It Takes Two to Make a Thing Go Right: Epistasis, Two-Component Response Systems, and Bacterial Adaptation
Abstract
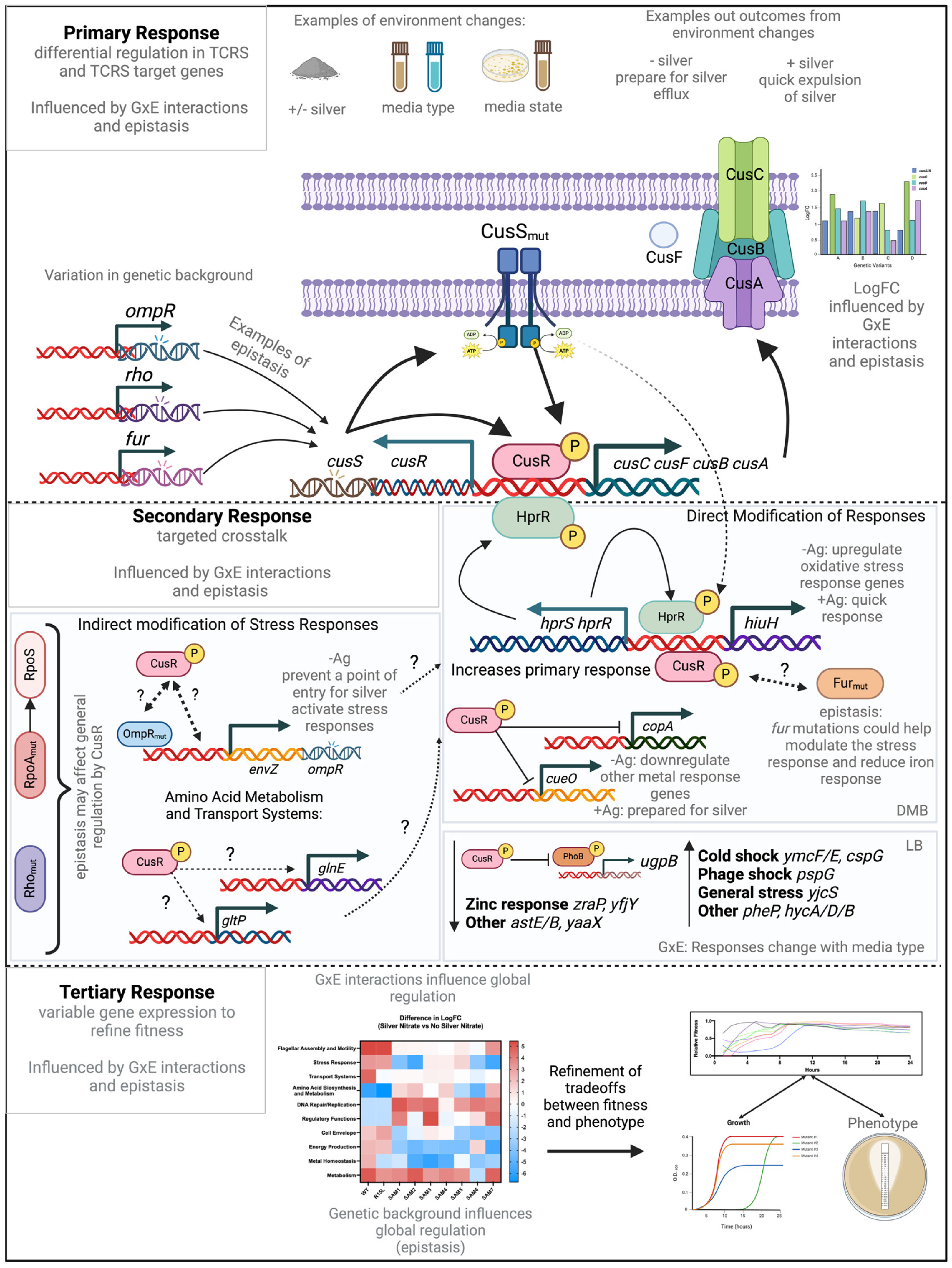
:1. Introduction
- How do cusS mutations, in combination with other genetic changes, influence the overall fitness and resistance of E. coli to silver?
- What are the specific gene expression patterns associated with these mutations, and how do they contribute to the observed phenotypes?
- How do epistatic interactions among these mutations shape the evolutionary trajectory of the bacteria in response to silver stress?
- How does our model of TCRS-driven bacterial adaptation need to be refined in light of these findings?
2. Materials and Methods
2.1. Bacterial Strains and Culturing
2.2. Whole Genome Illumina Sequencing of the SAM Populations
2.3. RNAseq and Differential Gene Expression
2.4. Twenty-Four-Hour Growth Assays
3. Results
3.1. DNA Sequencing and Mutation Analysis
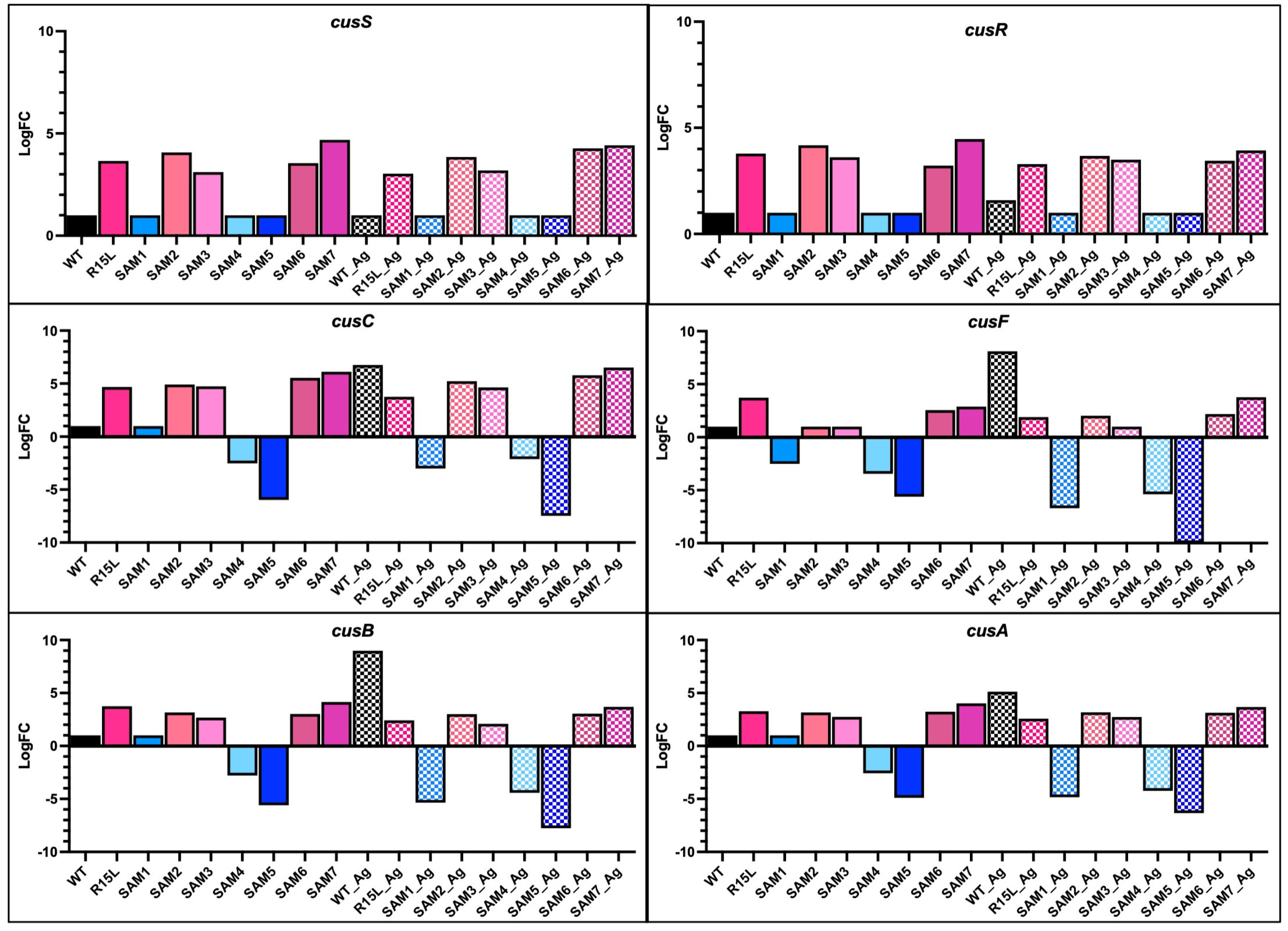
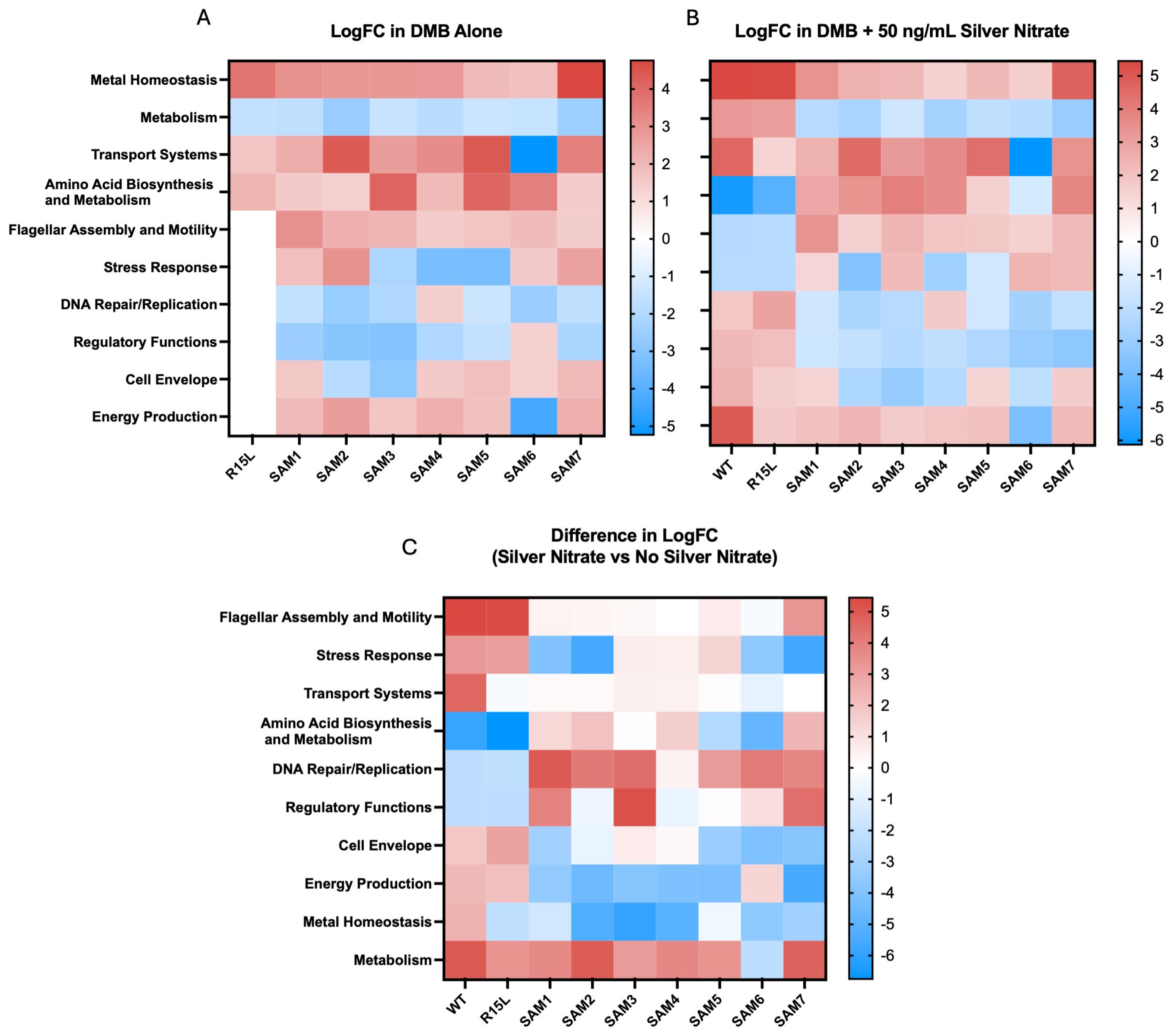
3.2. Gene Expression and Functional Implications
- Oxidative Stress Response Genes: Many SAM mutants, particularly those with cusS mutations, showed upregulation in genes associated with the oxidative stress response. For instance, msrP (methionine sulfoxide reductase) and huiH (hypothetical protein involved in stress response) were consistently upregulated. Also, msrP is known for its role in reducing methionine sulfoxide residues in proteins, which helps protect cells from oxidative damage caused by reactive oxygen species (ROS) [47]. The upregulation of these genes suggests that the mutants have developed a preemptive defense mechanism to mitigate oxidative damage associated with silver ion exposure, contributing to their enhanced survival under stress.
- Transport Systems Genes: The expression of various transport-related genes, beyond the cus operon, was significantly altered. SAM2 and SAM5, for instance, showed strong upregulation of zntA, a gene encoding a P-type ATPase that functions as a zinc efflux pump. This gene plays a dual role in protecting the cell from toxic levels of zinc and potentially providing cross-resistance to other heavy metals, including silver [48]. Additionally, copA, a gene encoding a copper-transporting ATPase, was variably expressed among the mutants. In SAM7, copA was downregulated, which might indicate a shift in the cell’s strategy to manage copper and silver ion homeostasis more efficiently under stress.
- Amino Acid Biosynthesis and Metabolism Genes: SAM3 and SAM6 exhibited notable upregulation in genes involved in amino acid biosynthesis and metabolism, such as gltB (glutamate synthase large subunit) and asd (aspartate-semialdehyde dehydrogenase). GltB plays a crucial role in the synthesis of glutamate, a key amino acid involved in nitrogen metabolism and as a precursor for other amino acids [49]. Asd is involved in the biosynthesis of lysine and other essential metabolites. The upregulation of these genes suggests an increased demand for amino acid synthesis, likely to support protein repair and synthesis during stress conditions. The prioritization of these metabolic pathways may help sustain cellular function and growth during silver exposure, providing a buffer against the detrimental effects of stress.
- Cell Envelope Integrity Genes: Several mutants, particularly SAM7, showed upregulation of genes involved in maintaining cell envelope integrity. For instance, murein (peptidoglycan synthesis genes) such as mrcA (penicillin-binding protein 1A) and lpoA (lipoprotein involved in peptidoglycan synthesis) were upregulated. These genes are critical for maintaining the structural integrity of the bacterial cell wall, especially under conditions where membrane integrity might be compromised by silver ions.
- Regulatory Functions Genes: The SAM mutants exhibited differential expression in several genes associated with regulatory functions. For example, rpoA (RNA polymerase alpha subunit) and rpoS (sigma factor S) were upregulated in SAM6, suggesting a heightened global stress response. RpoS is particularly important for the bacterial stress response, regulating the expression of numerous genes involved in survival during the stationary phase and under various environmental stresses. The upregulation of rpoS indicates that SAM6 might have an enhanced ability to manage multiple stressors, contributing to its superior fitness and resistance profile.
- Metabolic Suppression Genes: Interestingly, several mutants, especially SAM7, exhibited downregulation of genes involved in central metabolism, including those associated with glycolysis (pfkA, phosphofructokinase) and the TCA cycle (sdhA, succinate dehydrogenase). This downregulation may reflect a strategic metabolic shift to conserve energy and resources, redirecting them toward essential stress responses and repair mechanisms. This metabolic suppression likely serves as a trade-off, allowing the cells to prioritize survival overgrowth in the presence of silver.
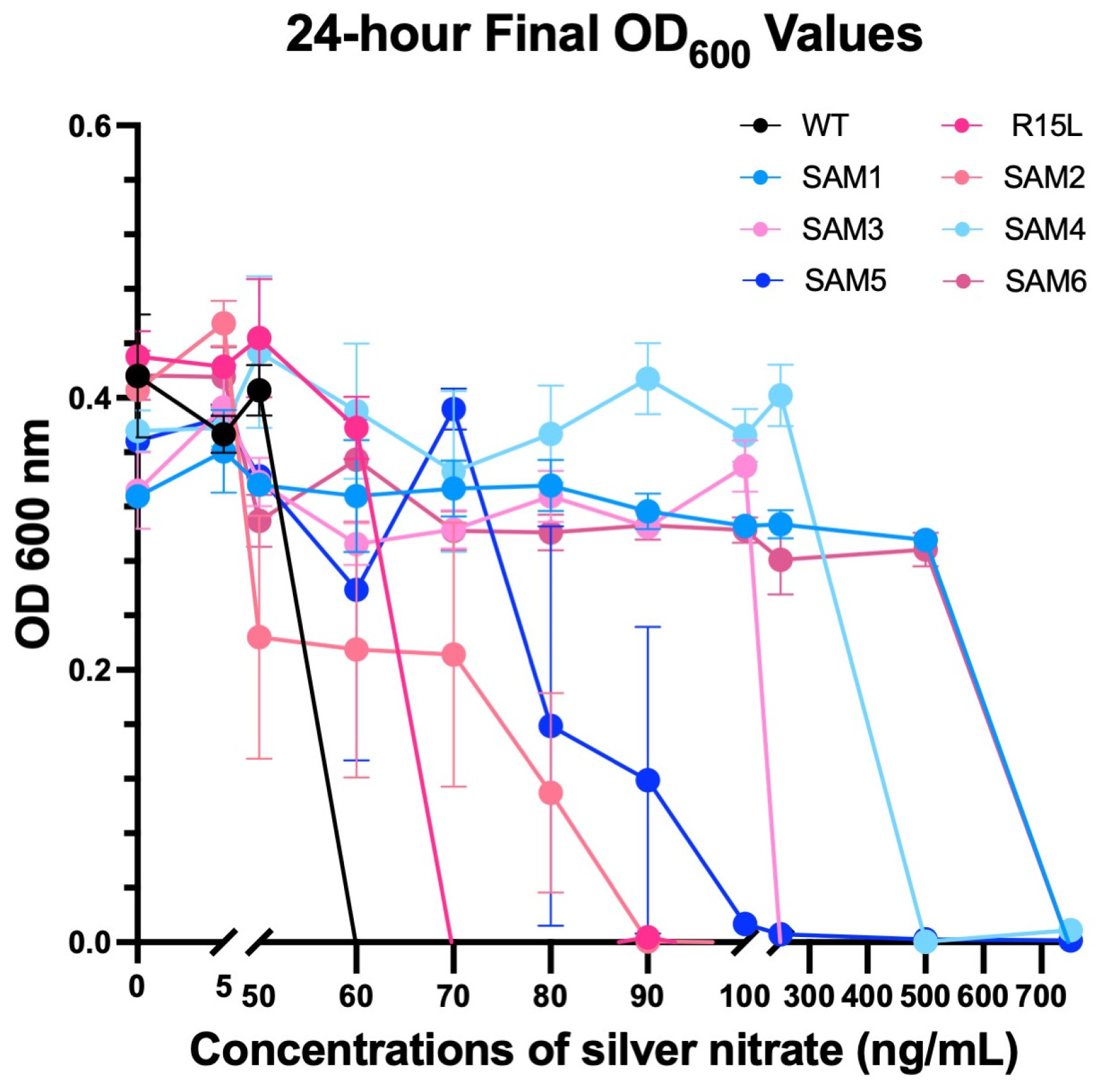
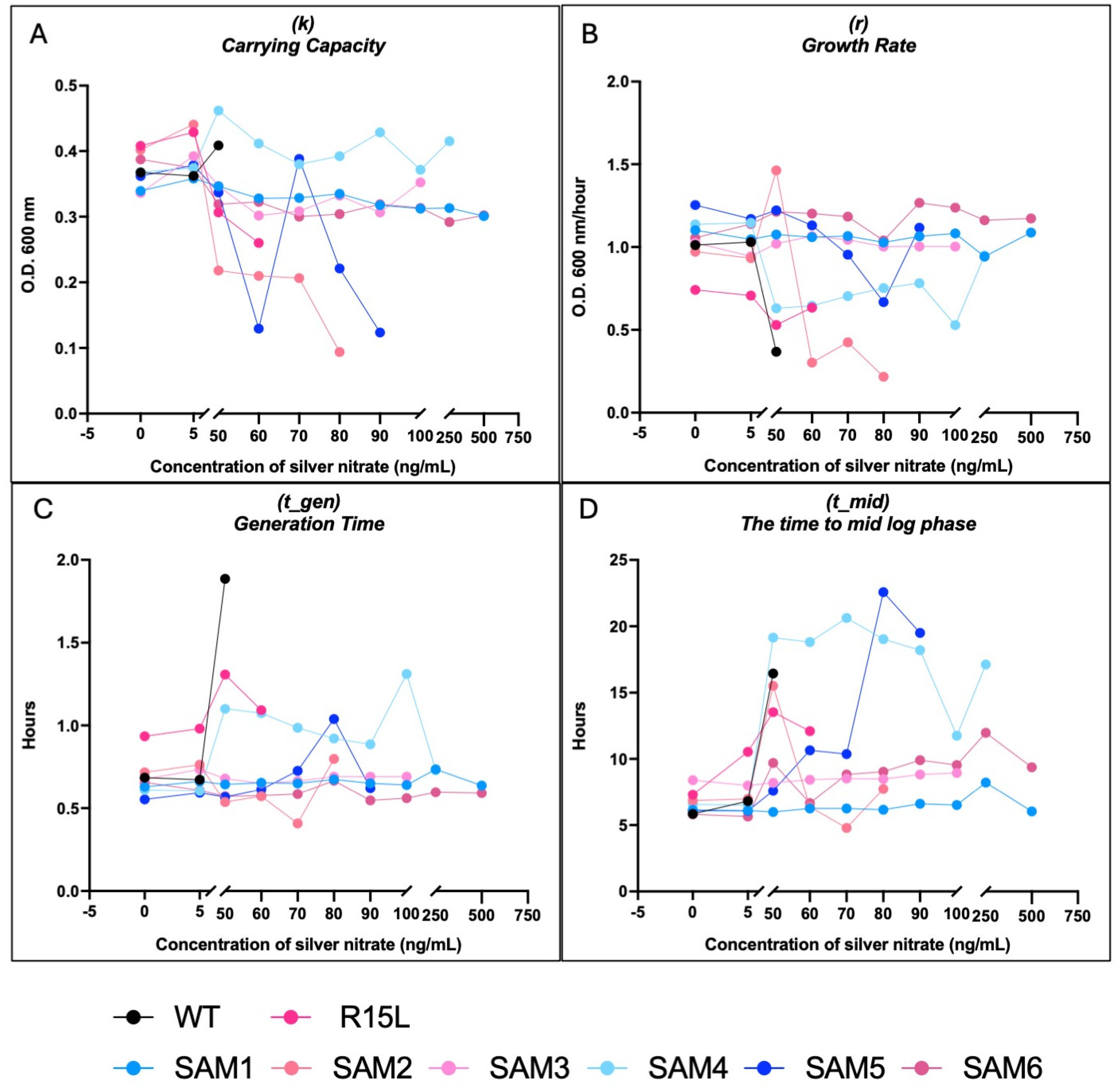
3.3. Comparative Fitness Analysis
4. Discussion
4.1. Role of Epistasis in Silver Resistance
4.2. New Adaptive Traits That Are Not Predictable from the Individual Effects of Each Mutation Alone
4.3. Mechanisms of Silver Resistance
4.4. Crosstalk and Adaptation in TCRS: Evaluating a Three-Step Model
4.5. Refinements and Modifications to the Model
4.5.1. Epistasis in GxE Interactions
4.5.2. Impact of Genetic Background
4.5.3. Multifunctional Pathways in GxE Dynamics
4.5.4. Adaptive Strategy Prioritization
4.6. Revised Model Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kassen, R. Distribution of fitness effects among beneficial mutations before selection in experimental populations of Bacteria. Nat. Genet. 2006, 38, 484. [Google Scholar] [CrossRef] [PubMed]
- Barrick, J.E.; Lenski, R.E. Genome dynamics during experimental evolution. Nat. Rev. Genet. 2013, 14, 827. [Google Scholar] [CrossRef]
- Elena, S.F.; Lenski, R.E. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat. Rev. Genet. 2003, 4, 457–469. [Google Scholar] [CrossRef]
- Phillips, P.C. Epistasis—The essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008, 9, 855–867. [Google Scholar] [CrossRef]
- Weinreich, D.M.; Lan, Y.; Wylie, C.S.; Heckendorn, R.B. Should evolutionary geneticists worry about higher-order epistasis? Curr. Opin. Genet. Dev. 2013, 23, 700–707. [Google Scholar] [CrossRef]
- Desai, M.M.; Fisher, D.S. Beneficial mutation–selection balance and the effect of linkage on positive selection. Genetics 2007, 176, 1759–1798. [Google Scholar] [CrossRef]
- Otto, S.P.; Feldman, M.W. Deleterious mutations, variable epistatic interactions, and the evolution of recombination. Theor. Popul. Biol. 1997, 51, 134–147. [Google Scholar] [CrossRef]
- Lehner, B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 2011, 27, 323–331. [Google Scholar] [CrossRef]
- Barillo, D.J.; Marx, D.E. Silver in medicine: A brief history BC 335 to present. Burns 2014, 40 (Suppl. S1), S3. [Google Scholar] [CrossRef]
- Rai, M.; Yadav, A.; Gade, A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef]
- Russell, A.D.; Hugo, W.B. 7 Antimicrobial activity and action of silver. Prog. Med. Chem. 1994, 31, 351–370. [Google Scholar]
- Haefeli, C.; Franklin, C.; Hardy, K. Plasmid-determined silver resistance in Pseudomonas stutzeri isolated from a silver mine. J. Bacteriol. 1984, 158, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Lemire, J.A.; Harrison, J.J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 2013, 11, 371–384. [Google Scholar] [CrossRef] [PubMed]
- McHugh, G.L.; Moellering, R.; Hopkins, C.; Swartz, M. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin: A new threat in burn units? Lancet 1975, 305, 235–240. [Google Scholar] [CrossRef] [PubMed]
- McNeilly, O.; Mann, R.; Hamidian, M.; Gunawan, C. Emerging concern for silver nanoparticle resistance in Acinetobacter baumannii and other bacteria. Front. Microbiol. 2021, 12, 652863. [Google Scholar]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Hoch, J.A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 2000, 3, 165–170. [Google Scholar] [CrossRef]
- Mascher, T.; Helmann, J.D.; Unden, G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 910–938. [Google Scholar] [CrossRef]
- Gupta, R.; Gupta, N. Two-component systems. In Fundamentals of Bacterial Physiology and Metabolism; Springer: Singapore, 2021; pp. 557–573. [Google Scholar]
- Koretke, K.K.; Lupas, A.N.; Warren, P.V.; Rosenberg, M.; Brown, J.R. Evolution of two-component signal transduction. Mol. Biol. Evol. 2000, 17, 1956–1970. [Google Scholar] [CrossRef]
- West, A.H.; Stock, A.M. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001, 26, 369–376. [Google Scholar] [CrossRef]
- Munson, G.P.; Lam, D.L.; Outten, F.W.; O’Halloran, T.V. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 2000, 182, 5864–5871. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Ishihama, A. Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 2005, 56, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Gudipaty, S.A.; McEvoy, M.M. The histidine kinase CusS senses silver ions through direct binding by its sensor domain. Biochim. Biophys. Acta. Proteins. Proteom. 2014, 1844, 1656–1661. [Google Scholar] [CrossRef]
- Capra, E.J.; Laub, M.T. Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 2012, 66, 325–347. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, Y.; Eguchi, Y.; Watanabe, T.; Okamoto, S.; Doi, A.; Utsumi, R. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 2010, 13, 232–239. [Google Scholar] [CrossRef]
- Podgornaia, A.I.; Laub, M.T. Pervasive degeneracy and epistasis in a protein-protein interface. Science 2015, 347, 673–677. [Google Scholar] [CrossRef]
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.Y.; Toufighi, K.; Mostafavi, S.; et al. The genetic landscape of a cell. Science 2010, 327, 425–431. [Google Scholar] [CrossRef]
- Chou, H.; Marx, C.J. Optimization of gene expression through divergent mutational paths. Cell Rep. 2012, 1, 133–140. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- Toprak, E.; Veres, A.; Michel, J.B.; Chait, R.; Hartl, D.L.; Kishony, R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2012, 44, 101–105. [Google Scholar] [CrossRef]
- Baquero, F.; Coque, T.M. Multilevel population genetics in antibiotic resistance. FEMS. Microbiol. Rev. 2011, 35, 705–706. [Google Scholar] [CrossRef] [PubMed]
- Barrick, J.E.; Lenski, R.E. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nat. Biotechnol. 2009, 461, 1243–1247. [Google Scholar] [CrossRef] [PubMed]
- Swallow, J.G.; Hayes, J.P.; Koteja, P.; Garland, T., Jr. Selection experiments and experimental evolution of performance and physiology. In Evolutionary and Genetic Biology; Regents of the University of California Press: Oakland, CA, USA, 2009; Chapter 12. [Google Scholar]
- Randall, C.P.; Gupta, A.; Jackson, N.; Busse, D.; O’Neill, A.J. Silver resistance in Gram-negative bacteria: A dissection of endogenous and exogenous mechanisms. J. Antimicrob. Chemother. 2015, 70, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.L., Jr.; Tajkarimi, M.; Cunningham, Q.; Campbell, A.; Nonga, H.; Harrison, S.H.; Barrick, J.E. Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front. Genet. 2015, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Tajkarimi, M.; Campbell, A.; Rhinehardt, K.; Thomas, M.; Akamu, E.J.; Boyd, S.; Turner, D.; Harrison, S.H.; Graves, J.L., Jr. Selection for ionic-silver confers silver nanoparticle resistance in Escherichia coli. JSM Nanotechnol. Nanomed. 2017, 5, 1047. [Google Scholar]
- Sanders, B.R.; Miller, J.E.; Ahmidouch, N.; Graves, J.L., Jr.; Thomas, M.D. Genotype-by-environment interactions govern fitness changes associated with adaptive mutations in two-component response systems. Front. Genet. 2024, 15, 1349507. [Google Scholar] [CrossRef]
- Sanders, B.R.; Townsend, S.E.; Ford, M.L.; Graves, J.L., Jr.; Thomas, M.D. Reporting off-target effects of recombinant engineering using the pORTMAGE system. J. Microbiol. Methods 2023, 204, 106627. [Google Scholar] [CrossRef] [PubMed]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. In Engineering and Analyzing Multicellular Systems: Methods and Protocols; Humana Press: New York, NY, USA, 2014; pp. 165–188. [Google Scholar]
- Illumina. BCL Convert: A proprietary Illumina Software for the Conversion of BCL Files to Basecalls. 2021. (v3.9.3). Available online: https://support-docs.illumina.com/SW/BCL_Convert/Content/SW/FrontPages/BCL_Convert.htm (accessed on 25 January 2024).
- Zhang, Y.; Park, C.; Bennett, C.; Thornton, M.; Kim, D. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021, 31, 1290–1295. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Sprouffske, K.; Wagner, A. Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016, 17, 172. [Google Scholar] [CrossRef]
- Franke, S.; Grass, G.; Nies, D.H. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 2001, 147, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Chacón, K.N.; Mealman, T.D.; McEvoy, M.M.; Blackburn, N.J. Tracking metal ions through a Cu/Ag efflux pump assigns the functional roles of the periplasmic proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 15373–15378. [Google Scholar] [CrossRef] [PubMed]
- Gennaris, A.; Ezraty, B.; Henry, C.; Agrebi, R.; Vergnes, A.; Oheix, E.; Bos, J.; Leverrier, P.; Espinosa, L.; Szewczyk, J.; et al. Repairing oxidized proteins in the bacterial envelope using respiratory chain electrons. Nature 2015, 528, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Rensing, C.; Mitra, B.; Rosen, B.P. The zntA gene of Escherichia coli encodes a Zn (II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 1997, 94, 14326–14331. [Google Scholar] [CrossRef]
- Goss, T.J.; Perez-Matos, A.; Bender, R.A. Roles of glutamate synthase gltBD and gltF in nitrogen metabolism of Escherichia coli and Klebsiella. aerogenes. J. Bacteriol. 2001, 183, 6607–6619. [Google Scholar] [CrossRef]
- Urano, H.; Yoshida, M.; Ogawa, A.; Yamamoto, K.; Ishihama, A.; Ogasawara, H. Cross-regulation between two common ancestral response regulators HprR and CusR in Escherichia coli. Microbiology 2017, 163, 243–252. [Google Scholar] [CrossRef]

| Location | Mutation | Frequency | Annotation | Gene | Description | |
|---|---|---|---|---|---|---|
| SAM1 | 847,955 | G→A | 1.000 | A17V (GCG→GTG) | glnH ← | glutamine transporter subunit |
| 4,188,510 | C→A | 0.877 | T1054N (ACC→AAC) | rpoC → | RNA polymerase, beta prime subunit | |
| 1,868,984 | C→A | 0.849 | N10K (AAC→AAA) | yeaH → | UPF0229 family protein | |
| 3,815,809 | Δ1 bp | 0.807 | intergenic (−41/+25) | pyrE ←/← rph | orotate phosphoribosyltransferase/ribonuclease PH (defective);enzyme; Degradation of RNA; RNase PH | |
| 3,831,168 | C→T | 0.732 | L238L (CTA→TTA) | yicH → | putative inner membrane-anchored periplasmic AsmA family protein | |
| SAM2 | 12,661 | C→G | 1.000 | R167G (CGT→GGT) | dnaK → | chaperone Hsp70, with co-chaperone DnaJ |
| 594,727 | C→A | 1.000 | R15L (CGC→CTC) | cusS ← | sensory histidine kinase in two-component regulatory system with CusR, senses copper ions | |
| 3,101,306 | G→T | 1.000 | I106I (ATC→ATA) | yggN ← | DUF2884 family putative periplasmic protein | |
| 3,133,461 | G→A | 1.000 | A166V (GCA→GTA) | yghS ← | putative ATP-binding protein | |
| 3,536,061 | Δ1 bp | 1.000 | coding (524/720 nt) | ompR ← | response regulator in two-component regulatory system with EnvZ | |
| 3,816,605 | Δ103 bp | 1.000 | [yicC] | [yicC] | ||
| 3,843,548 | A→C | 1.000 | I81R (ATA→AGA) | yicO ← | putative adenine permease | |
| 3,966,612 | C→T | 1.000 | R66C (CGT→TGT) | rho → | transcription termination factor | |
| 3,992,735 | C→A | 0.930 | S528 * (TCG→TAG) | cyaA → | adenylate cyclase | |
| 3,177,973 | IS1 (+) +9 bp | 0.552 | intergenic (−63/−134) | nudF ←/→ tolC | ADP-ribose pyrophosphatase/transport channel | |
| 915,226 | T→C | 0.446 | intergenic (−369/+126) | ybjE ←/← aqpZ | putative transporter/aquaporin Z | |
| 2,229,177 | A→G | 0.376 | intergenic (+112/+261) | yohP →/← dusC | uncharacterized protein/tRNA-dihydrouridine synthase C | |
| 4,296,060 | C→T | 0.282 | intergenic (+266/+376) | gltP →/← yjcO | glutamate/aspartate:proton symporter/Sel1 family TPR-like repeat protein | |
| SAM3 | 12,661 | C→G | 1.000 | R167G (CGT→GGT) | dnaK → | chaperone Hsp70, with co-chaperone DnaJ |
| 594,727 | C→A | 1.000 | R15L (CGC→CTC) | cusS ← | sensory histidine kinase in two-component regulatory system with CusR, senses copper ions | |
| 3,101,306 | G→T | 1.000 | I106I (ATC→ATA) | yggN ← | DUF2884 family putative periplasmic protein | |
| 3,133,461 | G→A | 1.000 | A166V (GCA→GTA) | yghS ← | putative ATP-binding protein | |
| 3,536,061 | Δ1 bp | 1.000 | coding (524/720 nt) | ompR ← | response regulator in two-component regulatory system with EnvZ | |
| 3,816,605 | Δ103 bp | 1.000 | [yicC] | [yicC] | ||
| 3,843,548 | A→C | 1.000 | I81R (ATA→AGA) | yicO ← | putative adenine permease | |
| 3,966,612 | C→T | 1.000 | R66C (CGT→TGT) | rho → | transcription termination factor | |
| 3,992,735 | C→A | 0.865 | S528 * (TCG→TAG) | cyaA → | adenylate cyclase | |
| 3,177,973 | IS1 (+) +9 bp | 0.648 | intergenic (−63/−134) | nudF ←/→ tolC | ADP-ribose pyrophosphatase/transport channel | |
| SAM4 | 3,992,735 | C→A | 0.865 | S528 * (TCG→TAG) | cyaA → | adenylate cyclase |
| 3,177,973 | IS1 (+) +9 bp | 0.648 | intergenic (−63/−134) | nudF ←/→ tolC | ADP-ribose pyrophosphatase/transport channel | |
| 3,360,120 | G→A | 0.163 | R308H (CGC→CAC) | gltD → | glutamate synthase, 4Fe-4S protein, small subunit | |
| 4,296,060 | C→T | 0.159 | intergenic (+266/+376) | gltP →/← yjcO | glutamate/aspartate:proton symporter/Sel1 family TPR-like repeat protein | |
| 2,725,169 | Δ1 bp | 0.147 | coding (578/1299 nt) | kgtP ← | alpha-ketoglutarate transporter | |
| 2,229,205 | G→C | 0.116 | intergenic (+140/+233) | yohP →/← dusC | uncharacterized protein/tRNA-dihydrouridine synthase C | |
| 3,815,859 | Δ82 bp | 0.533 | [rph]–[rph] | [rph], [rph] | ||
| 3,815,824 | G→T | 0.277 | intergenic (−56/+10) | pyrE ←/← rph | orotate phosphoribosyltransferase/ribonuclease PH (defective);enzyme; Degradation of RNA; RNase PH | |
| 3,198,033 | IS186 (+) +6 bp :: Δ2 bp | 0.258 | coding (1604–1609/2841 nt) | glnE ← | fused deadenylyltransferase/adenylyltransferase for glutamine synthetase | |
| 4,296,060 | C→T | 0.235 | intergenic (+266/+376) | gltP →/← yjcO | glutamate/aspartate:proton symporter/Sel1 family TPR-like repeat protein | |
| 3,815,809 | Δ1 bp | 0.138 | intergenic (−41/+25) | pyrE ←/← rph | orotate phosphoribosyltransferase/ribonuclease PH (defective);enzyme; Degradation of RNA; RNase PH | |
| 915,226 | T→C | 0.112 | intergenic (−369/+126) | ybjE ←/← aqpZ | putative transporter/aquaporin Z | |
| 594,727 | C→A | 0.055 | R15L (CGC→CTC) | cusS ← | sensory histidine kinase in two-component regulatory system with CusR, senses copper ions | |
| SAM5 | 3,197,294 | T→A | 0.408 | E783V (GAA→GTA) | glnE ← | fused deadenylyltransferase/adenylyltransferase for glutamine synthetase |
| 3,815,809 | Δ1 bp | 0.373 | intergenic (−41/+25) | pyrE ←/← rph | orotate phosphoribosyltransferase/ribonuclease PH (defective);enzyme; Degradation of RNA; RNase PH | |
| 3,815,859 | Δ82 bp | 0.367 | [rph]–[rph] | [rph], [rph] | ||
| 4,296,060 | C→T | 0.236 | intergenic (+266/+376) | gltP →/← yjcO | glutamate/aspartate:proton symporter/Sel1 family TPR-like repeat protein | |
| 3,992,588 | (ATCAGCC)2→1 | 0.235 | coding (1436–1442/2547 nt) | cyaA → | adenylate cyclase | |
| 1,907,503 | IS3 (–) +5 bp :: +T | 0.178 | coding (85–89/144 nt) | yobF ← | DUF2527 family heat-induced protein | |
| 4,181,669 | A→G | 0.164 | E142G (GAG→GGG) | rpoB → | RNA polymerase, beta subunit | |
| 3,485,966 | IS2 (+) +5 bp | 0.129 | intergenic (−148/−150) | yhfA ←/→ crp | OsmC family protein/cAMP-activated global transcription factor, mediator of catabolite repression | |
| 3,897,059 | C→A | 0.129 | L96I (CTC→ATC) | yieH → | phosphoenolpyruvate and 6-phosphogluconate phosphatase | |
| SAM6 | 594,727 | C→A | 1.000 | R15L (CGC→CTC) | cusS ← | sensory histidine kinase in two-component regulatory system with CusR, senses copper ions |
| 710,620 | C→A | 1.000 | K9N (AAG→AAT) | fur ← | ferric iron uptake regulon transcriptional repressor; autorepressor | |
| 3,440,186 | C→A | 1.000 | V282L (GTA→TTA) | rpoA ← | RNA polymerase, alpha subunit | |
| 3,536,041 | G→A | 1.000 | R182C (CGC→TGC) | ompR ← | response regulator in two-component regulatory system with EnvZ | |
| 3,966,604 | G→T | 1.000 | G63V (GGT→GTT) | rho → | transcription termination factor | |
| 4,232,641 | C→A | 0.318 | R198L (CGT→CTT) | lysC ← | lysine-sensitive aspartokinase 3 | |
| 1,213,820 | G→C | 0.279 | D80E (GAC→GAG) | bluR ← | repressor of blue light-responsive genes | |
| 3,359,461 | Δ1 bp | 0.226 | coding (264/1419 nt) | gltD → | glutamate synthase, 4Fe-4S protein, small subunit | |
| SAM7 | 594,727 | C→A | 1.000 | R15L (CGC→CTC) | cusS ← | sensory histidine kinase in two-component regulatory system with CusR, senses copper ions |
| 710,620 | C→A | 1.000 | K9N (AAG→AAT) | fur ← | ferric iron uptake regulon transcriptional repressor; autorepressor | |
| 1,428,765 | T→C | 1.000 | intergenic (−39/−30) | insH1 ←/→ lomR | IS5 transposase and trans-activator; IS, phage, Tn; Transposon-related functions; extrachromosomal; transposon related/pseudogene, Rac prophage lom homolog; Phage or Prophage Related; interrupted by IS5 and N-ter deletion | |
| 3,440,186 | C→A | 1.000 | V282L (GTA→TTA) | rpoA ← | RNA polymerase, alpha subunit | |
| 3,536,041 | G→A | 1.000 | R182C (CGC→TGC) | ompR ← | response regulator in two-component regulatory system with EnvZ | |
| 3,815,801 | Δ1 bp | 1.000 | intergenic (−33/+33) | pyrE ←/← rph | orotate phosphoribosyltransferase/ribonuclease PH (defective);enzyme; Degradation of RNA; RNase PH | |
| 3,966,604 | G→T | 1.000 | G63V (GGT→GTT) | rho → | transcription termination factor | |
| 1,212,080:1 | +C | 1.000 | intergenic (−77/+623) | iraM ←/← ycgX | RpoS stabilizer during Mg starvation, anti-RssB factor/DUF1398 family protein | |
| 1,213,820 | G→C | 0.627 | D80E (GAC→GAG) | bluR ← | repressor of blue light-responsive genes | |
| 3,359,461 | Δ1 bp | 0.590 | coding (264/1419 nt) | gltD → | glutamate synthase, 4Fe-4S protein, small subunit | |
| 4,296,060 | C→T | 0.248 | intergenic (+266/+376) | gltP →/← yjcO | glutamate/aspartate:proton symporter/Sel1 family TPR-like repeat protein |
| In DMB Alone | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Biological Function | Gene | R15L_logFC | SAM2_logFC | SAM3_logFC | SAM6_logFC | SAM7_logFC | |||||
| Metal Homeostasis (Zinc Transport) | znuA | 1.694 | Up | −5.107 | Down | −4.516 | Down | −3.553 | Down | −4.875 | Down |
| Metal Homeostasis (Copper Export) | copA | −1.723 | Down | −7.115 | Down | −6.099 | Down | −6.739 | Down | −6.873 | Down |
| Regulatory Functions (Redox Stress Response) | hprR | 3.078 | Up | 2.780 | Up | 4.052 | Up | 1.609 | Up | 3.787 | Up |
| Metal Homeostasis (Copper Detoxification) | cueO | −1.560 | Down | −5.212 | Down | −4.669 | Down | −6.070 | Down | −4.945 | Down |
| Membrane Proteins (Potential Stress Response or Transport) | shoB | 1.941 | Up | 3.827 | Up | 3.525 | Up | 4.512 | Up | 3.934 | Up |
| Energy Production (Anaerobic Respiration) | napH | 2.023 | Up | 4.341 | Up | 4.789 | Up | 4.199 | Up | 4.403 | Up |
| Bacteriophage Interaction (Phage Entry) | nfrB | 1.617 | Up | 2.174 | Up | 1.619 | Up | 2.803 | Up | 1.790 | Up |
| Cell Envelope (Peptidoglycan Remodeling) | mepM | 1.372 | Up | −3.552 | Down | −3.977 | Down | −3.465 | Down | −4.218 | Down |
| Metal Homeostasis (Copper/Silver Efflux) | cusA | 3.285 | Up | 3.167 | Up | 2.753 | Up | 3.237 | Up | 4.018 | Up |
| Metal Homeostasis (Iron Transport) | yfhH | 1.594 | Up | 2.597 | Up | 2.571 | Up | 3.292 | Up | 2.572 | Up |
| Metal Homeostasis (Copper/Silver Efflux) | cusB | 3.746 | Up | 3.164 | Up | 2.676 | Up | 3.026 | Up | 4.154 | Up |
| Transport Systems (Amino Acid Transport) | pheP | 2.550 | Up | 3.148 | Up | 2.715 | Up | 3.146 | Up | 3.517 | Up |
| Nucleotide Metabolism (Purine Salvage Pathway) | ghxP | 2.450 | Up | 5.917 | Up | 3.650 | Up | 2.340 | Up | 5.340 | Up |
| In Presence of 50 ng/mL Silver Nitrate | |||||||||||
| Biological Function | Gene | R15L_logFC | SAM2_logFC | SAM3_logFC | SAM6_logFC | SAM7_logFC | |||||
| Metal Homeostasis (Zinc Transport) | znuA | 5.344 | Up | −4.877 | Down | −4.602 | Down | −4.134 | Down | −5.168 | Down |
| Metal Homeostasis (Copper Export) | copA | 5.769 | Up | −6.581 | Down | −6.517 | Down | −5.676 | Down | −7.027 | Down |
| Regulatory Functions (Redox Stress Response) | hprR | −2.073 | Down | 2.971 | Up | 3.221 | Up | 2.544 | Up | 3.570 | Up |
| Metal Homeostasis (Copper Detoxification) | cueO | 5.807 | Up | −5.137 | Down | −4.975 | Down | −5.726 | Down | −5.079 | Down |
| Membrane Proteins (Potential Stress Response or Transport) | shoB | −1.593 | Down | 3.410 | Up | 3.196 | Up | 3.972 | Up | 3.797 | Up |
| Energy Production (Anaerobic Respiration) | napH | −3.244 | Down | 4.158 | Up | 4.843 | Up | 2.773 | Up | 4.864 | Up |
| Bacteriophage Interaction (Phage Entry) | nfrB | −2.437 | Down | 2.269 | Up | 1.867 | Up | 3.743 | Up | 2.115 | Up |
| Cell Envelope (Peptidoglycan Remodeling) | mepM | 2.915 | Up | −4.029 | Down | −3.910 | Down | −3.861 | Down | −4.885 | Down |
| Metal Homeostasis (Copper/Silver Efflux) | cusA | 2.293 | Up | 3.184 | Up | 2.735 | Up | 3.129 | Up | 3.674 | Up |
| Metal Homeostasis (Iron Transport) | yfhH | −2.223 | Down | 2.936 | Up | 2.577 | Up | 4.065 | Up | 2.348 | Up |
| Metal Homeostasis (Copper/Silver Efflux) | cusB | 2.827 | Up | 3.013 | Up | 2.097 | Up | 3.055 | Up | 3.690 | Up |
| Transport Systems (Amino Acid Transport) | pheP | 2.377 | Up | 2.968 | Up | 2.503 | Up | 2.918 | Up | 2.879 | Up |
| Nucleotide Metabolism (Purine Salvage Pathway) | ghxP | −1.647 | Down | 5.197 | Up | 4.370 | Up | 3.033 | Up | 4.724 | Up |
| Population | MIC (ng/mL) | Growth Metrics | Notable Mutations | Differential Expression | Mechanism of Silver Resistance | Notable Observations |
|---|---|---|---|---|---|---|
| SAM1 | 750 | r: 1.2 (50 ng/mL), 1.15 (750 ng/mL); t_gen: Short; t_mid: Stable; k: Strong resistance | glnH, rpoC | Moderate upregulation of zntA and metal transporters, downregulation of cus efflux system | Alternative metal transport, downregulates cus system | Highest relative fitness, strong adaptability, short lag phase |
| SAM2 | 90 | r: 1.0 (50 ng/mL), 0.2 (80 ng/mL); t_gen: Extended; t_mid: Lower fitness; k: Declines at high concentrations | cusS, rho, ompR | Downregulation of cus efflux genes, upregulation of zntA and dnaK | Moderate cus efflux activity, zinc efflux upregulation | Struggles at higher silver concentrations, fitness lower than WT at low silver |
| SAM3 | 250 | r: Steady; t_gen: Steady; t_mid: Moderate; k: Varies | cusS, rho, ompR | Upregulation of zntA, moderate cus efflux expression | Balanced cus efflux and metal transport systems | Moderate resistance, extended lag phases under high silver stress |
| SAM4 | 500 | r: Slower growth; t_gen: Significant increase; t_mid: ~20 h (above 50 ng/mL); k: Moderate resistance | glnE, rph | No cus efflux upregulation, upregulation of RNA processing genes | RNA processing, alternative metabolic adaptations | Adapts slowly, extended lag phase, lacks fixed cusS mutation |
| SAM5 | 100 | r: Stable; t_gen: ~10 h (80 ng/mL), ~20 h (90 ng/mL); t_mid: ~10–20 h; k: Moderate resistance | rph, glnE | Downregulation of central metabolism, upregulation of zntA | Metabolic adjustments, zinc efflux | Moderate fitness, struggles with adaptation under higher silver stress |
| SAM6 | 750 | r: 1.18 (50 ng/mL), 1.1 (750 ng/mL); t_gen: Short; t_mid: Fast; k: High carrying capacity | cusS, fur, rpoA, ompR, rho | Strong upregulation of cus efflux genes, msrQ and zinT for stress protection | High-efficiency cus efflux, stress defense upregulation | Superior resistance, robust response to high silver concentrations, short lag phase |
| SAM7 | 750 | r: 1.15 (50 ng/mL), 1.1 (750 ng/mL); t_gen: Short; t_mid: Fast; k: High carrying capacity | cusS, fur, rpoA, ompR, rho | Highest upregulation of cus efflux genes, rho and ompR for membrane stability | Robust cus efflux, membrane integrity | Similar to SAM6, strong genetic flexibility due to transposable elements |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanders, B.R.; Thomas, L.S.; Lewis, N.M.; Ferguson, Z.A.; Graves, J.L., Jr.; Thomas, M.D. It Takes Two to Make a Thing Go Right: Epistasis, Two-Component Response Systems, and Bacterial Adaptation. Microorganisms 2024, 12, 2000. https://doi.org/10.3390/microorganisms12102000
Sanders BR, Thomas LS, Lewis NM, Ferguson ZA, Graves JL Jr., Thomas MD. It Takes Two to Make a Thing Go Right: Epistasis, Two-Component Response Systems, and Bacterial Adaptation. Microorganisms. 2024; 12(10):2000. https://doi.org/10.3390/microorganisms12102000
Chicago/Turabian StyleSanders, Brittany R., Lauren S. Thomas, Naya M. Lewis, Zaria A. Ferguson, Joseph L. Graves, Jr., and Misty D. Thomas. 2024. "It Takes Two to Make a Thing Go Right: Epistasis, Two-Component Response Systems, and Bacterial Adaptation" Microorganisms 12, no. 10: 2000. https://doi.org/10.3390/microorganisms12102000
APA StyleSanders, B. R., Thomas, L. S., Lewis, N. M., Ferguson, Z. A., Graves, J. L., Jr., & Thomas, M. D. (2024). It Takes Two to Make a Thing Go Right: Epistasis, Two-Component Response Systems, and Bacterial Adaptation. Microorganisms, 12(10), 2000. https://doi.org/10.3390/microorganisms12102000





