Abstract
Many species of the genus Pseudomonas are known to be highly tolerant to solvents and other environmental stressors. Based on phylogenomic and comparative genomic analyses, several Pseudomonas species were recently transferred to a new genus named Halopseudomonas. Because of their unique enzymatic machinery, these strains are being discussed as novel biocatalysts in biotechnology. In order to test their growth parameters and stress tolerance, five Halopseudomonas strains were assessed regarding their tolerance toward different n-alkanols (1-butanol, 1-hexanol, 1-octanol, 1-decanol), as well as to salt stress and elevated temperatures. The toxicity of the solvents was investigated by their effects on bacterial growth rates and presented as EC50 concentrations. Hereby, all Halopseudomonas strains showed EC50 values up to two-fold lower than those previously detected for Pseudomonas putida. In addition, the activity of the cis-trans isomerase of unsaturated fatty acids (Cti), which is an urgent stress response mechanism known to be present in all Pseudomonas species, was monitored in the five Halopseudomonas strains. Although several of the tested species were known to contain the cti gene, no significant phenotypic activity could be detected in the presence of the assayed stressors. A bioinformatic analysis of eight cti-carrying Halopseudomonas strains examining promotor binding sites, binding motifs and signal peptides showed that most of the cti genes have a lipoprotein signal peptide and promotor regions and binding motifs that do not coincide with those of Pseudomonas. These insights represent putative reasons for the absence of the expected Cti activity in Halopseudomonas, which in turn has always been observed in cti-carrying Pseudomonas. The lack of Cti activity under membrane stress conditions when the cti gene is present has never been documented, and this could represent potential negative implications on the utility of the genus Halopseudomonas for some biotechnological applications.
1. Introduction
Certain bacteria possess exceptional capabilities regarding tolerance and degradation potential toward organic compounds present in ecosystems impacted by anthropogenic organic pollutants or crude oil contamination [1,2]. These specialized bacteria play a crucial role in mitigating the harmful effects of such contaminants and promoting environmental remediation [3]. Whole toolboxes of different adaptive mechanisms enable them to thrive in contaminated environments and efficiently degrade complex organic compounds. By harnessing their enzymatic potential and metabolic pathways [4,5], these bacteria contribute to the restoration of ecosystems affected by organic pollutants, providing a promising avenue for addressing environmental challenges associated with these contaminations. Among all microorganisms pertinent to this matter, the well-studied genus Pseudomonas stands out as very broadly active among the Gram-negative proteobacteria [6].
Among Pseudomonas sp., solvent-tolerant strains are highly regarded as ideal biocatalysts for producing various chemicals in biotechnology, e.g., propionate or p-hydroxybenzoate [7,8]. However, in Germany, many of these strains, especially those of P. putida (excluding P. putida KT2440), are classified as biosafety level 2, imposing costly safety measures and reducing their attractiveness to the biotechnological industry. This classification is questioned because these bacteria have no pathogenic characteristics in their genome, and their safety is demonstrably guaranteed. In contrast, the U.S. FDA considers the P. putida group non-pathogenic [9]. The ongoing debate over the classification of P. putida has prompted suggestions of a new species, P. alloputida, which includes well-studied strains such as P. putida KT2440, P. putida S12 and P. putida DOT-T1E [10]. Apart from regulatory and taxonomic concerns, there is a crucial need to identify Pseudomonas or other bacterial strains with high tolerance to solvents and other stressors for establishing biotechnological production platforms [11,12].
In environments with toxic pollutants, fluctuating temperatures, or high levels of osmotic pressure, numerous species within the genus Halopseudomonas have been discovered that potentially exhibit tolerance to stress [13]. Previously classified within the Pseudomonas genus as part of the Pseudomonas pertucinogena lineage, these bacteria were recently reclassified as Halopseudomonas (also known as Neopseudomonas) within the Pseudomonadaceae family due to significant molecular and metabolic distinctions from other Pseudomonas species [14].
Genomic analyses have uncovered genes responsible for encoding various interesting enzymes in Halopseudomonas, including esterases and dehalogenases, as well as the potential for biosynthesis of polyhydroxyalkanoates (PHA), among others [13,15,16]. Additionally, there is speculation about the involvement of Halopseudomonas in breaking down human-made pollutants like oil or plastic waste [13,17,18,19,20]. In particular, in marine crude oil drilling sites, species like H. aestusnigri have been found to cope with alkanes and alkane alcohols. Despite these insights, information on the solvent tolerance of Halopseudomonas species has been limited. As they are potential candidates for the role of biotechnological workhorses [21], there is a necessity to elucidate their robustness against solvents. Testing their tolerance toward different n-alkanols (1-butanol, 1-hexanol, 1-octanol, 1-decanol) is suitable as a kind of benchmark test, and it has been applied already to novel bacterial strains, as well as genome-reduced strains, to compare them to their wildtype counterparts [22].
The purpose of this study is to evaluate the robustness of five members of the newly established Halopseudomonas genus against n-alkanols, heat shock and osmotic stress, and document their stress response. Therefore, the effective n-alkanol concentration for reducing cell growth by 50% compared to the control (EC50) was determined for five Halopseudomonas strains. Among the five mentioned strains, three carry the gene for the cis-trans isomerase (Cti) of membrane fatty acids, with its activity being a unique urgent stress response mechanism for stabilizing the bacterial membrane and a potential taxonomic marker for the genus Pseudomonas [11,23]. Only very few bacterial genera carry the gene for the periplasmic Cti, a cytochrome c-type protein with a heme-binding motif [23,24]. Given that the gene is constitutively transcribed in Pseudomonas spp., it was suggested that the protein remains in the periplasmic region until the fluidity of the membrane requires it to move toward its cis-configurated unsaturated fatty acid substrates [24]. After its action, the membrane is reduced in fluidity and the Cti returns to its original periplasmic location. The assessment of Cti activity in the recent genera Halopseudomonas is still pending. Therefore, the role of this enzyme in the robustness of the strains, which has yet to be determined, was investigated in a total of eight Halopseudomonas strains bioinformatically. Of these, three selected strains were also tested for a Cti phenotype upon exposure to n-alkanols.
2. Materials and Methods
2.1. Bacterial Strains and Media
Unless otherwise indicated, a modified Hartmans [25] mineral salt medium was used to grow the cultures, usually with 18 mM sebacic acid (decanedioic acid). The medium was prepared with the following solutions:
Mineral Salts Solution (100×)—In 1 L of distilled water, the following salts were added: 100 g of NH4Cl, 70 g of Na2HPO4 × 2H2O, 30 g of NaCl and 28 g of KH2PO4.
Trace Elements Solution (500×)—In 1 L of distilled water, the following compounds were added: 73 mL of HCl (37%), 50 g of MgSO4 × 7H2O, 5 g of FeSO4 × 7H2O, 5 g of EDTA, 3.25 g of H3BO3, 3.2 g of ZnCl2, 2.5 g of MnSO4 × H2O, 0.5 g of CaCl2 × 6H2O, 0.3 g of BaCl2, 0.18 g of CoSO4 × 7H2O and 0.18 g of CuSO4 × 5H2O.
The rifampicin-resistant Halopseudomonas strains (H. aestusnigri VGXO14R, H. oceani KX20R, H. litoralis 2SM5R and H. bauzanensis BZ93R) were grown in a modified Hartmans mineral medium (0.3% NaCl) with 18 mM sebacic acid (decanedioic acid) as a carbon source at 30 °C and 150 rpm at a pH of 6.9 [21]. A spontaneous rifampicin-resistant mutant of H. pachastrellae was isolated as follows: H. pachastrellae was streak-plated on an LB-Agar plate (5 g/L yeast extract, 10 g/L tryptone, 10 g/L glucose and 20 g/L agar) and incubated at 30 °C for 48 h. The colonies formed were then streak-plated on another 25 mL LB-Agar plate with 25 μg/mL of the rifampicin. After 48 h, this process was repeated. Once the final colonies were formed, the bacterium was grown under the conditions previously described [21]. The day before each experiment, 50 mL overnight cultures (around 15 h) were prepared in 500 mL DURAN wide neck Erlenmeyer flasks containing the aforementioned modified Hartmans mineral medium, achieving a final optical density (OD560) of 2–3. For the subsequent experiments, a 50 mL culture was grown at an OD560 of 0.1–0.2 in 250 mL screw cap glass bottles (30 °C, 150 rpm, pH of 6.9). The growing conditions were the same as for the overnight culture. The bottles were kept in an orbital shaking water bath (GFL 1092) throughout the experiment (30 °C and 180 rpm).
For each experiment, the medium was prepared in bulk and distributed in each of the 250 mL screw cap glass bottles. Depending on the achieved overnight OD, the amount of inoculum was chosen to make up an initial OD of 0.1–0.2. Rifampicin solution was added to each bottle to reach a final concentration of 25 μg/mL. The rifampicin solution was always protected from the light with tin foil and stored at −20 °C. Sebacic acid (≥99%), rifampicin (≥97%), dimethyl sulfoxide (DMSO, ≥99.9%) as well as all applied n-alkanols, NaCl and acetone were obtained from Merck (Darmstadt, Germany).
In addition, Pseudomonas taiwanensis VLB120 was used as a benchmark for solvent tolerance and as a positive control for Cti activity. The cultivation followed a standard procedure using mineral salt medium [25] and disodium succinate (24.7 mM) as a carbon and energy source at 30 °C and 150 rpm [22].
The monitoring of bacterial growth was carried out by measuring the optical density at 560 nm in an Agilent Cary 100 UV–Vis Spectrophotometer (Waldbronn, Germany) every hour. The release of a red pigment was observed to be absorbed at the same wavelength. In an attempt to reduce its effect, a 1:10 dilution with a 50 mM phosphate buffer (phosphate buffer blank) was made in a single-use polystyrene semi-micro cuvette. This reduced the contribution of the released red pigment to the real OD.
2.2. Growth Under Solvent, Osmotic and Temperature Stress
The bacterial stress experiments were conducted with 1M NaCl (osmotic stress); 45 °C (heat shock); 0–100 mM 1-butanol; 0–7 mM 1-hexanol; 0–0.7 mM 1-octanol; and 0–0.25 mM 1-decanol (solvent stress) under the aforementioned growth conditions (modified Hartmans mineral medium with 18 mM sebacic acid at 30 °C and 150 rpm, pH of 6.9). The concentrations of the n-alkanols mentioned were chosen according to their log Poctanol/water (partitioning coefficient between octanol and water [26]). The stressed bacterial cultures were compared to a control without the stressor. The stressors were added and exposed, respectively, to exponentially growing bacteria. After 3 h in the presence of the stressors, the cultures were centrifuged at 10,000× g for 15 min in a Hermle Z 383 K table cooling centrifuge. The pellets were then resuspended in 1.75 mL of the aforementioned phosphate buffer and centrifuged at 14,000× g for 7 min, using 2 mL Eppendorf tubes. The buffer was then removed, and the pellets were frozen at −20 °C to store them for the later extraction of membrane lipids. Growth inhibition caused by the toxic compounds was measured by comparing the percentage difference in the growth rates between intoxicated cultures with that of the control [27]. Growth rates in the first three hours after stressor addition and growth inhibition were calculated as published earlier [22].
2.3. Extraction of Membrane Lipids
Lipid extraction was conducted according to Bligh and Dyer [28]. Using the method of Morrison, Smith [29], fatty acid methyl esters (FAMEs) were synthesized by 15 min incubation at 95 °C in 0.6 mL of boron trifluoride–methanol complex (20% MeOH; Sigma Aldrich, Munich, Germany). FAME analysis was performed using gas chromatography with a flame ionization detector (GC-FID, Agilent Technologies, Waldbronn, Germany 6890N Network GC System, 7683B Series Injector). The instrument used a CP-Sil 88 column (Varian CP7488, Palo Alto, CA, USA) in the stationary phase and helium as a carrier gas. The peak areas of the FAMEs were used to determine their relative amounts. The fatty acids were identified by a co-injection of authentic reference compounds obtained from Supelco (Bellefonte, PA, USA). Equation 1 was used to calculate the degree of saturation (DoS) of the FAMEs. It is the quotient of the sum of the saturated FAMEs with the sum of the unsaturated FAMEs.
Degree of Saturation = (% C16:0 + % C18:0)/sum of unsaturated fatty acids
The trans/cis ratio was calculated by taking the sum of the FAME of palmitoleic acid C16:1 ∆9cis) and cis-vaccenic acid (C18:1 ∆11cis) as the divisor and the sum of their corresponding trans configuration as the dividend (Equation (2)).
trans/cis ratio = (% C16:1∆9trans + % C18:1∆11trans)/(% C16:1∆9cis + % C18:1∆11cis)
2.4. Bioinformatic Analysis
The cti genes were identified by BLAST analysis, using the cti sequence of Pseudomonas taiwanensis VLB120 as a query to search the genomes of Halopseudomonas species. The binding motifs conservation (HMM logo) was studied. The derived protein sequences of the following strains were aligned and analyzed: H. aestusnigri VGXO14R, H. oceani KX20R and H. pachastrellae JCM 12285, P. abyssi MT5, H. gallaeciensis V113, H. pelagia CL-AP6, H. sabulinigri JCM 13963 and H. sp. RR6. The prediction of the signal peptides was performed with the use of the online tool SignalP 6.0 described by Nielsen and colleagues [30]. It has to be mentioned that P. abyssi MT5 was also proposed to be a member of the genus Halopseudomonas. However, due to the lack of an available strain type in culture collections, this could not be implemented yet [14].
3. Results and Discussion
3.1. Growth of Five Halopseudomonas Strains in Mineral Medium
The observed reduction in growth rates under stress conditions in comparison to unchallenged cells is an established parameter to quantify the impact of a stressor [22]. As a benchmark, the growth rates of all five tested Halopseudomonas strains on sebacic acid in mineral media under the applied cultivation conditions were determined. Among the five species, the growth rates were within the same order of magnitude and comparable to previously published results [21]. H. aestusnigri and H. oceani achieved the highest growth rates among the cultivated strains (Table 1).

Table 1.
Growth rates of the five tested Halopseudomonas strains on sebacic acid in mineral media evaluated during the first five hours of growth; n = 2 for H. oceani [31], H. bauzanensis [32] and H. aestusnigri [33], n ≥ 1 for H. pachastrellae [34] and H. litoralis [35].
3.2. Distribution of Cti Genes Among Halopseudomonas
Having been classified as Pseudomonadales and until recently even belonging to the genus Pseudomonas, the genomes of Halopseudomonas species were searched for genes encoding homologs of fatty acid cis/trans isomerase. This enzyme is characteristic among Pseudomonas species, and it is a key trait of their robustness against hydrophobic chemicals, being an urgent response mechanism against membrane destabilization [23]. Notably, putative cti genes were present in some investigated Halopseudomonas species but are apparently not a shared feature of the genus Halopseudomonas (Table 2) The Halopseudomonas strains carrying a cti gene are listed in Table 2. Since a gene for proteins homologous to the Cti of P. taiwanensis VLB120 was found in the genomes of H. aestusnigri, H. oceani and H. pachastrellae, Cti activity was determined in five different Halopseudomonas species (Table 2) based on the incubation at elevated temperatures, and osmotic and solvent stress (1-butanol, 1-hexanol, 1-octanol, 1-decanol).

Table 2.
Overview of the strains used in the study for bioinformatic analyses, including the five analyzed for tolerance toward n-alkanols. P. abyssi MT5 was also proposed to be a member of the genus Halopseudomonas. However, due to the lack of an available strain type in culture collections, this is still to be implemented [13].
3.3. Growth Under Solvent, Osmotic and Temperature Stress
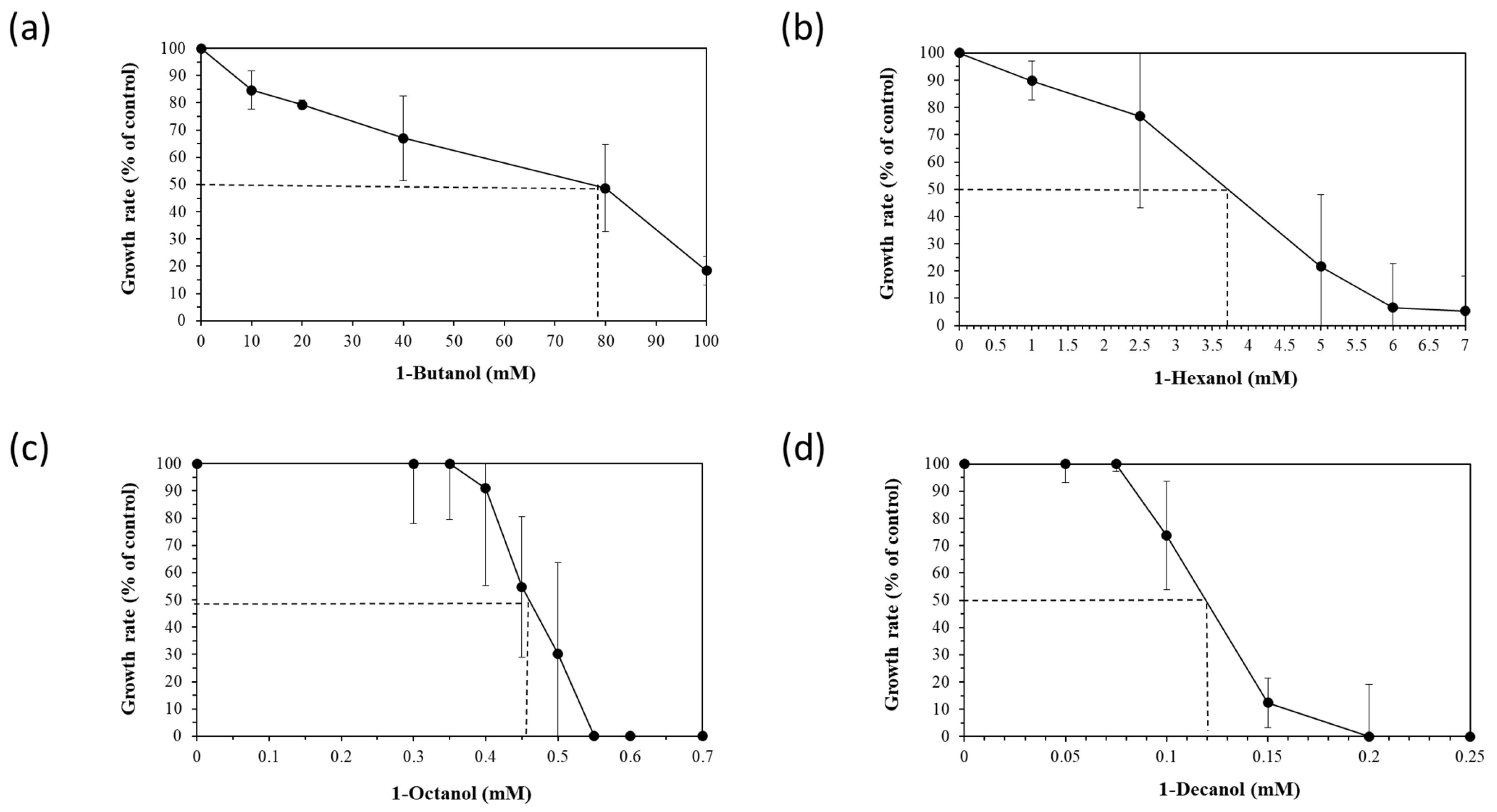
To assess the robustness of the five selected Halopseudomonas strains against a chemical stressor, the effect of different concentrations of four n-alkanols (1-butanol, 1-hexanol, 1-octanol and 1-decanol), added during the early exponential (logarithmic) growth phase, was tested. The exponential growth phase is usually chosen for stressor addition to provide a controlled and consistent environment to study the immediate and specific responses of actively dividing cells that do not face nutrient limitations yet. Relative growth inhibition was calculated between the growth rates of the control and stressed cultures and is showcased for H. aestusnigri in Figure 1. The EC50 (effective concentration for reducing cell growth by 50% compared to the control) was deduced from the growth inhibition for each n-alkanol (dotted line in Figure 1).
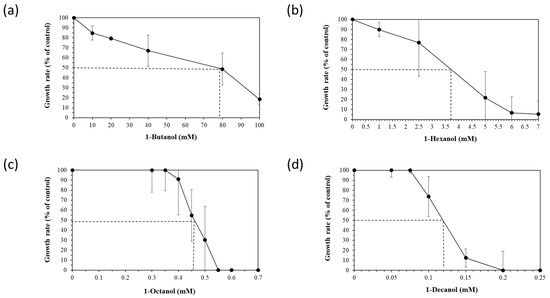
Figure 1.
Growth rates (black circles) and trans/cis ratios (white diamonds) of H. aestusnigri incubated with different n-alkanols: (a) 1-butanol, (b) 1-hexanol, (c) 1-octanol and (d) 1-decanol. The EC50 (effective concentration for reducing cell growth by 50% compared to the control) is represented by a dotted line. n = 3.
The approximate EC50 values for all strains tested are given in Table 3. A difference between the cti carriers (H. aestusnigri, H. pachastrellae and H. oceani) and the non-cti-carriers (H. bauzanensis and H. litoralis) can be observed, because the latter have comparatively lower EC50 values. However, there are exceptions to this trend, such as H. litoralis exposed to 1-butanol and 1-decanol, which showed similar EC50 values compared to the cti carriers. Simultaneously, H. oceani exhibits a lower tolerance toward 1-butanol and 1-octanol, but a higher tolerance toward 1-decanol. The EC50 values of the Halopseudomonas strains were at least half compared to those of P. taiwanensis for every n-alkanol except for 1-hexanol (Table 3).

Table 3.
EC50 values expressed in mM of each strain toward the four n-alkanols tested. C4 = 1-Butanol; C6 = 1-Hexanol; C8 = 1-Octanol; C10 = 1-Decanol. For H. pachastrellae, H. oceani and H. litoralis, n ≥ 1. For H. aestusnigri, H. litoralis and P. taiwanensis, n = 3.
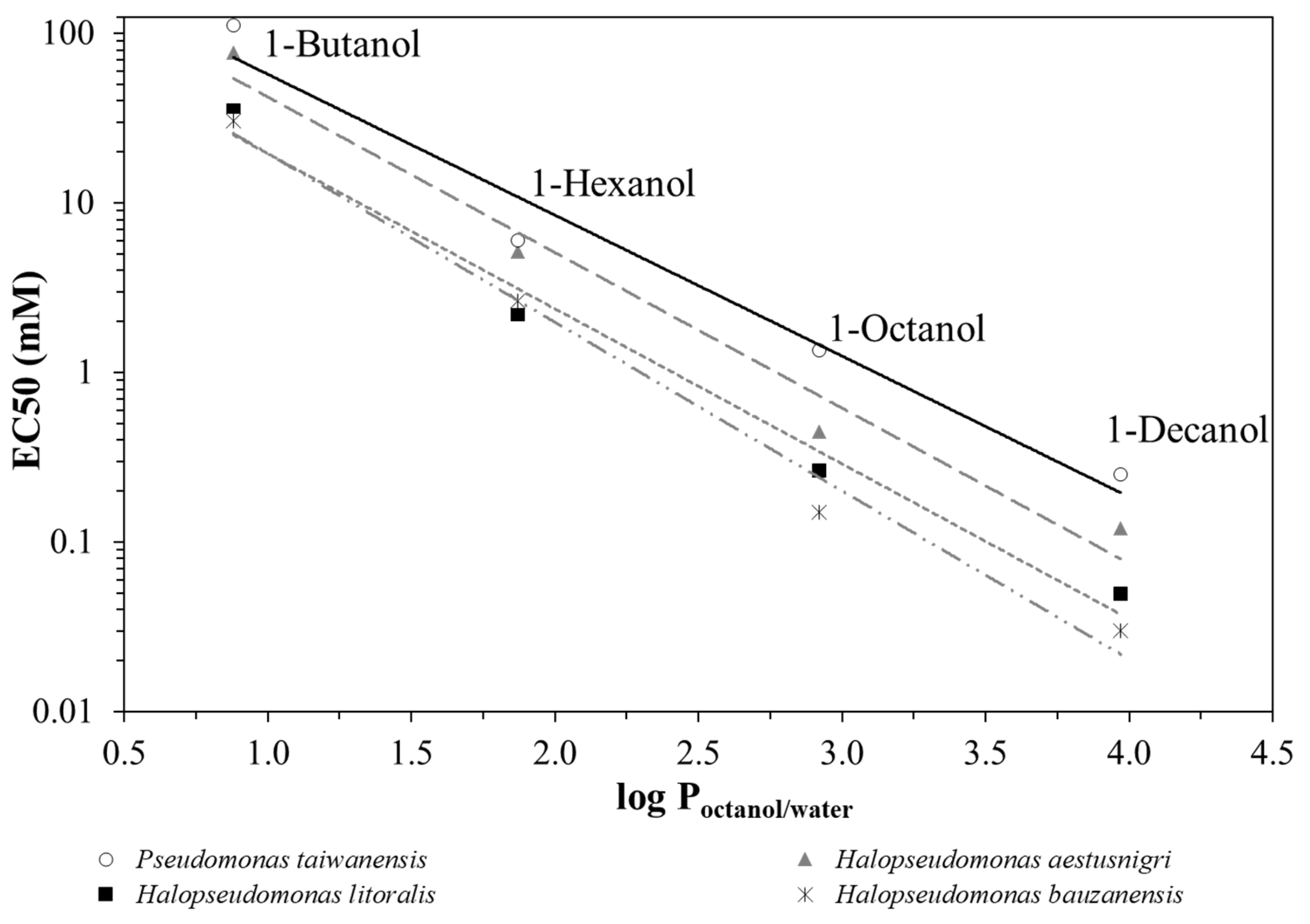
By plotting the EC50 values as a function of the partitioning coefficient between octanol and water, log Poctanol/water [26] of the respective alkanol (Figure 2), it can be seen that P. taiwanensis VLB120 outperforms the Halopseudomonas strains regarding its robustness against n-alkanols of increasing hydrophobicity. H. aestusnigri can be considered the most tolerant among the strains tested. Typically, substances with a log Poctanol/water between 1 and 4, such as long-chain alkanols, aromatics and esters, exhibit toxicity to microorganisms even at very low concentrations because they tend to accumulate in the cytoplasmic membrane [36,37,38].
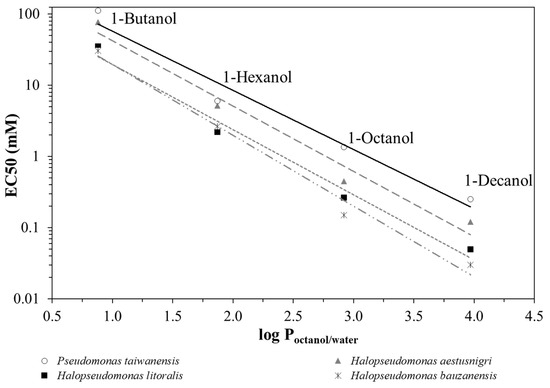
Figure 2.
Correlation between EC50 and log Poctanol/water values for 1-butanol, 1-hexanol, 1-octanol and 1-decanol. As examples, H. bauzanensis (mix of dashed and dotted grey trendline), H. aestusnigri (long dashed grey trendline), H. litoralis (short dashed grey trendline) and P. taiwanensis VLB120 (solid black trendline) are shown.
After the toxicity tests, the membrane fatty acids were extracted to calculate the activity of the Cti. The phospholipid fatty acid profile of the strains showed the presence of the following fatty acids: C16:0, C16:1trans, C16:1cis, 17cyclo, C18:0, C18:1trans and C18:1cis. In accordance with their percentage distribution, the degree of saturation and the trans/cis ratio of each species’ control were calculated (Table 4). The trans/cis ratio in the controls without solvent was either zero or very close to zero for all Halopseudomonas strains. Remarkably, despite the incubation with four n-alkanols, elevated temperature, or osmotic stress, the trans/cis ratio remained stable in the Halopseudomonas strains, whereas a clear increase occurred in strain VLB120. Maximum trans/cis ratios are shown in Table 4. The trans/cis ratios of H. litoralis and H. bauzanensis, both of which do not have a cti homolog, were also measured routinely, and the values were always close to zero, regardless of the presence or absence of a solvent.

Table 4.
Maximum values of trans/cis ratios of P. taiwanensis, H. aestusnigri, H. litoralis, H. oceani and H. pachastrellae cells under osmotic stress (1 M NaCl), temperature stress (45 °C) and upon exposure to the four n-alkanols tested. C4 = 1-butanol (0–100 mM); C6 = 1-hexanol (0–7 mM); C8 = 1-octanol (0–0.7 mM); C10 = 1-decanol (0–0.25 mM); control = without stressor. n ≥ 2.
The results from Table 4 show that the Cti had neglectable or no activity upon solvent exposure in all tested Halopseudomonas strains. This was made clear through the comparison with P. taiwanensis VLB120, which was used as positive control. Strains H. litoralis and H. bauzanensis did not show any Cti activity, which was expected due to the cti gene’s absence. Increased trans/cis ratios of more than 1 upon exposure to a stressor are documented for various cti-carrying bacteria, like for Vibrio cholerae under starvation stress [39], as well as for Pseudomonas putida S12 [27], and for Alcanivorax borkumensis SK2 under solvent stress [40]. Also, for Methylococcus capsulatus Bath, a small increase in trans/cis ratios was found upon solvent exposure [41]. Hence, we tested other stressors known to trigger cis/trans isomerization [42]. However, Cti activity was observed neither under osmotic nor heat stress conditions (Table 4). Nevertheless, an extremely low or non-existent increase in trans/cis ratios like those shown by the Halopseudomonas strains investigated here had never been observed before. In fact, such a discrepancy between a Cti genotype and phenotype had never been described, to our knowledge. This discrepancy is also consistent with the fact that all tested strains of the genus Halopseudomonas were significantly less solvent-tolerant than all strains of the closely related genus Pseudomonas. However, this leaves open the question of why Halopseudomonas strains do not show Cti activity even though they possess the corresponding cti gene.
3.4. Binding Motifs Comparison and Signal Peptides
In light of these experimental results, we tried to find mutual differences between Halopseudomonas Cti and Cti with proven activity from Pseudomonas spp. Therefore, we included all of the Cti identifiable in the Halopseudomonas clade, thereby adding another five strains (P. abyssi MT5, H. gallaeciensis V113, H. pelagia CL-AP6, H. sabulinigri JCM 13963 and H. sp. RR6) to the Halopseudomonas strains that were assessed phenotypically. Using the Multiple Sequence Alignment tool from Uniprot and a hidden Markov model, the similarities in common amino acid sequences between the tested Halopseudomonas strains become apparent, as well as their differences compared to the Pseudomonas strains [43] (Figure S1).
Even though the Cti proteins in all strains investigated so far do not have a high degree of sequence identity, their mode of action is the same. Hence, a heme binding motif should be present, which will allocate the ferrous cation, as described previously [24,43,44]. In addition, three putative phosphorylation sites (PGSTEAL, SRTPSG and DMNRYENL) are expected. Within the known Ctis of Pseudomonas, there is a certain sequence variation. By comparing the sequences with the reference Cti of P. aeruginosa PAO1, P. capeferrum TDA1 [45] shows 65.32% sequence identity, and H. aestusnigri and H. oceani as well as H. pachastrellae show around 49% sequence identity (query coverage for all between 97 and 100%, and E value for all 0.00) (Figure S1, Supplementary Materials). For Pseudomonads, this was also shown in the work of Mauger and colleagues, but the mentioned motifs, however, were found to be conserved [43]. The heme binding site and the proline residue are conserved in the Halopseudomonas strains as well. However, there is an observable difference in the Halopseudomonas strains compared to the Pseudomonas strains at the putative phosphorylation sites, more prominently at the expected PGSTEAL and DMNRYENL sites (conserved as XGREQL and DINRYQNL, respectively). The differences in the phosphorylation sites could be an explanation for Cti inactivity. Support for the hypothesis of Cti inactivity because of the non-conserved phosphorylation sites comes from another study with the cti-carrying Methylococcus capsulatus Bath [41], whose diminished Cti activity could be related to the different phosphorylation sites (SSPKERL, SRTPPG, DLSRTDNP). However, there may be other factors influencing this matter. For instance, as mentioned in the introduction, the Cti is a periplasmic protein, hence the need for a signal peptide that will allow the cell to recognize the protein’s translocation paths.
In order to direct the Cti to its final cellular compartment, namely the periplasm, a signal peptide is required, which is usually a short peptide sequence, typically 15–30 amino acids long and often located at the N-terminus. Therefore, the signal peptides of the Ctis were compared using the online software SignalP 6.0 [30]. It predicts the presence of signal peptides and the location of their cleavage sites. Table 5 gives an overview of the results. Figure S2 (Supplementary Materials) shows the software’s output for the Cti of H. aestusnigri, but it is representative of six out of eight Halopseudomonas strains (except for H. oceani and H. pelagia), and it detected the presence of a Sec-dependent lipoprotein signal peptide cleaved by the signal peptidase II (SPase II) [46], resulting in a membrane-anchored periplasmic Cti. In contrast to this, in the putative protein of the cti gene in H. oceani, P. capeferrum TDA1 and P. aeruginosa PAO1, a Sec-dependent signal peptide cleaved by the signal peptidase I (SPase I) is present (Table 5). Hence, all Halopseudomonas strains bear a signal peptide in their potential Cti sequence, but only in H. oceani and H. pelagia is the Cti not anchored in the membrane after membrane translocation. Hence, constrained flexibility of the enzyme by n-terminal covalent attachment to the membrane could be a reason for the very low or absent activity of the Cti in Halopseudomonas strains, but not in H. oceani and H. pelagia. Nevertheless, enzymes such as thioesterases or peptidases might also hydrolyze the bond between the Cti and the membrane and thereby release a membrane-anchored Cti.

Table 5.
Halopseudomonas strains’ Cti location prediction using the SignalP 6.0 software. The signal peptide is predicted to either be Sec-dependent and cleaved by the signal peptidase I (Sec/SPI) or a lipoprotein signal peptide and cleaved by the signal peptidase II (Sec/SPII).
4. Conclusions
Five strains of the barely explored genus Halopseudomonas were subjected to a series of toxicity tests in the presence of four n-alkanols. A rapid decrease in OD560 was consistently observed upon incubation with the n-alkanols tested. The EC50 values for each n-alkanol were considerably lower than those observed with the selected tolerant Pseudomonas strains.
Remarkably, the cis-trans isomerase of unsaturated fatty acids, the activity of which is considered a typical immediate response trait of Pseudomonads leading to a fast rigidification of the membrane, appears not be conserved among Halopseudomonas strains, but encoded only in the genomes of some species. However, even the latter strains were not able to elevate their trans/cis ratio in response to alkanols, or to other stressors including salt and elevated temperature. This is the first study showing a discrepancy between a cti genotype and missing Cti activity in the phenotype. It is hypothesized that a membrane-anchored Cti and non-functional phosphorylation sites could be responsible for this. The lower membrane stress tolerance compared to other Pseudomonas might have negative implications on the utility of the genus Halopseudomonas as whole-cell biocatalysts in certain biotechnological applications besides the described utilization of aliphatic plastics [15,19]. The insights first gained here into the physiology of Halopseudomonas support a niche adoption of these species compared to their robust versatile relatives among the Pseudomonadaceae clade.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12112116/s1, Figure S1: Multiple Sequence Alignment of the cis-trans isomerase amino acid sequences belonging to H. pelagia, H. sabulinigri, Halospeudomonas sp. RR6, H. pachastrellae, P. abyssi, H. gallaeciensis, H. aestusnigri and H. oceani. Figure S2: H. aestusnigri SignalP 6.0 signal peptide prediction output (representative of all mentioned Halopseudomonas strains excluding H. oceani). The first 70 amino acids are shown (x-axis) and plotted against the signal peptide’s probability. The dark green dotted line is the cleavage site (CS). To the right of the CS, the software does not find a signal peptide, thus it predicts “OTHER”, which means it is the protein itself. A blue line indicates the presence of a characteristic cysteine residue, signalizing the presence of a Sec-dependent lipoprotein signal peptide. To the left of the CS, the lipoprotein signal peptide is predicted. It is divided into two lines (red and orange), which correspond to two constitutive regions of the signal peptide (N and H, respectively). The signal peptide is predicted to be Sec-dependent and cleaved by the signal peptidase II (SPase II), as indicated by the legend in the upper right corner.
Author Contributions
Conceptualization and methodology, H.J.H., C.C.C.R.d.C., S.T. and C.E.; software, S.B. and P.D.M.A.S.M.; validation, all authors; formal analysis, S.B., P.D.M.A.S.M., S.T. and C.E.; investigation, S.B. and P.D.M.A.S.M.; resources, H.J.H., S.T. and L.K.; data curation, S.B., P.D.M.A.S.M. and C.E.; writing—original draft preparation, S.B., P.D.M.A.S.M. and C.E.; writing—review and editing, all authors; visualization, S.B. and P.D.M.A.S.M.; supervision, H.J.H., C.C.C.R.d.C., S.T., L.K. and C.E.; project administration, H.J.H. and C.E.; funding acquisition, H.J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the German Federal Ministry of Education and Research (BMBF) via the project NO-STRESS [grant numbers 031B0852B (to S.T. and L.K.) and 031B085C (to C.E. and H.J.H.)].
Data Availability Statement
The original contributions presented in the study are partially included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- de Carvalho, C.C.C.R.; da Fonseca, M.M. Degradation of hydrocarbons and alcohols at different temperatures and salinities by Rhodococcus erythropolis DCL14. FEMS Microbiol. Ecol. 2005, 51, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Segura, A.; Molina, L.; Fillet, S.; Krell, T.; Bernal, P.; Munoz-Rojas, J.; Ramos, J.-L. Solvent tolerance in Gram-negative bacteria. Curr. Opin. Biotechnol. 2012, 23, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Muriel-Millan, L.F.; Millan-Lopez, S.; Pardo-Lopez, L. Biotechnological applications of marine bacteria in bioremediation of environments polluted with hydrocarbons and plastics. Appl. Microbiol. Biotechnol. 2021, 105, 7171–7185. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Li, L.-Z.; Wu, Y.; Tian, W.; Zhang, L.-P.; Xu, L.; Shen, Q.-R.; Shen, B. Isolation of an alkane-degrading Alcanivorax sp. strain 2B5 and cloning of the alkB gene. Bioresour. Technol. 2010, 101, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, J.; Liang, R.; Liu, J. Characterization of the medium- and long-chain n-alkanes degrading Pseudomonas aeruginosa strain SJTD-1 and its alkane hydroxylase genes. PLoS ONE 2014, 9, e105506. [Google Scholar] [CrossRef]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef]
- Ma, C.; Mu, Q.; Xue, Y.; Xue, Y.; Yu, B.; Ma, Y. One major facilitator superfamily transporter is responsible for propionic acid tolerance in Pseudomonas putida KT2440. Microb. Biotechnol. 2021, 14, 386–391. [Google Scholar] [CrossRef]
- Verhoef, S.; Ballerstedt, H.; Volkers, R.J.; de Winde, J.H.; Ruijssenaars, H.J. Comparative transcriptomics and proteomics of p-hydroxybenzoate producing Pseudomonas putida S12: Novel responses and implications for strain improvement. Appl. Microbiol. Biot. 2010, 87, 679–690. [Google Scholar] [CrossRef]
- Kampers, L.F.C.; Volkers, R.J.M.; dos Santos, V.A.P.M. Pseudomonas putida KT2440 is HV1 certified, not GRAS. Microb. Biotechnol. 2019, 12, 845–848. [Google Scholar] [CrossRef]
- Keshavarz-Tohid, V.; Vacheron, J.; Dubost, A.; Prigent-Combaret, C.; Taheri, P.; Tarighi, S.; Taghavi, S.M.; Moënne-Loccoz, Y.; Muller, D. Genomic, phylogenetic and catabolic re-assessment of the Pseudomonas putida clade supports the delineation of Pseudomonas alloputida sp. nov., Pseudomonas inefficax sp. nov., Pseudomonas persica sp. nov., and Pseudomonas shirazica sp. nov. Syst. Appl. Microbiol. 2019, 42, 468–480. [Google Scholar] [CrossRef]
- Bitzenhofer, N.L.; Kruse, L.; Thies, S.; Wynands, B.; Lechtenberg, T.; Rönitz, J.; Kozaeva, E.; Wirth, N.T.; Eberlein, C.; Jaeger, K.-E.; et al. Towards robust Pseudomonas cell factories to harbour novel biosynthetic pathways. Essays Biochem. 2021, 65, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; de Lorenzo, V. Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab. Eng. 2018, 50, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Bollinger, A.; Thies, S.; Katzke, N.; Jaeger, K.E. The biotechnological potential of marine bacteria in the novel lineage of Pseudomonas pertucinogena. Microb. Biotechnol. 2018, 13, 19–31. [Google Scholar] [CrossRef]
- Rudra, B.; Gupta, R.S. Phylogenomic and comparative genomic analyses of species of the family Pseudomonadaceae: Proposals for the genera Halopseudomonas gen. nov. and Atopomonas gen. nov., merger of the genus Oblitimonas with the genus Thiopseudomonas and transfer of some misclassified species of the genus Pseudomonas into other genera. Int. J. Syst. Evol. Microbiol. 2021, 71, 005011. [Google Scholar] [CrossRef]
- Molitor, R.; Bollinger, A.; Kubicki, S.; Loeschcke, A.; Jaeger, K.E.; Thies, S. Agar plate-based screening methods for the identification of polyester hydrolysis by Pseudomonas species. Microb. Biotechnol. 2020, 13, 274–284. [Google Scholar] [CrossRef]
- Zukic, E.; Mokos, D.; Weber, M.; Stix, N.; Ditrich, K.; Ferrario, V.; Müller, H.; Willrodt, C.; Gruber, K.; Daniel, B.; et al. Biocatalytic Heteroaromatic Amide Formation in Water Enabled by a Catalytic Tetrad and Two Access Tunnels. ACS Catal. 2024, 14, 8913–8921. [Google Scholar] [CrossRef]
- Villela, H.; Modolon, F.; Schultz, J.; Delgadillo-Ordoñez, N.; Carvalho, S.; Soriano, A.U.; Peixoto, R.S. Genome analysis of a coral-associated bacterial consortium highlights complementary hydrocarbon degradation ability and other beneficial mechanisms for the host. Sci. Rep. 2023, 13, 12273. [Google Scholar] [CrossRef]
- Gomila, M.; Mulet, M.; Lalucat, J.; Garcia-Valdes, E. Draft Genome Sequence of the Marine Bacterium Pseudomonas aestusnigri VGXO14(T). Genome Announc. 2017, 5, e00765-17. [Google Scholar] [CrossRef]
- de Witt, J.; Molitor, R.; Gätgens, J.; Northumberland, C.O.d.P.; Kruse, L.; Polen, T.; Wynands, B.; van Goethem, K.; Thies, S.; Jaeger, K.; et al. Biodegradation of poly(ester-urethane) coatings by Halopseudomonas formosensis. Microb. Biotechnol. 2024, 17, e14362. [Google Scholar] [CrossRef]
- Avilan, L.; Lichtenstein, B.R.; König, G.; Zahn, M.; Allen, M.D.; Oliveira, L.; Clark, M.; Bemmer, V.; Graham, R.; Austin, H.P.; et al. Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering. ChemSusChem 2023, 16, e202202277. [Google Scholar] [CrossRef]
- Kruse, L.; Loeschcke, A.; de Witt, J.; Wierckx, N.; Jaeger, K.E.; Thies, S. Halopseudomonas species: Cultivation and molecular genetic tools. Microb. Biotechnol. 2024, 17, e14369. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, M.J.C.; Schmidgall, T.; Pohl, J.; Wagner, G.; Wynands, B.; Wierckx, N.; Heipieper, H.J.; Eberlein, C. Assessment of New and Genome-Reduced Strains Regarding Their Robustness as Chassis in Biotechnological Applications. Microorganisms 2023, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Eberlein, C.; Baumgarten, T.; Starke, S.; Heipieper, H.J. Immediate response mechanisms of Gram-negative solvent-tolerant bacteria to cope with environmental stress: Isomerization of unsaturated fatty acids and outer membrane vesicle secretion. Appl. Microbiol. Biotechnol. 2018, 102, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- von Wallbrunn, A.; Richnow, H.H.; Neumann, G.; Meinhardt, F.; Heipieper, H.J. Mechanism of cis-trans isomerization of unsaturated fatty acids in Pseudomonas putida. J. Bacteriol. 2003, 185, 1730–1733. [Google Scholar] [CrossRef]
- Hartmans, S.; Smits, J.P.; van der Werf, M.J.; Volkering, F.; de Bont, J.A.M. Metabolism of Styrene Oxide and 2-Phenylethanol in the Styrene-Degrading Xanthobacter Strain 124x. Appl. Environ. Microbiol. 1989, 55, 2850–2855. [Google Scholar] [CrossRef]
- Laane, C.; Boeren, S.; Vos, K.; Veeger, C. Rules for optimization of biocatalysis in organic solvents. Biotechnol. Bioeng. 1987, 30, 81–87. [Google Scholar] [CrossRef]
- Heipieper, H.J.; Loffeld, B.; Keweloh, H.; de Bont, J.A.M. The Cis/Trans Isomerization of Unsaturated Fatty-Acids in Pseudomonas putida S12—An Indicator for Environmental-Stress Due to Organic-Compounds. Chemosphere 1995, 30, 1041–1051. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Phys. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Morrison, W.R.; Smith, L.M. Preparation of Fatty Acid Methyl Esters + Dimethylacetals from Lipids with Boron Fluoride-Methanol. J. Lipid Res. 1964, 5, 600–608. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Wang, M.Q.; Sun, L. Pseudomonas oceani sp. nov., isolated from deep seawater. Int. J. Syst. Evol. Microbiol. 2016, 66, 4250–4255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.C.; Liu, H.C.; Zhou, Y.G.; Schinner, F.; Margesin, R. Pseudomonas bauzanensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 2333–2337. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sánchez , D.; Mulet, M.; Rodríguez, A.C.; David, Z.; Lalucat, J.; García-Valdés, E. Pseudomonas aestusnigri sp. nov., isolated from crude oil-contaminated intertidal sand samples after the Prestige oil spill. Syst. Appl. Microbiol. 2014, 37, 89–94. [Google Scholar] [CrossRef]
- Romanenko, L.A.; Uchino, M.; Falsen, E.; Frolova, G.M.; Zhukova, N.V.; Mikhailov, V.V. Pseudomonas pachastrellae sp. nov., isolated from a marine sponge. Int. J. Syst. Evol. Microbiol. 2005, 55, 919–924. [Google Scholar] [CrossRef]
- Pascual, J.; Lucena, T.; Ruvira, M.A.; Giordano, A.; Gambacorta, A.; Garay, E.; Arahal, D.R.; Pujalte, M.J.; Macián, M.C. Pseudomonas litoralis sp. nov., isolated from Mediterranean seawater. Int. J. Syst. Evol. Microbiol. 2012, 62, 438–444. [Google Scholar] [CrossRef]
- Heipieper, H.J.; de Bont, J.A.M. Adaptation of Pseudomonas putida S12 to ethanol and toluene at the level of fatty acid composition of membranes. Appl. Environ. Microbiol. 1994, 60, 4440–4444. [Google Scholar] [CrossRef]
- Sikkema, J.; de Bont, J.A.M.; Poolman, B. Mechanisms of Membrane Toxicity of Hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [CrossRef]
- Weber, F.J.; de Bont, J.A.M. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1996, 1286, 225–245. [Google Scholar] [CrossRef]
- Guckert, J.B.; Hood, M.A.; White, D.C. Phospholipid Ester-Linked Fatty-Acid Profile Changes during Nutrient Deprivation of Vibrio cholerae Increases in the Trans Cis Ratio and Proportions of Cyclopropyl Fatty-Acids. Appl. Environ. Microbiol. 1986, 52, 794–801. [Google Scholar] [CrossRef]
- Naether, D.J.; Slawtschew, S.; Stasik, S.; Engel, M.; Olzog, M.; Wick, L.Y.; Timmis, K.N.; Heipieper, H.J. Adaptation of the Hydrocarbonoclastic Bacterium Alcanivorax borkumensis SK2 to Alkanes and Toxic Organic Compounds: A Physiological and Transcriptomic Approach. Appl. Environ. Microbiol. 2013, 79, 4282–4293. [Google Scholar] [CrossRef]
- Loeffler, C.; Eberlein, C.; Mausezahl, I.; Kappelmeyer, U.; Heipieper, H.J. Physiological evidence for the presence of a cis-trans isomerase of unsaturated fatty acids in Methylococcus capsulatus Bath to adapt to the presence of toxic organic compounds. FEMS Microbiol. Lett. 2010, 308, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Heipieper, H.J.; Meulenbeld, G.; van Oirschot, Q.; de Bont, J.A.M. Effect of Environmental Factors on the trans/cis Ratio of Unsaturated Fatty Acids in Pseudomonas putida S12. Appl. Environ. Microbiol. 1996, 62, 2773–2777. [Google Scholar] [CrossRef] [PubMed]
- Mauger, M.; Ferreri, C.; Chatgilialoglu, C.; Seemann, M. The bacterial protective armor against stress: The isomerase of unsaturated fatty acids, a cytochrome-c type enzyme. J. Inorg. Biochem. 2021, 224, 111564. [Google Scholar] [CrossRef] [PubMed]
- Holtwick, R.; Keweloh, H.; Meinhardt, F. cis/trans isomerase of unsaturated fatty acids of Pseudomonas putida P8:: Evidence for a heme protein of the cytochrome c Type. Appl. Environ. Microbiol. 1999, 65, 2644–2649. [Google Scholar] [CrossRef]
- Espinosa, M.J.C.; Blanco, A.C.; Schmidgall, T.; Atanasoff-Kardjalieff, A.K.; Kappelmeyer, U.; Tischler, D.; Pieper, D.H.; Heipieper, H.J.; Eberlein, C. Toward Biorecycling: Isolation of a Soil Bacterium That Grows on a polyurethane Oligomer and Monomer. Front. Microbiol. 2020, 11, 404. [Google Scholar] [CrossRef]
- Buddelmeijer, N. The molecular mechanism of bacterial lipoprotein modification—How, when and why? FEMS Microbiol. Rev. 2015, 39, 246–261. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).