| Gene | Positive Control | Primer | Sequence 5′-3′ | Size (bp) | Reference |
|---|---|---|---|---|---|
| blaTEM | Kpnasey | F | ATGAGTATTCAACATTTCCGTG | 861 | [27] |
| R | TTACCAATGCTTAATCAGTGAG | ||||
| blaCTX | Kpnasey | F | CCCATGGTTAAAAAATCACTGC | 944 | [26] |
| R | CAGCGCTTTTGCCGTCTCCG | ||||
| blaSHV | Kpnasey | F | ATTTGTCGCTTCTTTACTCGC | 1051 | [28] |
| R | TTTATGGCGTTACCTTTGACC | ||||
| blaOXA-48 | E. coli CMUL64 | F | TTGGTGGCATCGATTATCGG | 744 | [29] |
| R | GAGCACTTCTTTTGTGATGGC | ||||
| blaKPC | Kpnasey | F | ATGTCACTGTATCGCCGTCT | 893 | Jean.M. ROLAIN Laboratory [23] |
| R | TTTTCAGAGCCTTACTGCCC |
| Strains | ERT MIC (mg/L) | IMP MIC (mg/L) | Carba Gene | bla Gene Types | Samples | Department | Age (Year) | Gender |
|---|---|---|---|---|---|---|---|---|
| KP13 | 0.75 | 0.25 | OXA-48 | blaCTX-M; blaTEM; blaSHV | Respiratory sample * | Surgery | 63 | M |
| KP34 | 1 | 2 | OXA-48 | blaCTX-M; blaTEM; blaSHV | Blood | ICU | 19 | M |
| KP46 | 0.75 | 0.75 | OXA-48 | blaCTX-M; blaTEM | Pus | Miscellaneous * | 2 | M |
| KP86 | 0.75 | 2 | OXA-48 | blaTX-M; blaTEM | Blood | Neonatal service | 25days | M |
| KP87 | 32 | 2 | - | blaSHV | Blood | Surgery | 32 | M |
| KP103 | 32 | 32 | OXA-48 | Blood | Neonatal service | 23 days | F | |
| KP111 | 0.75 | 0.5 | OXA-48 | Pus | Neonatal service | 14 days | F | |
| KP117 | 1 | 0.25 | - | Blood | Miscellaneous ** | 32 | F | |
| KP118 | 0.75 | 0.38 | OXA-48 | Urine | ICU | 43 | F | |
| KP121 | 1 | 1 | OXA-48 | Urine | ICU | 70 | M | |
| KP122 | 2 | 0.25 | - | blaCTX-M blaTEM blaSHV | Urine | ICU | 41 | F |
| KP124 | 0.5 | 1 | OXA-48 | Blood | ICU | 70 | M |
| Strains | Resistance Phenotype | ColMIC (mg/L) | bla Gene Types | mcr Genes | mgrB Mutations | Department | Samples | Age | Gender |
|---|---|---|---|---|---|---|---|---|---|
| KP23 | CT, AMX, AMC, CRO, FEP, TPZ, AK, FF, SXT, TET | 16 | blaCTX-M, blaTEM, blaSHV | mcr-8 | 2 insertions 1 substitution | Internal medicine | Respiratory sample * | 48 | F |
| KP24 | CT, AMX, AMC, CRO, FEP, TPZ, AK GN, FF, SXT | 4 | blaCTX-M, blaTEM, blaSHV | - | 1 insertion | Pediatrics | Pus | 12 | M |
| KP27 | CT, AMX, AMC, CRO, FEP, TPZ, AK, GN, FF, SXT, TET | 4 | blaCTX-M, blaTEM, blaSHV | - | 3 insertions | Miscellaneous * | Respiratory ** | 46 | M |
| KP33 | CT, AMX, AMC, CRO, FEP, TPZ, FF, SXT, TET | 8 | blaCTX–M, blaSHV | mcr-8 | 2 insertions | Miscellaneous * | Biological fluid * | 11 months | F |
| KP46 | CT, AMX, AMC, CRO, FEP, TPZ, MEC, ETP, AK, GN, CIP, FF, SXT, TET | 16 | blaCTX-M, blaTEM, blaOXA-48- | - | 3 insertions | Neonatology | Blood | 25 days | M |
| KP86 | CT, AMX, AMC, CRO, FEP, TPZ, MEC, ETP, AK, GN, CIP, FF, SXT, TET | 4 | blaCTX–M, blaTEM, blaOXA-48- | - | 2 insertions | Miscellaneous * | Pus | 2 | M |
| KP115 | CT, AMX, AMC, CRO, FEP, TPZ, AK, GN, CIP, FF, SXT, TET | 23 | blaCTX-M, blaTEM, blaSHV | - | 1 insertion | ICU | Urine | 27 | F |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rihane, R.; Hecini-Hannachi, A.; Bentchouala, C.; Benlabed, K.; Diene, S.M. Molecular Characterization of Carbapenem and Colistin Resistance in Klebsiella pneumoniae Isolates Obtained from Clinical Samples at a University Hospital Center in Algeria. Microorganisms 2024, 12, 1942. https://doi.org/10.3390/microorganisms12101942
Rihane R, Hecini-Hannachi A, Bentchouala C, Benlabed K, Diene SM. Molecular Characterization of Carbapenem and Colistin Resistance in Klebsiella pneumoniae Isolates Obtained from Clinical Samples at a University Hospital Center in Algeria. Microorganisms. 2024; 12(10):1942. https://doi.org/10.3390/microorganisms12101942
Chicago/Turabian StyleRihane, Riyane, Abla Hecini-Hannachi, Chafia Bentchouala, Kaddour Benlabed, and Seydina M. Diene. 2024. "Molecular Characterization of Carbapenem and Colistin Resistance in Klebsiella pneumoniae Isolates Obtained from Clinical Samples at a University Hospital Center in Algeria" Microorganisms 12, no. 10: 1942. https://doi.org/10.3390/microorganisms12101942






