A Causal Relationship between Type 2 Diabetes and Candidiasis through Two-Sample Mendelian Randomization Analysis
Abstract
:1. Introduction
2. Materials and Methods
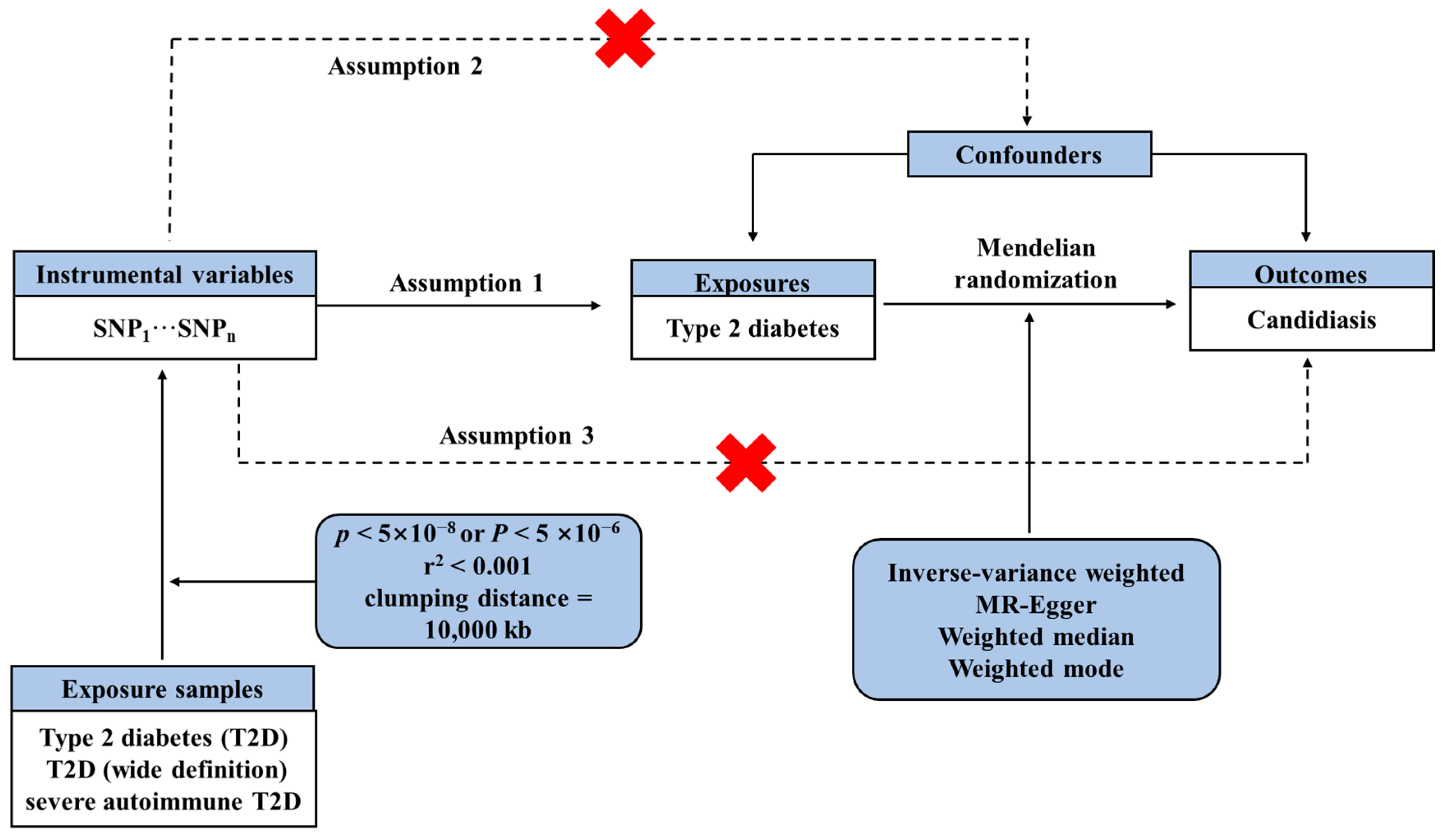
2.1. Study Design
2.2. GWAS Data Sources
2.3. Selection Criteria for IVs Selection
2.4. Mendelian Randomization Analysis
2.5. Sensitivity Analysis
3. Results
3.1. IVs Selection
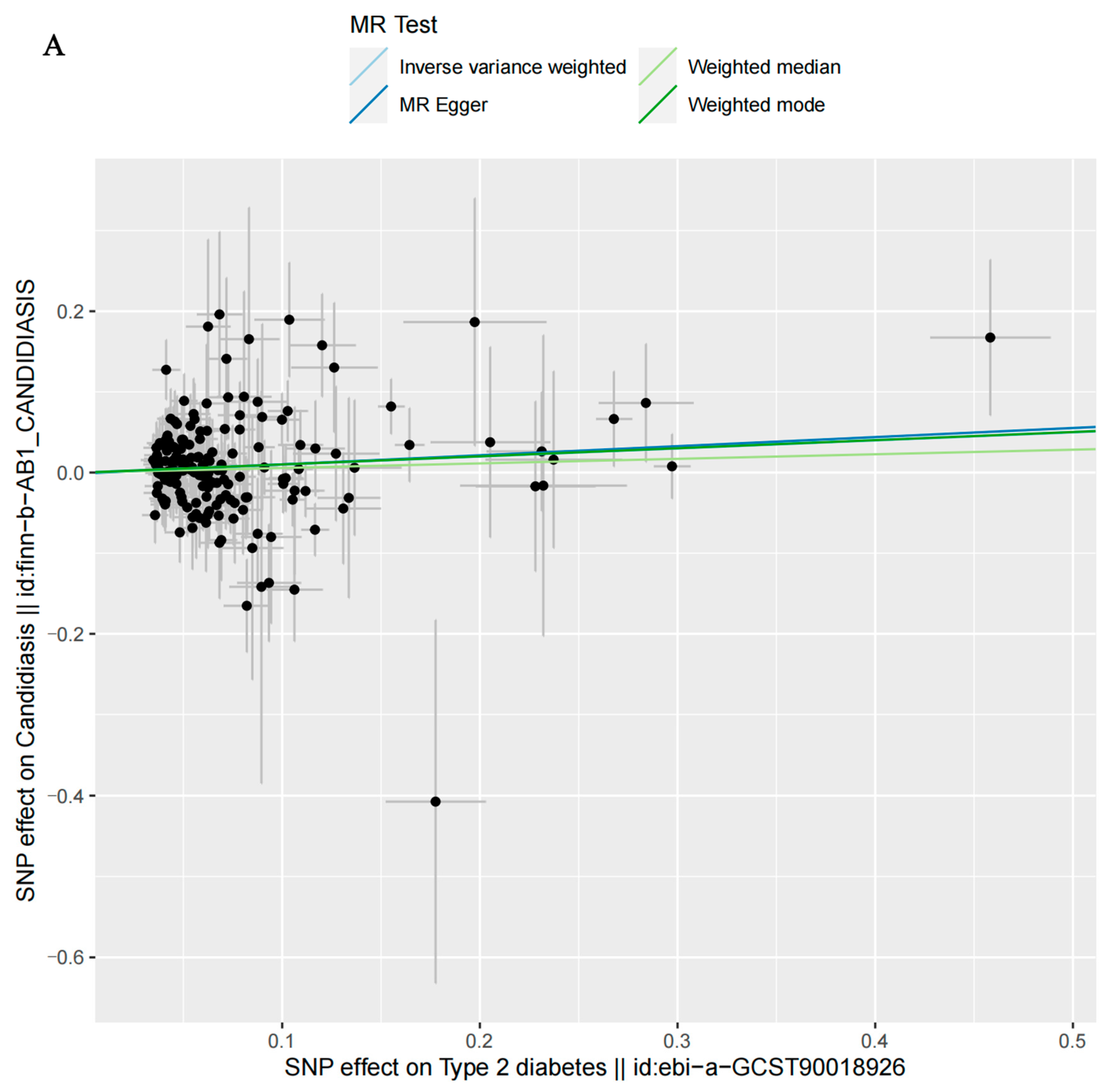
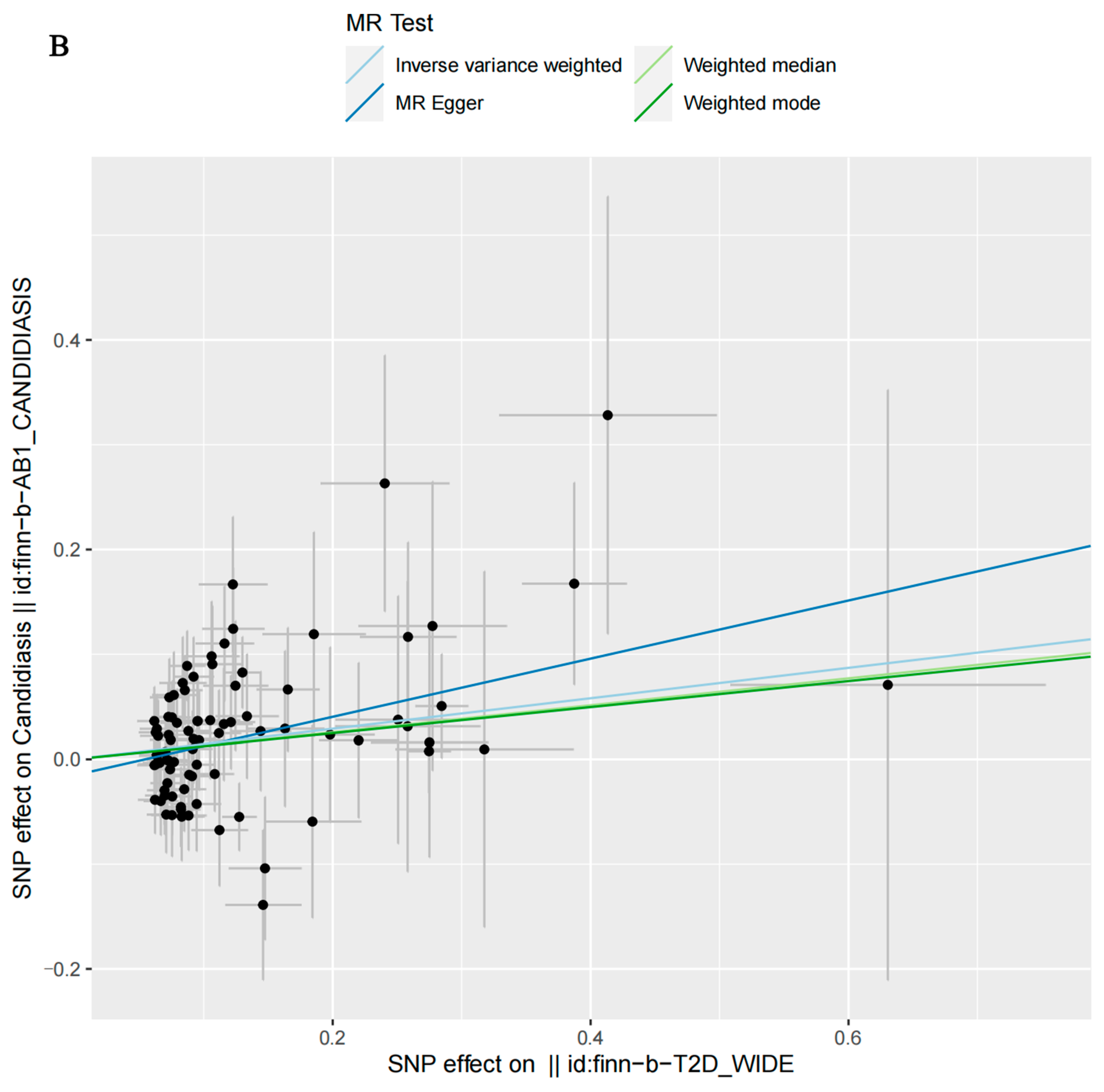
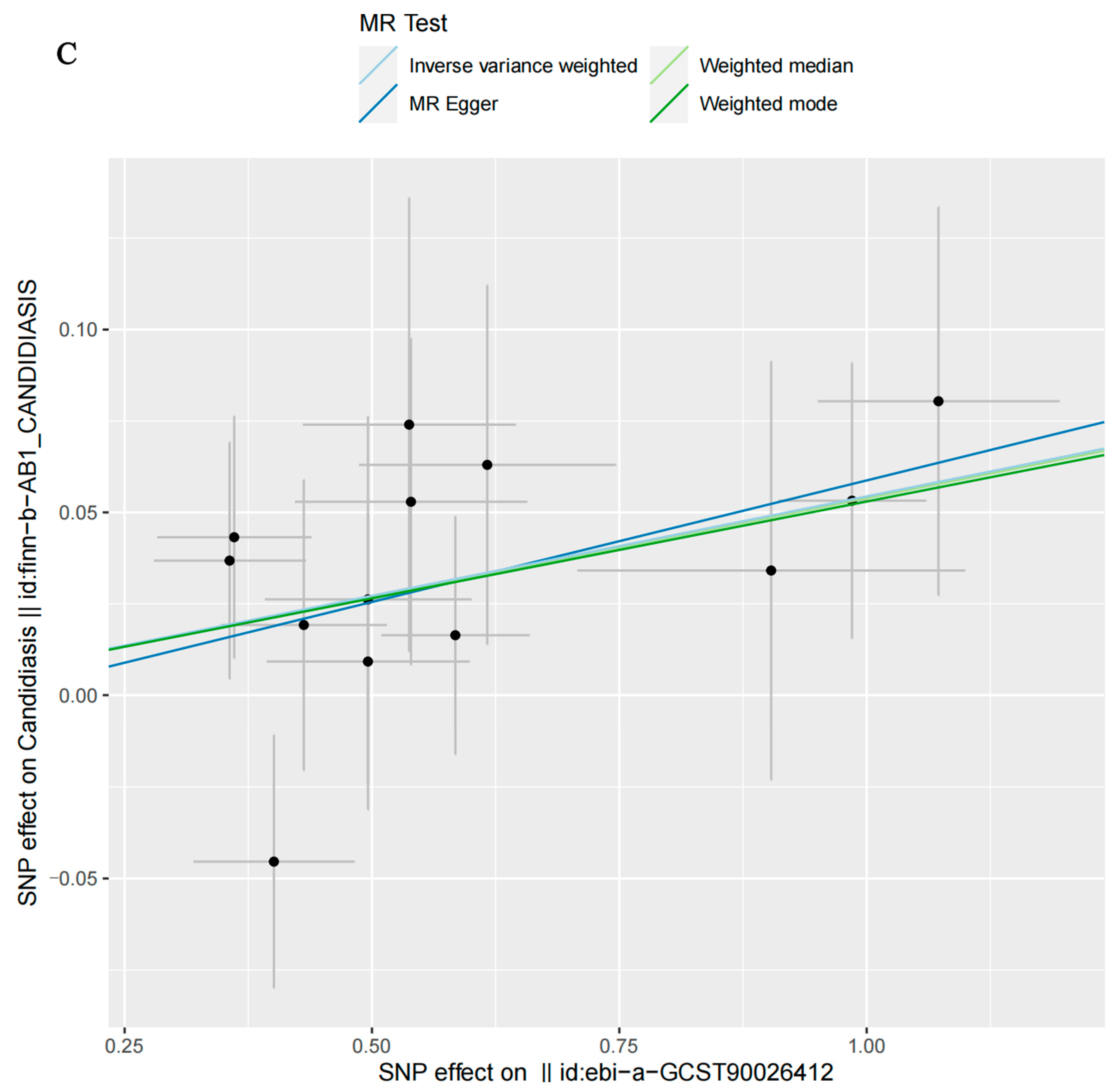
3.2. MR Analysis
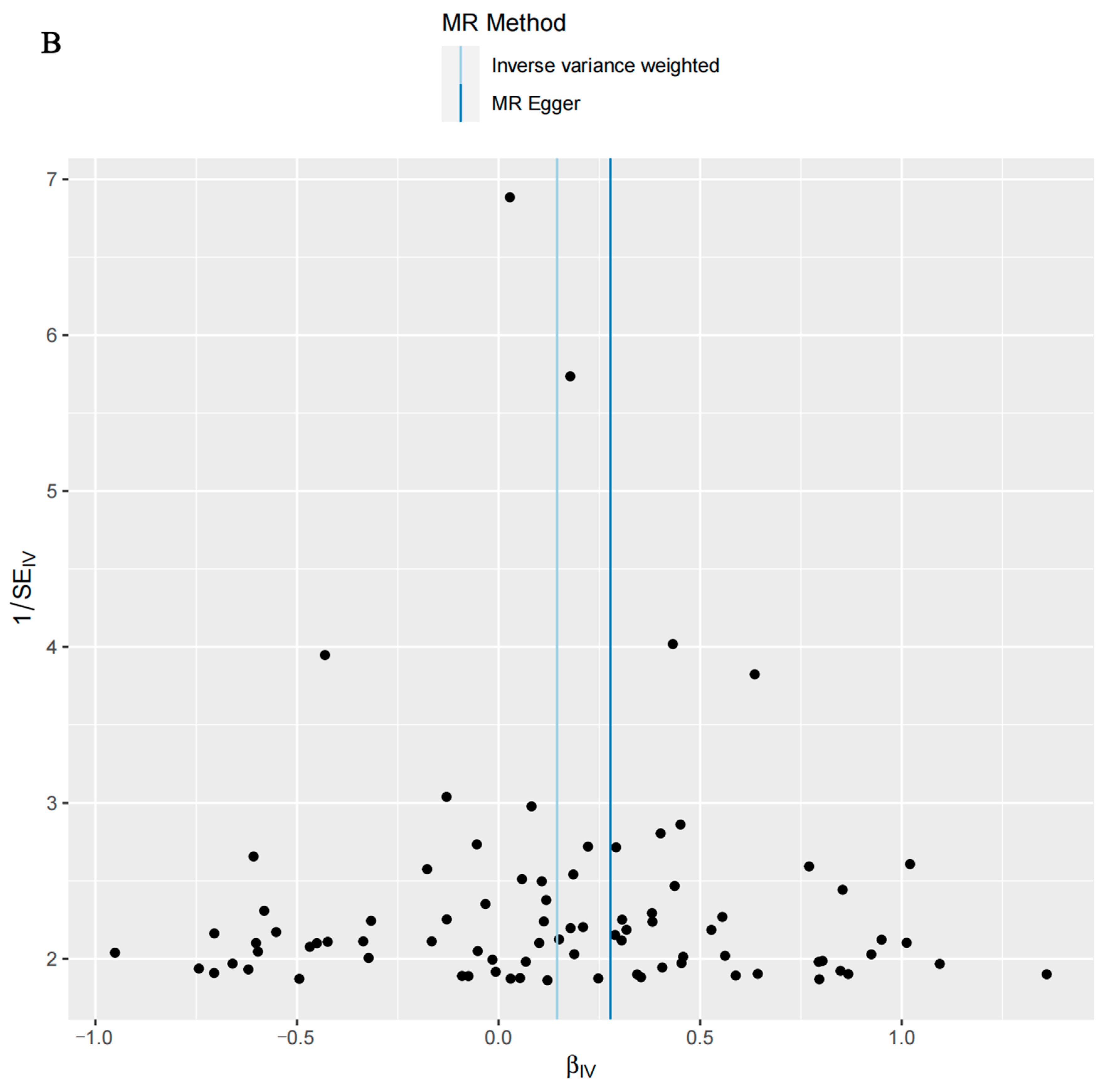
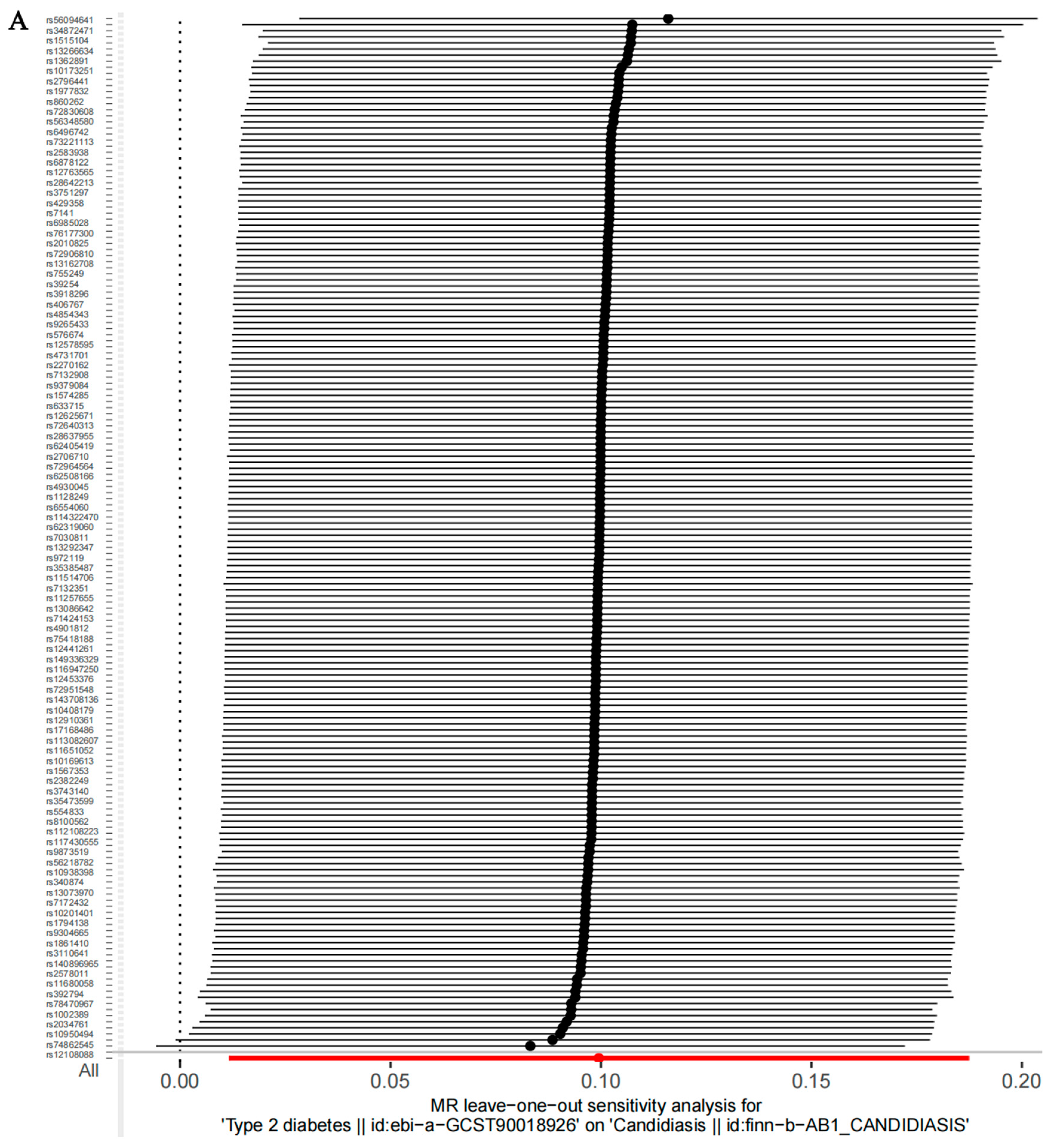
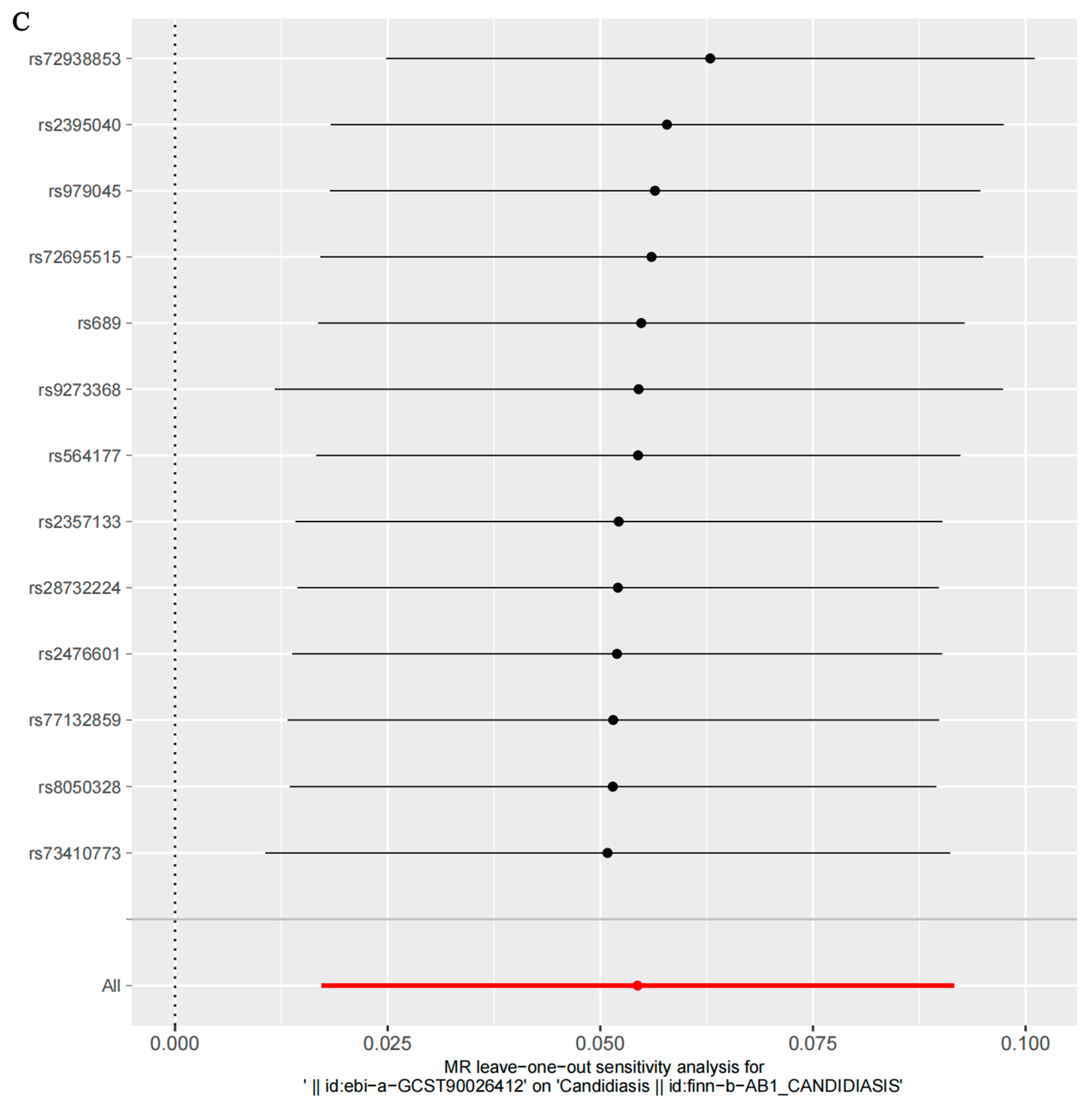
3.3. Sensitivity Analysis Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, H.; Hong, T.; Jiang, Y.; Whiteway, M.; Zhang, S. Candidiasis: From cutaneous to systemic, new perspectives of potential targets and therapeutic strategies. Adv. Drug Deliv. Rev. 2023, 199, 114960. [Google Scholar] [CrossRef]
- Denning, D.W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 2024, 24, E428–E438. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lu, H.; Jiang, Y. Natural Polyketides Act as Promising Antifungal Agents. Biomolecules 2023, 13, 1572. [Google Scholar] [CrossRef]
- Xiong, J.; Wang, L.; Feng, Y.; Zhen, C.; Hang, S.; Yu, J.; Lu, H.; Jiang, Y. Geldanamycin confers fungicidal properties to azole by triggering the activation of succinate dehydrogenase. Life Sci. 2024, 348, 122699. [Google Scholar] [CrossRef]
- Lu, H.; Li, W.; Whiteway, M.; Wang, H.; Zhu, S.; Ji, Z.; Feng, Y.; Yan, L.; Fang, T.; Li, L.; et al. A Small Molecule Inhibitor of Erg251 Makes Fluconazole Fungicidal by Inhibiting the Synthesis of the 14alpha-Methylsterols. mBio 2023, 14, e0263922. [Google Scholar] [CrossRef]
- Feng, Y.; Lu, H.; Whiteway, M.; Jiang, Y. Understanding fluconazole tolerance in Candida albicans: Implications for effective treatment of candidiasis and combating invasive fungal infections. J. Glob. Antimicrob. Resist. 2023, 35, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Mendonca, A.; Santos, H.; Franco-Duarte, R.; Sampaio, P. Fungal infections diagnosis—Past, present and future. Res. Microbiol. 2022, 173, 103915. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Li, Y.; Li, P.; Wang, M.; Wang, J.; Tang, Z.; Wang, T.; Luo, L.; Wang, C.; Wang, T.; et al. Research progress in the relationship between type 2 diabetes mellitus and intestinal flora. Biomed. Pharmacother. 2019, 117, 109138. [Google Scholar] [CrossRef]
- Tinajero, M.G.; Malik, V.S. An Update on the Epidemiology of Type 2 Diabetes: A Global Perspective. Endocrinol. Metab. Clin. North. Am. 2021, 50, 337–355. [Google Scholar] [CrossRef]
- Lao, M.; Li, C.; Li, J.; Chen, D.; Ding, M.; Gong, Y. Opportunistic invasive fungal disease in patients with type 2 diabetes mellitus from Southern China: Clinical features and associated factors. J. Diabetes Investig. 2020, 11, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Goswami, D.; Goswami, R.; Banerjee, U.; Dadhwal, V.; Miglani, S.; Lattif, A.A.; Kochupillai, N. Pattern of Candida species isolated from patients with diabetes mellitus and vulvovaginal candidiasis and their response to single dose oral fluconazole therapy. J. Infect. 2006, 52, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, H.; Nagao, A.; Watanabe, S.; Honjo, J. Incidence and risk of vaginal candidiasis associated with sodium-glucose cotransporter 2 inhibitors in real-world practice for women with type 2 diabetes. J. Diabetes Investig. 2019, 10, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Sampath, A.; Weerasekera, M.; Dilhari, A.; Gunasekara, C.; Bulugahapitiya, U.; Fernando, N.; Samaranayake, L. Type 2 diabetes mellitus and oral Candida colonization: Analysis of risk factors in a Sri Lankan cohort. Acta Odontol. Scand. 2019, 77, 508–516. [Google Scholar] [CrossRef]
- Martorano-Fernandes, L.; Dornelas-Figueira, L.M.; Marcello-Machado, R.M.; Silva, R.B.; Magno, M.B.; Maia, L.C.; Del Bel Cury, A.A. Oral candidiasis and denture stomatitis in diabetic patients: Systematic review and meta-analysis. Braz. Oral Res. 2020, 34, e113. [Google Scholar] [CrossRef] [PubMed]
- Mussi, M.C.M.; Fernandes, K.S.; Gallottini, M.H.C. A call for further research on the relation between type 2 diabetes and oral candidiasis. Oral Surg. Oral. Med. Oral Pathol. Oral Radiol. 2022, 134, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, L.; Jha, G.; Malasevskaia, I.; Goud, H.K.; Hassan, A. The Interplay Between Sugar and Yeast Infections: Do Diabetics Have a Greater Predisposition to Develop Oral and Vulvovaginal Candidiasis? Cureus 2021, 13, e13407. [Google Scholar] [CrossRef]
- Hostetter, M.K. Handicaps to host defense. Effects of hyperglycemia on C3 and Candida albicans. Diabetes 1990, 39, 271–275. [Google Scholar] [CrossRef]
- Khanna, M.; Challa, S.; Kabeil, A.S.; Inyang, B.; Gondal, F.J.; Abah, G.A.; Minnal Dhandapani, M.; Manne, M.; Mohammed, L. Risk of Mucormycosis in Diabetes Mellitus: A Systematic Review. Cureus 2021, 13, e18827. [Google Scholar] [CrossRef]
- Greenland, S.; Morgenstern, H. Confounding in health research. Annu. Rev. Public. Health 2001, 22, 189–212. [Google Scholar] [CrossRef]
- Birney, E. Mendelian Randomization. Cold Spring Harb. Perspect. Med. 2022, 12, a041302. [Google Scholar] [CrossRef]
- Burgess, S.; Davey Smith, G.; Davies, N.M.; Dudbridge, F.; Gill, D.; Glymour, M.M.; Hartwig, F.P.; Kutalik, Z.; Holmes, M.V.; Minelli, C.; et al. Guidelines for performing Mendelian randomization investigations: Update for summer 2023. Wellcome Open Res. 2019, 4, 186. [Google Scholar] [CrossRef] [PubMed]
- Kurki, M.I.; Karjalainen, J.; Palta, P.; Sipila, T.P.; Kristiansson, K.; Donner, K.M.; Reeve, M.P.; Laivuori, H.; Aavikko, M.; Kaunisto, M.A.; et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 2023, 613, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021, 53, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Mansour Aly, D.; Dwivedi, O.P.; Prasad, R.B.; Karajamaki, A.; Hjort, R.; Thangam, M.; Akerlund, M.; Mahajan, A.; Udler, M.S.; Florez, J.C.; et al. Genome-wide association analyses highlight etiological differences underlying newly defined subtypes of diabetes. Nat. Genet. 2021, 53, 1534–1542. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, S.G.; Collaboration, C.C.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Wan, X.; Zheng, H.; Xu, K.; Xie, J.; Yu, H.; Wang, J.; Xu, P. No Evidence of a Genetic Causal Relationship between Ankylosing Spondylitis and Gut Microbiota: A Two-Sample Mendelian Randomization Study. Nutrients 2023, 15, 1057. [Google Scholar] [CrossRef] [PubMed]
- Mounier, N.; Kutalik, Z. Bias correction for inverse variance weighting Mendelian randomization. Genet. Epidemiol. 2023, 47, 314–331. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Hartwig, F.P.; Davey Smith, G.; Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017, 46, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.F.; Minelli, C.; Sheehan, N.A.; Thompson, J.R. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat. Med. 2015, 34, 2926–2940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Mendelian Randomization Study Implies Causal Linkage Between Telomere Length and Juvenile Idiopathic Arthritis in a European Population. J. Inflamm. Res. 2022, 15, 977–986. [Google Scholar] [CrossRef]
- Goncalves, B.; Ferreira, C.; Alves, C.T.; Henriques, M.; Azeredo, J.; Silva, S. Vulvovaginal candidiasis: Epidemiology, microbiology and risk factors. Crit. Rev. Microbiol. 2016, 42, 905–927. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Fatima, Z.; Hameed, S. Predisposing factors endorsing Candida infections. Infez. Med. 2015, 23, 211–223. [Google Scholar] [PubMed]
- Bassyouni, R.H.; Wegdan, A.A.; Abdelmoneim, A.; Said, W.; AboElnaga, F. Phospholipase and Aspartyl Proteinase Activities of Candida Species Causing Vulvovaginal Candidiasis in Patients with Type 2 Diabetes Mellitus. J. Microbiol. Biotechnol. 2015, 25, 1734–1741. [Google Scholar] [CrossRef] [PubMed]
- Berbudi, A.; Rahmadika, N.; Tjahjadi, A.I.; Ruslami, R. Type 2 Diabetes and its Impact on the Immune System. Curr. Diabetes Rev. 2020, 16, 442–449. [Google Scholar] [CrossRef]
- Sabina, J.; Brown, V. Glucose sensing network in Candida albicans: A sweet spot for fungal morphogenesis. Eukaryot. Cell 2009, 8, 1314–1320. [Google Scholar] [CrossRef]
- Dornelas Figueira, L.M.; Ricomini Filho, A.P.; da Silva, W.J.; Del Be, L.C.A.A.; Ruiz, K.G.S. Glucose effect on Candida albicans biofilm during tissue invasion. Arch. Oral Biol. 2020, 117, 104728. [Google Scholar] [CrossRef] [PubMed]
- Van Ende, M.; Wijnants, S.; Van Dijck, P. Sugar Sensing and Signaling in Candida albicans and Candida glabrata. Front. Microbiol. 2019, 10, 99. [Google Scholar] [CrossRef]
- Saud, B.; Bajgain, P.; Paudel, G.; Shrestha, V.; Bajracharya, D.; Adhikari, S.; Dhungana, G.; Awasthi, M.S. Fungal Infection among Diabetic and Nondiabetic Individuals in Nepal. Interdiscip. Perspect. Infect. Dis. 2020, 2020, 7949868. [Google Scholar] [CrossRef] [PubMed]
- Halimi, A.; Mortazavi, N.; Memarian, A.; Zahedi, M.; Niknejad, F.; Sohrabi, A.; Sarraf, S.J. The relation between serum levels of interleukin 10 and interferon-gamma with oral candidiasis in type 2 diabetes mellitus patients. BMC Endocr. Disord. 2022, 22, 296. [Google Scholar] [CrossRef]
- Graves, D.T.; Correa, J.D.; Silva, T.A. The Oral Microbiota Is Modified by Systemic Diseases. J. Dent. Res. 2019, 98, 148–156. [Google Scholar] [CrossRef]
- Chen, T.; Qin, Y.; Chen, M.; Zhang, Y.; Wang, X.; Dong, T.; Chen, G.; Sun, X.; Lu, T.; White, R.A., 3rd; et al. Gestational diabetes mellitus is associated with the neonatal gut microbiota and metabolome. BMC Med. 2021, 19, 120. [Google Scholar] [CrossRef]
- Hu, Y.; Peng, J.; Li, F.; Wong, F.S.; Wen, L. Evaluation of different mucosal microbiota leads to gut microbiota-based prediction of type 1 diabetes in NOD mice. Sci. Rep. 2018, 8, 15451. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cao, R.; Liu, L.; Lv, Y.; Qi, X.; Yuan, Z.; Fan, X.; Yu, C.; Guan, Q. Correlation between Gut Microbiota and Testosterone in Male Patients with Type 2 Diabetes Mellitus. Front. Endocrinol. 2022, 13, 836485. [Google Scholar] [CrossRef] [PubMed]
- Diviccaro, S.; Falvo, E.; Piazza, R.; Cioffi, L.; Herian, M.; Brivio, P.; Calabrese, F.; Giatti, S.; Caruso, D.; Melcangi, R.C. Gut microbiota composition is altered in a preclinical model of type 1 diabetes mellitus: Influence on gut steroids, permeability, and cognitive abilities. Neuropharmacology 2023, 226, 109405. [Google Scholar] [CrossRef]
- Du, F.; Ma, J.; Gong, H.; Bista, R.; Zha, P.; Ren, Y.; Gao, Y.; Chen, D.; Ran, X.; Wang, C. Microbial Infection and Antibiotic Susceptibility of Diabetic Foot Ulcer in China: Literature Review. Front. Endocrinol. 2022, 13, 881659. [Google Scholar] [CrossRef]



| Exposures or Outcome | Number of Case | Number of Control | Ancestry | GWAS ID |
|---|---|---|---|---|
| Type 2 diabetes | 38,841 | 451,248 | European | ebi-a-GCST90018926 |
| Type 2 diabetes (wide definition) | 17,268 | 184,778 | European | finn-b-T2D_WIDE |
| Severe autoimmune type 2 diabetes | 452 | 2744 | European | ebi-a-GCST90026412 |
| Candidiasis | 2015 | 214,816 | European | finn-b-AB1_CANDIDIASIS |
| Exposure | Methods | No. of SNPs | p-Value | OR | OR_LCI95 | OR_UCI95 |
|---|---|---|---|---|---|---|
| T2D | MR Egger | 170 | 0.2100 | 1.1193 | 0.8984 | 1.3402 |
| Weighted median | 170 | 0.4490 | 1.0583 | 0.9473 | 1.1693 | |
| Inverse-variance-weighted | 170 | 0.0264 | 1.1046 | 0.9096 | 1.2996 | |
| Weighted mode | 170 | 0.2775 | 1.1057 | 0.9087 | 1.3027 | |
| T2D, wide definition | MR Egger | 84 | 0.0154 | 1.3197 | 0.7760 | 1.8634 |
| Weighted median | 84 | 0.0778 | 1.1370 | 0.8853 | 1.3887 | |
| Inverse-variance-weighted | 84 | 0.0031 | 1.1562 | 0.8718 | 1.4406 | |
| Weighted mode | 84 | 0.2801 | 1.1320 | 0.8890 | 1.3751 | |
| Severe autoimmune T2D | MR Egger | 13 | 0.2270 | 1.0688 | 0.9384 | 1.1991 |
| Weighted median | 13 | 0.0285 | 1.0554 | 0.9498 | 1.1610 | |
| Inverse-variance-weighted | 13 | 0.0041 | 1.0559 | 0.9493 | 1.1625 | |
| Weighted mode | 13 | 0.0984 | 1.0544 | 0.9506 | 1.1582 |
| Exposures | Outcomes | No. of SNPs | Cochran’s Heterogeneity Test | Pleiotropy Test | ||
|---|---|---|---|---|---|---|
| Single-SNP IVW | MR-Egger Intercept | |||||
| Q | p-Value | Intercept | p-Value | |||
| T2D | Candidiasis | 170 | 193.1 | 0.09842 | −0.0011 | 0.865 |
| T2D (wide definition) | Candidiasis | 84 | 101.2 | 0.08546 | −0.015 | 0.194 |
| Severe autoimmune T2D | Candidiasis | 13 | 6.423 | 0.8933 | −0.0078 | 0.807 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, J.; Lu, H.; Jiang, Y. A Causal Relationship between Type 2 Diabetes and Candidiasis through Two-Sample Mendelian Randomization Analysis. Microorganisms 2024, 12, 1984. https://doi.org/10.3390/microorganisms12101984
Xiong J, Lu H, Jiang Y. A Causal Relationship between Type 2 Diabetes and Candidiasis through Two-Sample Mendelian Randomization Analysis. Microorganisms. 2024; 12(10):1984. https://doi.org/10.3390/microorganisms12101984
Chicago/Turabian StyleXiong, Juan, Hui Lu, and Yuanying Jiang. 2024. "A Causal Relationship between Type 2 Diabetes and Candidiasis through Two-Sample Mendelian Randomization Analysis" Microorganisms 12, no. 10: 1984. https://doi.org/10.3390/microorganisms12101984
APA StyleXiong, J., Lu, H., & Jiang, Y. (2024). A Causal Relationship between Type 2 Diabetes and Candidiasis through Two-Sample Mendelian Randomization Analysis. Microorganisms, 12(10), 1984. https://doi.org/10.3390/microorganisms12101984






