Safety Assessment and Evaluation of Probiotic Potential of Lactobacillus bulgaricus IDCC 3601 for Human Use
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Culture Conditions
2.2. Whole-Genome Sequencing of L. bulgaricus IDCC 3601
2.3. Determination of the Minimum Inhibitory Concentration (MIC)
2.4. Production of Biogenic Amines (BAs)
2.5. Hemolytic Activity
2.6. Carbohydrates Utilization and Extracellular Enzyme Activities
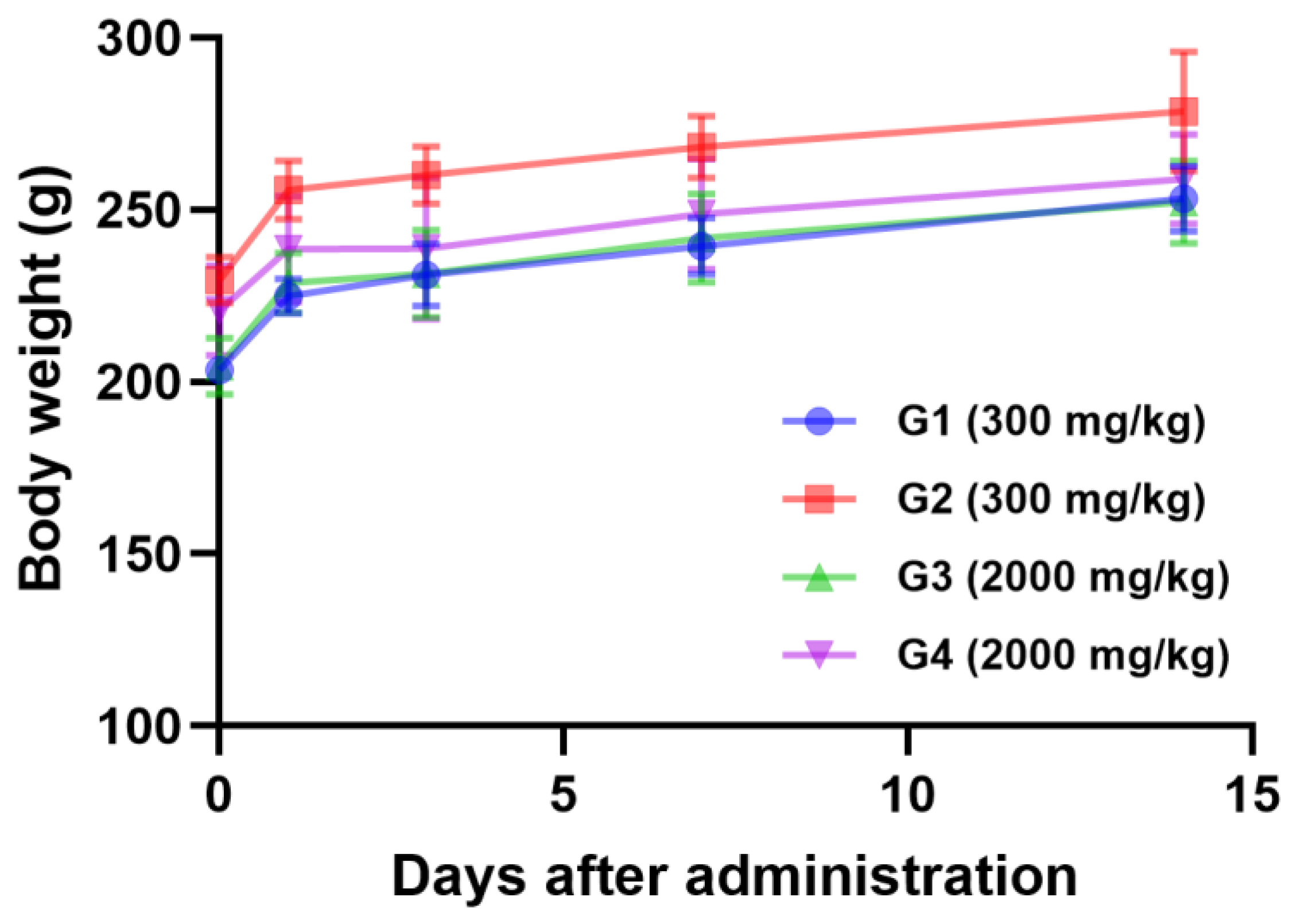
2.7. Acute Oral Toxicity Study of L. bulgaricus IDCC 3601
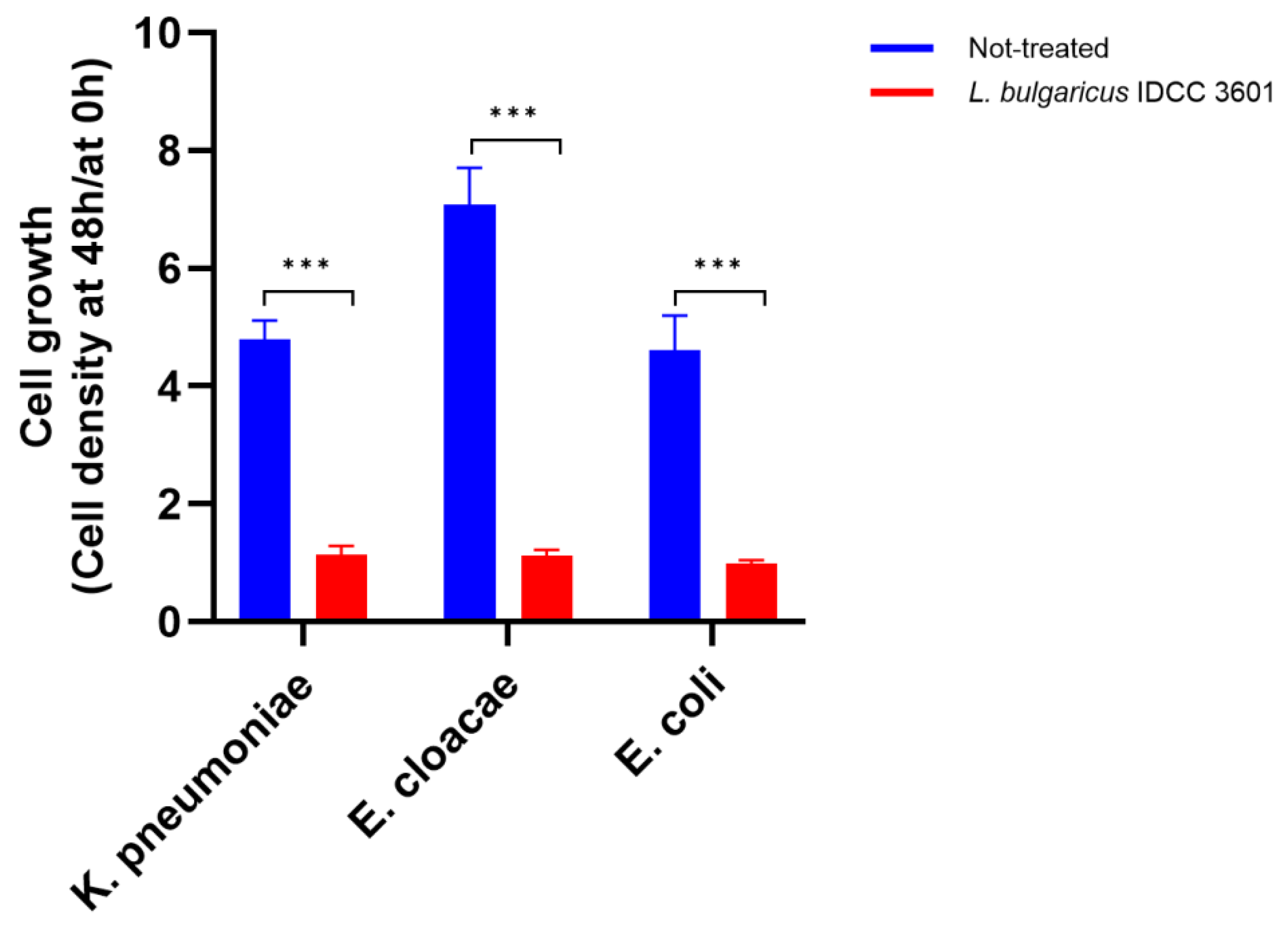
2.8. Antibacterial Activity against Enterobacteriaceae
2.9. Statistical Analysis
3. Results and Discussion
3.1. Genomic Analysis of L. bulgaricus IDCC 3601
3.2. Antibiotic Susceptibility
3.3. Determination of Biogenic Amines (BAs)
3.4. Hemolytic Property
3.5. Carbohydrate Utilization and Extracellular Enzymatic Activities
3.6. Acute Oral Toxicity Test
3.7. Antibacterial Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quinto, E.J.; Jiménez, P.; Caro, I.; Tejero, J.; Mateo, J.; Girbés, T. Probiotic lactic acid bacteria: A review. Food Nutr. Sci. 2014, 5, 1765. [Google Scholar] [CrossRef]
- Collins, J.K.; Thornton, G.; Sullivan, G.O. Selection of probiotic strains for human application. Int. Dairy J. 1998, 8, 487–490. [Google Scholar] [CrossRef]
- Das, D.; Goyal, A. Lactic acid bacteria in food industry. In Microorganisms in Sustainable Agriculture and Biotechnology; Satyanarayana, T., Johri, B., Eds.; Springer: Dordrecht, The Netherlands, 13 December 2012; pp. 757–772. [Google Scholar]
- Gemechu, T. Review on lactic acid bacteria function in milk fermentation and preservation. Afr. J. Food Sci. 2015, 9, 170–175. [Google Scholar]
- Mazahreh, A.S.; Ershidat, O.T.M. The benefits of lactic acid bacteria in yogurt on the gastrointestinal function and health. Pak. J. Nutr. 2009, 8, 1404–1410. [Google Scholar]
- Masood, M.I.; Qadir, M.I.; Shirazi, J.H.; Khan, I.U. Beneficial effects of lactic acid bacteria on human beings. Crit. Rev. Microbiol. 2011, 37, 91–98. [Google Scholar] [CrossRef]
- Gupta, R.; Jeevaratnam, K.; Fatima, A. Lactic Acid Bacteria: Probiotic Characteristic, Selection Criteria, and its Role in Human Health (A Review). Int. J. Emerg. Technol. Innov. Res. 2018, 5, 411–424. [Google Scholar]
- Abedin, M.M.; Chourasia, R.; Phukon, L.C.; Sarkar, P.; Ray, R.C.; Singh, S.P.; Rai, A.K. Lactic acid bacteria in the functional food industry: Biotechnological properties and potential applications. Crit. Rev. Food Sci. Nutr. 2023, 8, 1–19. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.; Haller, D.; Hammerman, C.; Heimbach, J.T.; Hörmannsperger, G.; Huys, G. Safety assessment of probiotics for human use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef]
- Didari, T.; Solki, S.; Mozaffari, S.; Nikfar, S.; Abdollahi, M. A systematic review of the safety of probiotics. Expert Opin. Drug Saf. 2014, 13, 227–239. [Google Scholar] [CrossRef]
- Binda, S.; Hill, C.; Johansen, E.; Obis, D.; Pot, B.; Sanders, M.E.; Tremblay, A.; Ouwehand, A.C. Criteria to qualify microorganisms as “probiotic” in foods and dietary supplements. Front. Microbiol. 2020, 11, 1662. [Google Scholar] [CrossRef]
- EFSA. Guidance on the characterization of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, 5206. [Google Scholar]
- Qureshi, N.; Gu, Q.; Li, P. Whole genome sequence analysis and in vitro probiotic characteristics of a Lactobacillus strain Lactobacillus paracasei ZFM54. J. Appl. Microbiol. 2020, 129, 422–433. [Google Scholar] [CrossRef]
- FAO/WHO. Guidelines for the Evaluation of Probiotics in Food; Report of a Joint FAO/WHO: London, ON, Canada, 2002. [Google Scholar]
- Mazzantini, D.; Calvigioni, M.; Celandroni, F.; Lupetti, A.; Ghelardi, E. Spotlight on the compositional quality of probiotic formulations marketed worldwide. Front. Microbiol. 2021, 20, 693973. [Google Scholar] [CrossRef]
- Fusco, V.; Fanelli, F.; Chieffi, D. Recent and advanced DNA-based technologies for the authentication of probiotic, protected designation of origin (PDO) and protected geographical indication (PGI) fermented foods and beverages. Foods 2023, 12, 3782. [Google Scholar] [CrossRef]
- Lidbeck, A.; Nord, C.E. Lactobacilli and the normal human anaerobic microflora. Clin. Infect. Dis. 1993, 16, S181–S187. [Google Scholar] [CrossRef]
- Fisberg, M.; Machado, R. History of yogurt and current patterns of consumption. Nutr. Rev. 2015, 73, 4–7. [Google Scholar] [CrossRef]
- Agrawal, R. Probiotics: An emerging food supplement with health benefits. Food Biotechnol. 2005, 19, 227–246. [Google Scholar] [CrossRef]
- Chen, Y.; Li, R.; Chang, Q.; Dong, Z.; Yang, H.; Xu, C. Lactobacillus bulgaricus or Lactobacillus rhamnosus suppresses NF-κB signaling pathway and protects against AFB1-induced hepatitis: A novel potential preventive strategy for aflatoxicosis? Toxins 2019, 11, 17. [Google Scholar] [CrossRef]
- Silveira, D.S.C.; Veronez, L.C.; Lopes-Júnior, L.C.; Anatriello, E.; Brunaldi, M.O.; Pereira-da-Silva, G. Lactobacillus bulgaricus inhibits colitis-associated cancer via a negative regulation of intestinal inflammation in azoxymethane/dextran sodium sulfate model. World J. Gastroenterol. 2020, 26, 6782–6794. [Google Scholar] [CrossRef]
- Mohtashami, M.; Mohamadi, M.; Azimi-Nezhad, M.; Saeidi, J.; Nia, F.F.; Ghasemi, A. Lactobacillus bulgaricus and Lactobacillus plantarum improve diabetic wound healing through modulating inflammatory factors. Biotechnol. Appl. Biochem. 2021, 68, 1421–1431. [Google Scholar] [CrossRef]
- Araya, M.; Morelli, L.; Reid, G.; Sanders, M.; Stanton, C.; Pineiro, M.; Ben Embarek, P. Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; World Health Organization: Geneva, Switzerland; Food and Agriculture Organization of the United Nations: Québec City, QC, Canada, 2002. [Google Scholar]
- Approved Standard M100–S20; Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute: Malvern, PA, USA, January 2010.
- Ban, O.H.; Bang, W.Y.; Jeon, H.J.; Jung, Y.H.; Yang, J.; Kim, D.H. Potential of Bifidobacterium lactis IDCC 4301 isolated from breast milk-fed infant feces as a probiotic and functional ingredient. Food Sci. Nutr. 2023, 11, 1952–1964. [Google Scholar] [CrossRef]
- Wu, J.J.; Zhou, Q.Y.; Liu, D.M.; Xiong, J.; Liang, M.H.; Tang, J.; Xu, Y.Q. Evaluation of the safety and probiotic properties of Lactobacillus gasseri LGZ1029 based on whole genome analysis. LWT 2023, 184, 114759. [Google Scholar] [CrossRef]
- Todorov, S.D.; Perin, L.M.; Carneiro, B.M.; Rahal, P.; Holzapfel, W.; Nero, L.A. Safety of Lactobacillus plantarum ST8Sh and its bacteriocin. Probiotics Antimicrob. Proteins 2017, 9, 334–344. [Google Scholar] [CrossRef]
- Kwon, Y.J.; Chun, B.H.; Jung, H.S.; Chu, J.; Joung, H.; Park, S.Y.; Kim, B.K.; Jeon, C.O. Safety assessment of Lactiplantibacillus (formerly Lactobacillus) plantarum Q180. J. Microbiol. Biotechnol. 2021, 31, 1420–1429. [Google Scholar] [CrossRef]
- Azevedo, I.; Barbosa, J.; Albano, H.; Nogueira, T.; Teixeira, P. Lactic acid bacteria isolated from traditional and innovative alheiras as potential biocontrol agents. Food Microbiol. 2024, 119, 104450. [Google Scholar] [CrossRef]
- Jaimee, G.; Halami, P.M. Emerging resistance to aminoglycosides in lactic acid bacteria of food origin—An impending menace. Appl. Microbiol. Biotechnol. 2016, 100, 1137–1151. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2018, 85, e01738-18. [Google Scholar] [CrossRef]
- Das, S.; Mishra, B.K.; Hati, S. Techno-functional characterization of indigenous Lactobacillus isolates from the traditional fermented foods of Meghalaya, India. Curr. Res. Food Sci. 2020, 11, 9–18. [Google Scholar] [CrossRef]
- Lim, E.S.; Lee, N.G. Control of histamine-forming bacteria by probiotic lactic acid bacteria isolated from fish intestine. Korean J. Microbiol. 2016, 52, 352–364. [Google Scholar] [CrossRef]
- Vesković-Moračanin, S.; Stefanović, S.; Borović, B.; Nastasijevic, I.; Milijasevic, M.; Stojanova, M.; Đukić, D. Assessment of biogenic amine production by lactic acid bacteria isolated from Serbian traditionally fermented foods. Acta Agric. Serb. 2022, 27, 49–55. [Google Scholar] [CrossRef]
- Elsanhoty, R.M.; Ramadan, M.F. Genetic screening of biogenic amines production capacity from some lactic acid bacteria strains. Food Control 2016, 68, 220–228. [Google Scholar] [CrossRef]
- Suvarna, S.; Dsouza, J.; Ragavan, M.L.; Das, N. Potential probiotic characterization and effect of encapsulation of probiotic yeast strains on survival in simulated gastrointestinal tract condition. Food Sci. Biotechnol. 2018, 27, 745–753. [Google Scholar] [CrossRef]
- Meng, L.; Zhu, X.; Tuo, Y.; Zhang, H.; Li, Y.; Xu, C.; Mu, G.; Jiang, S. Reducing antigenicity of β-lactoglobulin, probiotic properties and safety evaluation of Lactobacillus plantarum AHQ-14 and Lactobacillus bulgaricus BD0390. Food Biosci. 2021, 42, 101137. [Google Scholar] [CrossRef]
- Berlowska, J.; Cieciura, W.; Borowski, S.; Dudkiewicz, M.; Binczarski, M.; Witonska, I.; Otlewska, A.; Kregiel, D. Simultaneous saccharification and fermentation of sugar beet pulp with mixed bacterial cultures for lactic acid and propylene glycol production. Molecules 2016, 21, 1380. [Google Scholar] [CrossRef]
- Azarnia, S.; Lee, B.; St-Gelais, D.; Kilcawley, K.; Noroozi, E. Effect of free and encapsulated recombinant aminopeptidase on proteolytic indices and sensory characteristics of Cheddar cheese. LWT-Food Sci. Technol. 2011, 44, 570–575. [Google Scholar] [CrossRef]
- El-Soda, M.; Macedo, A.; Olson, N. Aminopeptidase and dipeptidyl aminopeptidase activities of several cheese related microorganisms. Milchwissenschaft 1991, 46, 223–226. [Google Scholar]
- Zhang, W.; Wang, C.; Huang, C.Y.; Yu, Q.; Liu, H.C.; Zhang, C.W.; Pei, X.F.; Xu, X.; Wang, G.Q. Analysis of β-galactosidase production and their genes of two strains of Lactobacillus bulgaricus. Biotechnol. Lett. 2012, 34, 1067–1071. [Google Scholar] [CrossRef]
- Liew, S.Y.; Sivasothy, Y.; Shaikh, N.N.; Isa, D.M.; Lee, V.S.; Choudhary, M.I.; Awang, K. β-Glucuronidase inhibitors from Malaysian plants. J. Mol. Struct. 2020, 1221, 128743. [Google Scholar] [CrossRef]
- Sohn, H.; Chang, Y.H.; Yune, J.H.; Jeong, C.H.; Shin, D.M.; Kwon, H.C.; Kim, D.H.; Hong, S.W.; Hwang, H.; Jeong, J.Y.; et al. Probiotic properties of Lactiplantibacillus plantarum LB5 isolated from Kimchi based on nitrate reducing capability. Foods 2020, 9, 1777. [Google Scholar] [CrossRef]
- Kim, G.S.; Bae, C.M.; Oh, S.K.; Ban, O.H.; Yang, J. Multi-Layer Coated Probiotics Having Improved Productivity and Stability. Korean Patent KR 10-2430949, 4 August 2022. [Google Scholar]
- Bang, W.Y.; Kim, H.; Chae, S.A.; Yang, S.Y.; Ban, O.H.; Kim, T.Y.; Kwon, H.S.; Jung, Y.H.; Yang, J. A quadruple coating of probiotics for enhancing intestinal adhesion and competitive exclusion of Salmonella typhimurium. J. Med. Food 2022, 25, 213–218. [Google Scholar] [CrossRef]
- Lee, J.H.; Shin, J.; Park, S.H.; Cha, B.; Hong, J.T.; Lee, D.H.; Kwon, K.S. Role of Probiotics in preventing carbapenem-resistant Enterobacteriaceae colonization in the intensive care unit: Risk factors and microbiome analysis study. Microorganisms 2023, 11, 2970. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Limbago, B.M.; Patel, J.B.; Kallen, A.J. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin. Infect. Dis. 2011, 53, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Aghamohammad, S.; Rohani, M. Antibiotic resistance and the alternatives to conventional antibiotics: The role of probiotics and microbiota in combating antimicrobial resistance. Microbiol. Res. 2023, 267, 127275. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Lai, C.C.; Huang, H.L.; Huang, W.Y.; Toh, H.S.; Weng, T.C.; Chuang, T.C.; Lu, Y.C.; Tang, H.J. Antimicrobial activity of Lactobacillus species against carbapenem-resistant Enterobacteriaceae. Front. Microbiol. 2019, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Bar, N.; Harris, N.D.; Rill, R.L. Purification and properties of an antimicrobial substance produced by Lacfobacillus bulgaricus. J. Food Sci. 1987, 52, 411–415. [Google Scholar] [CrossRef]
- Mogna, L.; Deidda, F.; Nicola, S.; Amoruso, A.; Del Piano, M.; Mogna, G. In vitro inhibition of Klebsiella pneumoniae by Lactobacillus delbrueckii subsp. delbrueckii LDD01 (DSM 22106): An innovative strategy to possibly counteract such infections in humans? J. Clin. Gastroenterol. 2016, 50, S136–S139. [Google Scholar] [CrossRef]
- Savino, F.; Cordisco, L.; Tarasco, V.; Locatelli, E.; Di Gioia, D.; Oggero, R.; Matteuzzi, D. Antagonistic effect of Lactobacillus strains against gas-producing coliforms isolated from colicky infants. BMC Microbiol. 2011, 11, 157. [Google Scholar] [CrossRef]
- Mandras, N.; Tullio, V.; Furneri, P.M.; Roana, J.; Allizond, V.; Scalas, D.; Petronio Petronio, G.; Fuochi, V.; Banche, G.; Cuffini, A.M. Key roles of human polymorphonuclear cells and ciprofloxacin in Lactobacillus species infection control. Antimicrob. Agents Chemother. 2015, 60, 1638–1641. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.D.; Braughton, K.R.; Whitney, A.R.; Voyich, J.M.; Schwan, T.G.; Musser, J.M.; DeLeo, F.R. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc. Natl. Acad. Sci. USA 2003, 100, 10948–10953. [Google Scholar] [CrossRef]
- Tajdozian, H.; Seo, H.; Jeong, Y.; Ghorbanian, F.; Park, C.E.; Sarafraz, F.; Rahim, M.A.; Lee, Y.; Kim, S.; Lee, S.; et al. Efficacy of lyophilized Lactobacillus sakei as a potential candidate for preventing carbapenem-resistant Klebsiella infection. Ann. Microbiol. 2024, 74, 28. [Google Scholar] [CrossRef]
- Ramos-Ramos, J.C.; Lázaro-Perona, F.; Arribas, J.R.; García-Rodríguez, J.; Mingorance, J.; Ruiz-Carrascoso, G.; Borobia, A.M.; Paño-Pardo, J.R.; Herruzo, R.; Arnalich, F. Proof-of-concept trial of the combination of lactitol with Bifidobacterium bifidum and Lactobacillus acidophilus for the eradication of intestinal OXA-48-producing Enterobacteriaceae. Gut Pathogens. 2020, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
| AMP d | VAN | GEN | KAN | STR | ERY | CLI | TET | CHL | |
|---|---|---|---|---|---|---|---|---|---|
| Cut-off value (µg/mL) a | 2 | 2 | 16 | 16 | 16 | 1 | 4 | 4 | 4 |
| L. bulgaricus IDCC 3601 | <0.125/S b | 0.25/S | 32/R c | 64–128/R | 16/S | <0.125/S | <0.125/S | 1/S | 2/S |
| No. | Substrate | Result | No. | Substrate | Result | No. | Substrate | Result |
|---|---|---|---|---|---|---|---|---|
| 1 | Glycerol | – | 18 | Mannitol | – | 35 | d-Raffinose | – |
| 2 | Erythritol | – | 19 | Sorbitol | – | 36 | Amidon | – |
| 3 | d-Arabinose | – | 20 | α-Methyl-D-mannoside | – | 37 | Glycogene | – |
| 4 | l-Arabinose | – | 21 | α-Methyl-D-glucoside | – | 38 | Xylitol | – |
| 5 | Ribose | – | 22 | N-Acethyl-glucosamine | +W a | 39 | Gentibiose | – |
| 6 | d-Xylose | – | 23 | Amygdaline | – | 40 | d-Turanose | – |
| 7 | l-Xylose | – | 24 | Arbutine | – | 41 | d-Lyxose | – |
| 8 | Adonitol | – | 25 | Esculine | + | 42 | d-Tagatose | – |
| 9 | β-Methyl-xylose | – | 26 | Salicine | – | 43 | d-Fucose | – |
| 10 | Galactose | – | 27 | Cellobiose | – | 44 | l-Fucose | – |
| 11 | d-Glucose | + | 28 | Maltose | – | 45 | d-Arabitol | – |
| 12 | d-Fructose | + | 29 | Lactose | + | 46 | l-Arabitol | – |
| 13 | d-Mannose | + | 30 | Melibiose | – | 47 | Gluconate | – |
| 14 | l-Sorbose | – | 31 | Sucrose | – | 48 | 2-Keto-gluconate | – |
| 15 | Rhamnose | – | 32 | Trehalose | – | 49 | 5-Keto-gluconate | – |
| 16 | Dulcitol | – | 33 | Inuline | – | |||
| 17 | Inositiol | – | 34 | Melizitose | – |
| No. | Enzyme | Result | No. | Substrate | Result |
|---|---|---|---|---|---|
| 1 | Alkaline phosphatase | – | 11 | Naphthol-AS-BI-phosphohydrolase | + |
| 2 | Esterase | + | 12 | α-galactosidase | – |
| 3 | Esterase lipase | – | 13 | β-galactosidase | + |
| 4 | Lipase | – | 14 | β-glucuronidase | – |
| 5 | Leucine arylamidase | + | 15 | α-glucosidase | – |
| 6 | Valine arylamidase | + | 16 | β-glucosidase | – |
| 7 | Cystine arylamidase | + | 17 | N-acetyl-β-glucosaminidase | – |
| 8 | Trypsin | – | 18 | α-mannosidase | – |
| 9 | α-chymotrypsin | – | 19 | α-fucosidase | – |
| 10 | Acid phosphatase | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.; Bang, W.-Y.; Lee, H.-B.; Yang, S.-Y.; Lee, K.-S.; Kang, H.-J.; Hong, S.-M.; Yang, J. Safety Assessment and Evaluation of Probiotic Potential of Lactobacillus bulgaricus IDCC 3601 for Human Use. Microorganisms 2024, 12, 2063. https://doi.org/10.3390/microorganisms12102063
Lee M, Bang W-Y, Lee H-B, Yang S-Y, Lee K-S, Kang H-J, Hong S-M, Yang J. Safety Assessment and Evaluation of Probiotic Potential of Lactobacillus bulgaricus IDCC 3601 for Human Use. Microorganisms. 2024; 12(10):2063. https://doi.org/10.3390/microorganisms12102063
Chicago/Turabian StyleLee, Minjee, Won-Yeong Bang, Han-Bin Lee, Soo-Yeon Yang, Kyu-Shik Lee, Hae-Ji Kang, Sun-Mee Hong, and Jungwoo Yang. 2024. "Safety Assessment and Evaluation of Probiotic Potential of Lactobacillus bulgaricus IDCC 3601 for Human Use" Microorganisms 12, no. 10: 2063. https://doi.org/10.3390/microorganisms12102063
APA StyleLee, M., Bang, W.-Y., Lee, H.-B., Yang, S.-Y., Lee, K.-S., Kang, H.-J., Hong, S.-M., & Yang, J. (2024). Safety Assessment and Evaluation of Probiotic Potential of Lactobacillus bulgaricus IDCC 3601 for Human Use. Microorganisms, 12(10), 2063. https://doi.org/10.3390/microorganisms12102063






