Halophilic Pectinase-Producing Bacteria from Arthrocnemum macrostachyum Rhizosphere: Potential for Fruit–Vegetable Juice Processing
Abstract
:1. Introduction
2. Methods
2.1. Reagents and Chemicals
2.2. Isolation of Bacteria from Rhizospheric Soil
2.3. Screening for Pectinase Activity
2.4. Molecular Identification Through 16S rDNA Sequencing
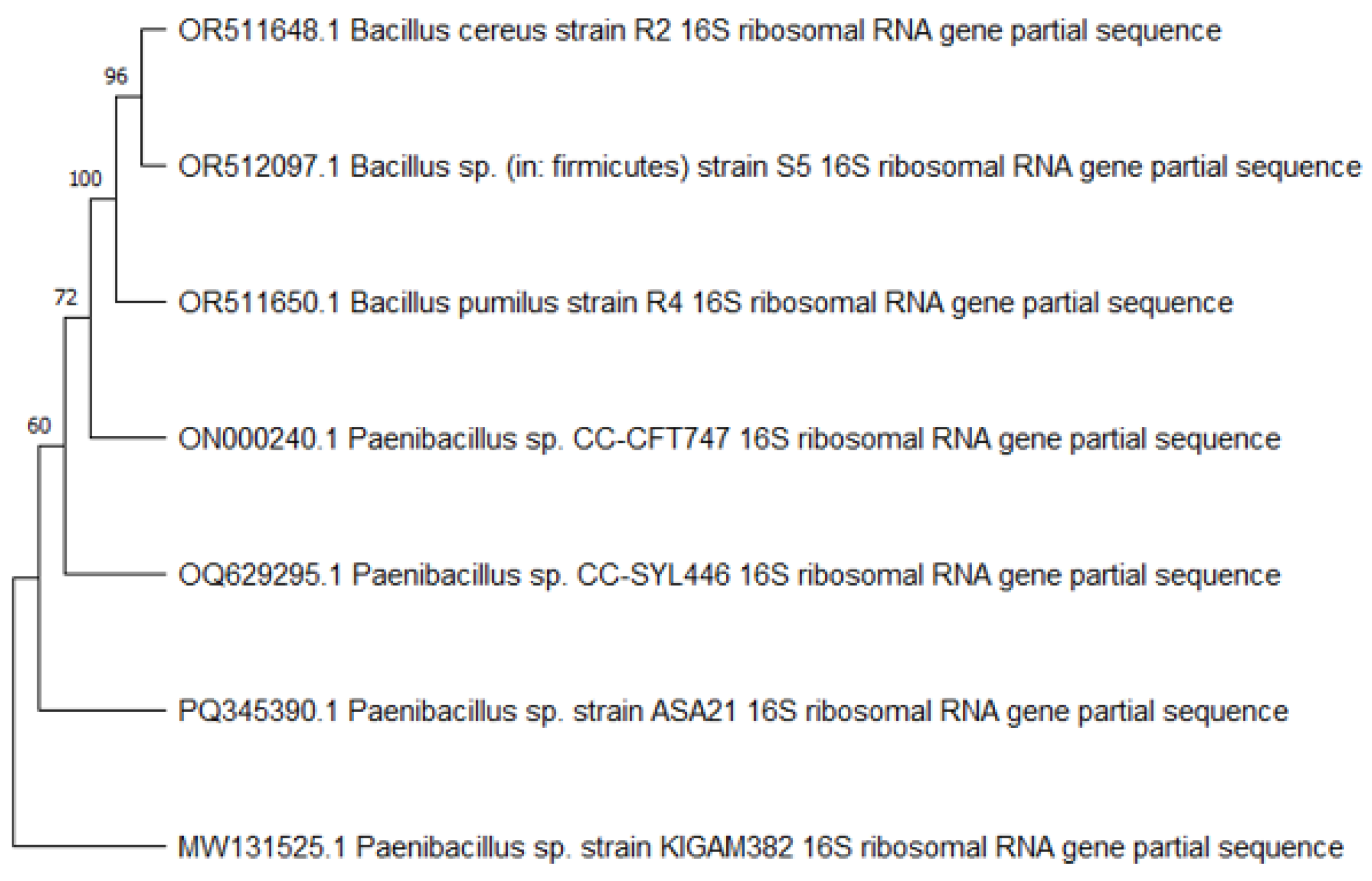
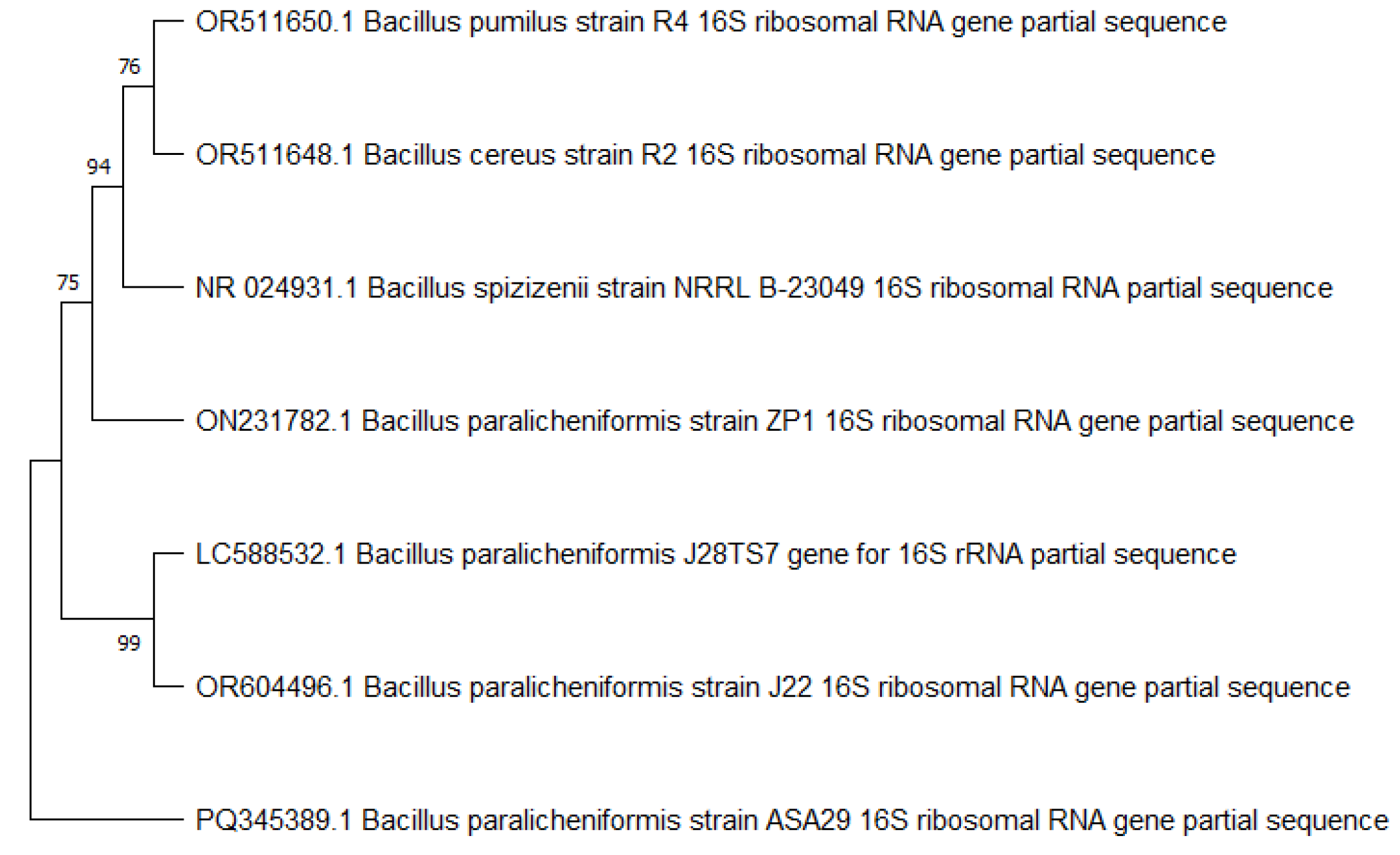
2.5. Phylogenetic Analysis
2.6. Investigating Salt Tolerance
2.7. Growth and Pectinase Production Kinetics
2.8. Juice Preparation
2.9. Characterization of Detox Juice Formulation
2.9.1. Juice Yield
2.9.2. Total Soluble Solids (°Bx)
2.9.3. Estimation of Total Phenolic Content
2.9.4. Estimation of Free Radical Scavenging Activity
2.10. Statistical Analysis
3. Results
3.1. Isolation and Screening of Halophilic Bacterial Strains for Pectinase Production
3.2. Identification of Promising Strains
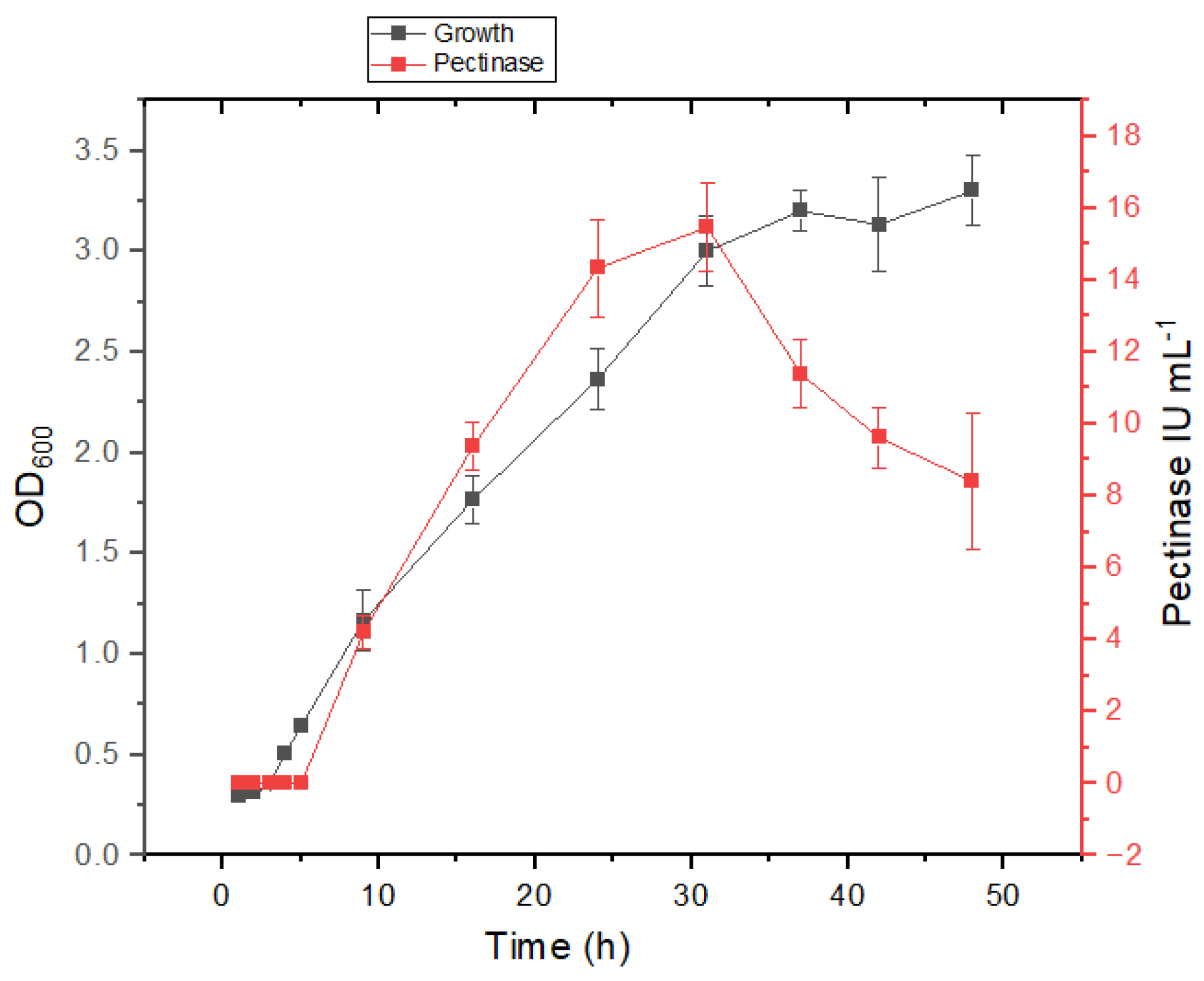
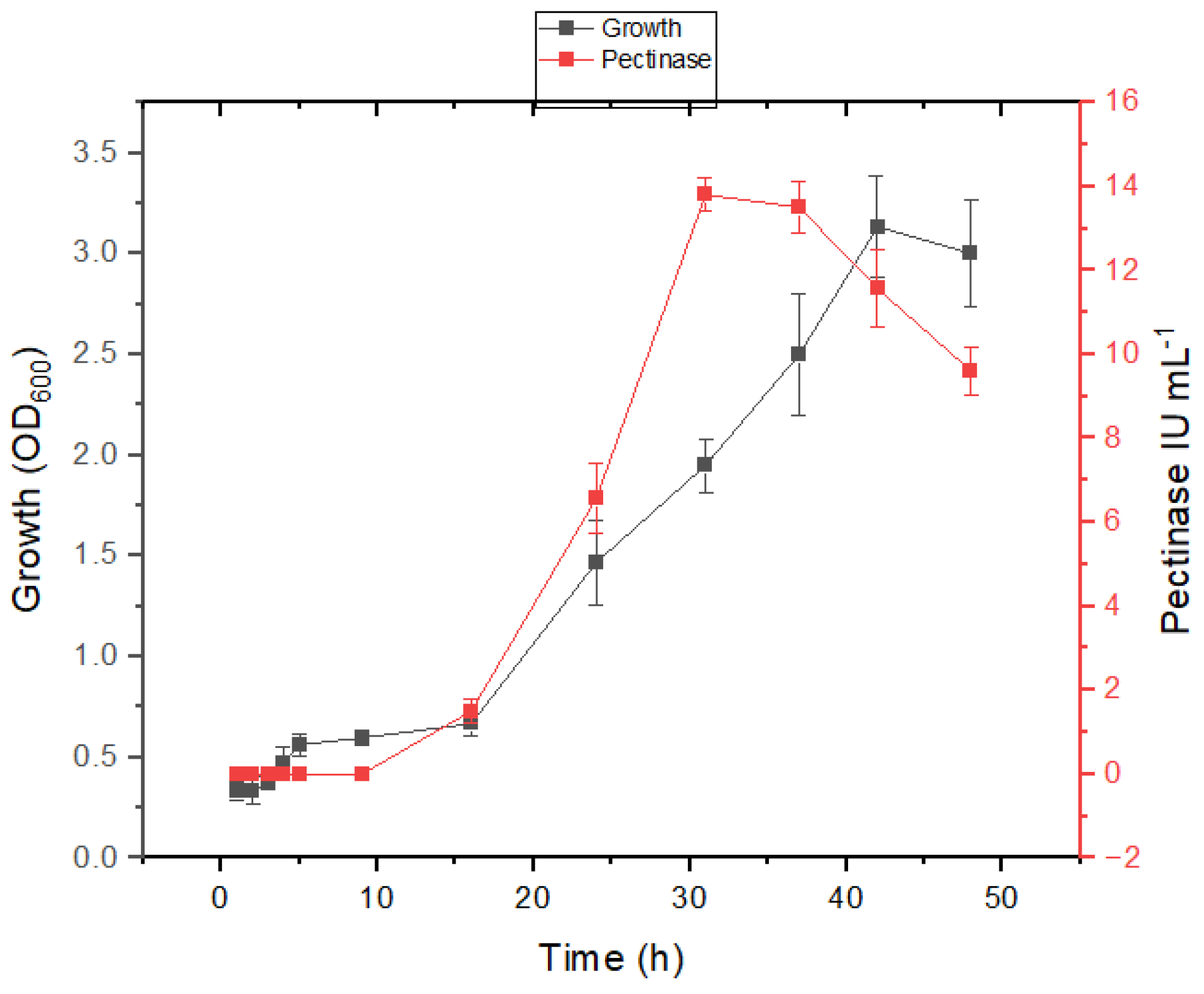
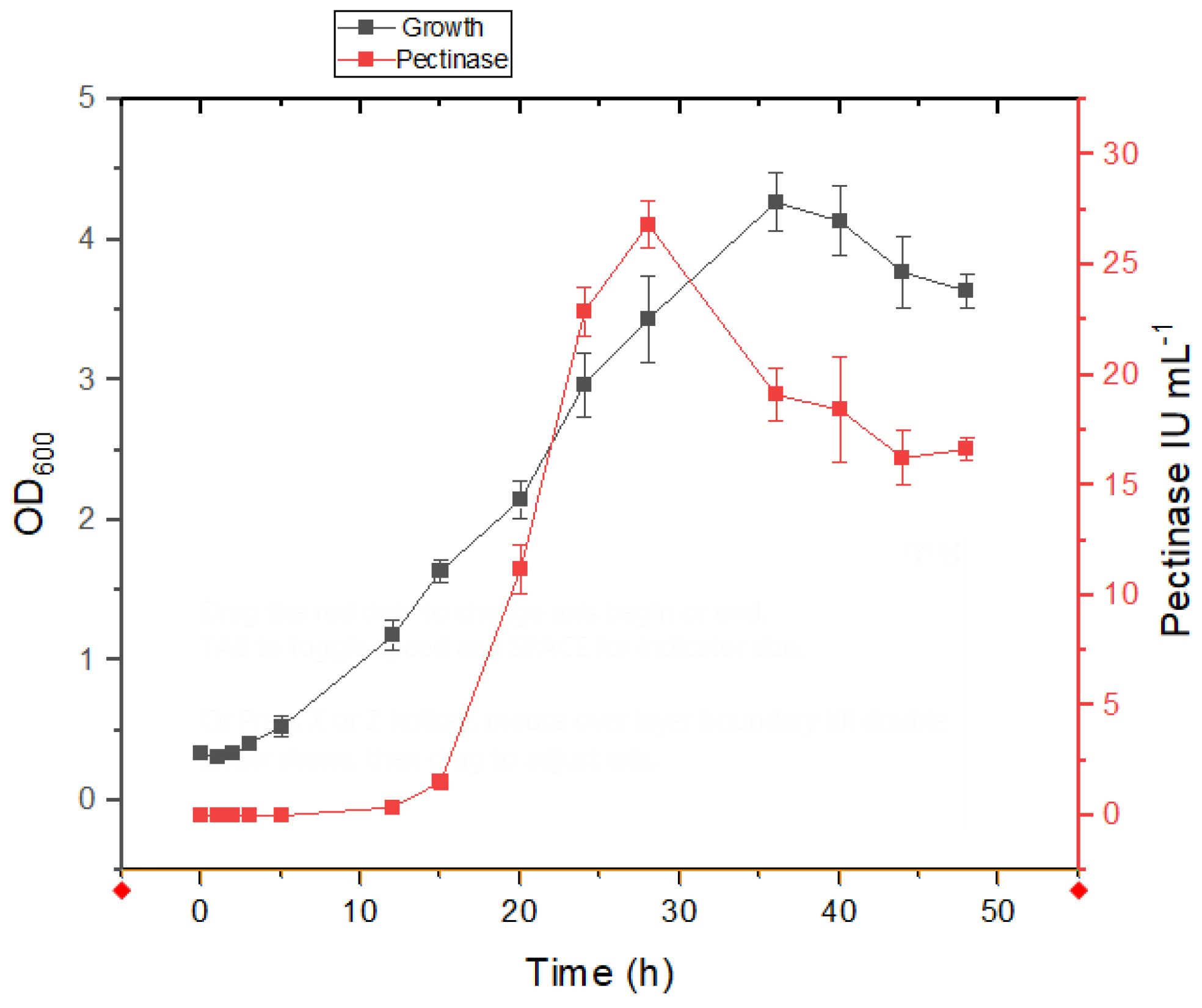
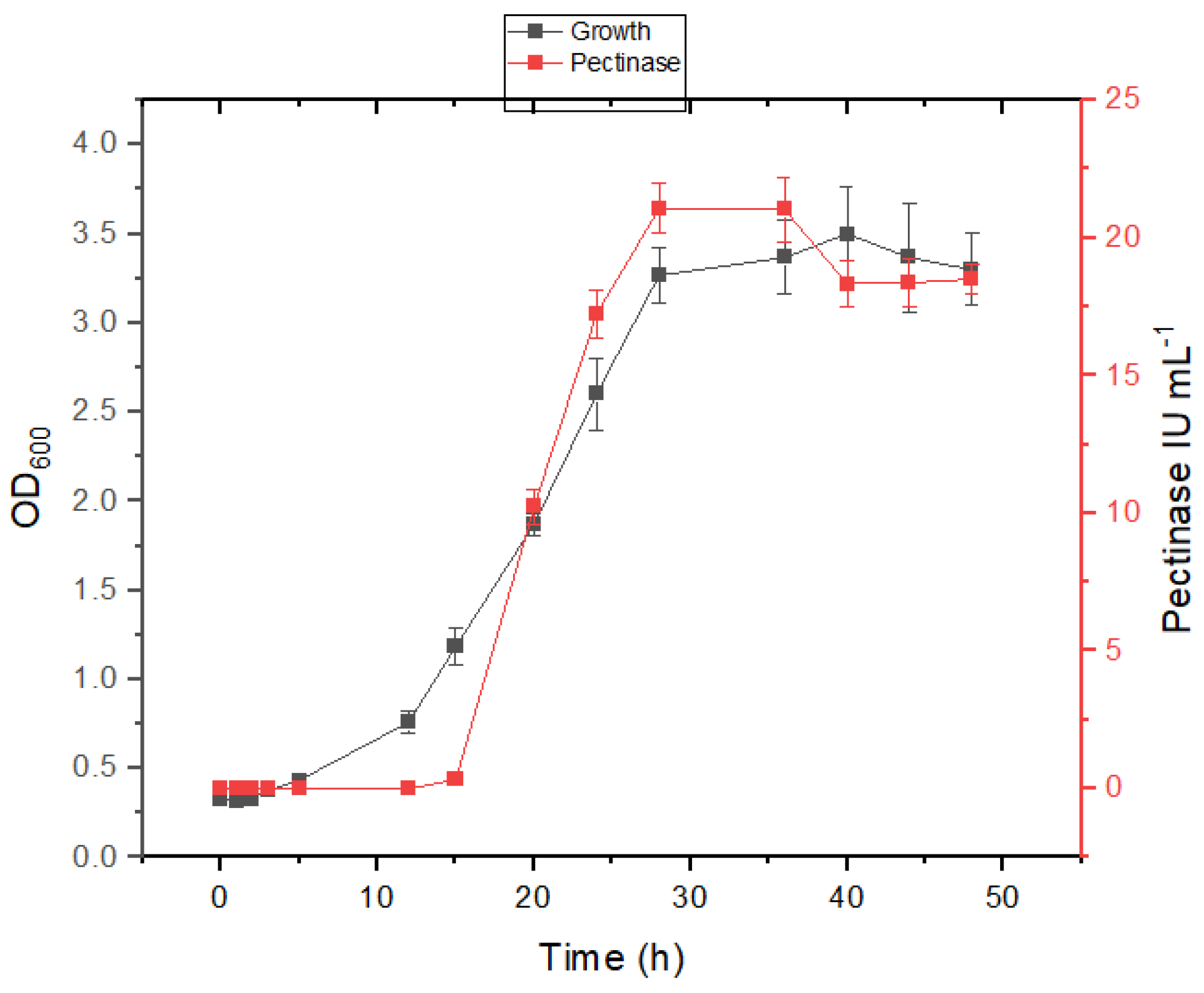
3.3. Growth and Pectinase Production Kinetics in Presence of Salt
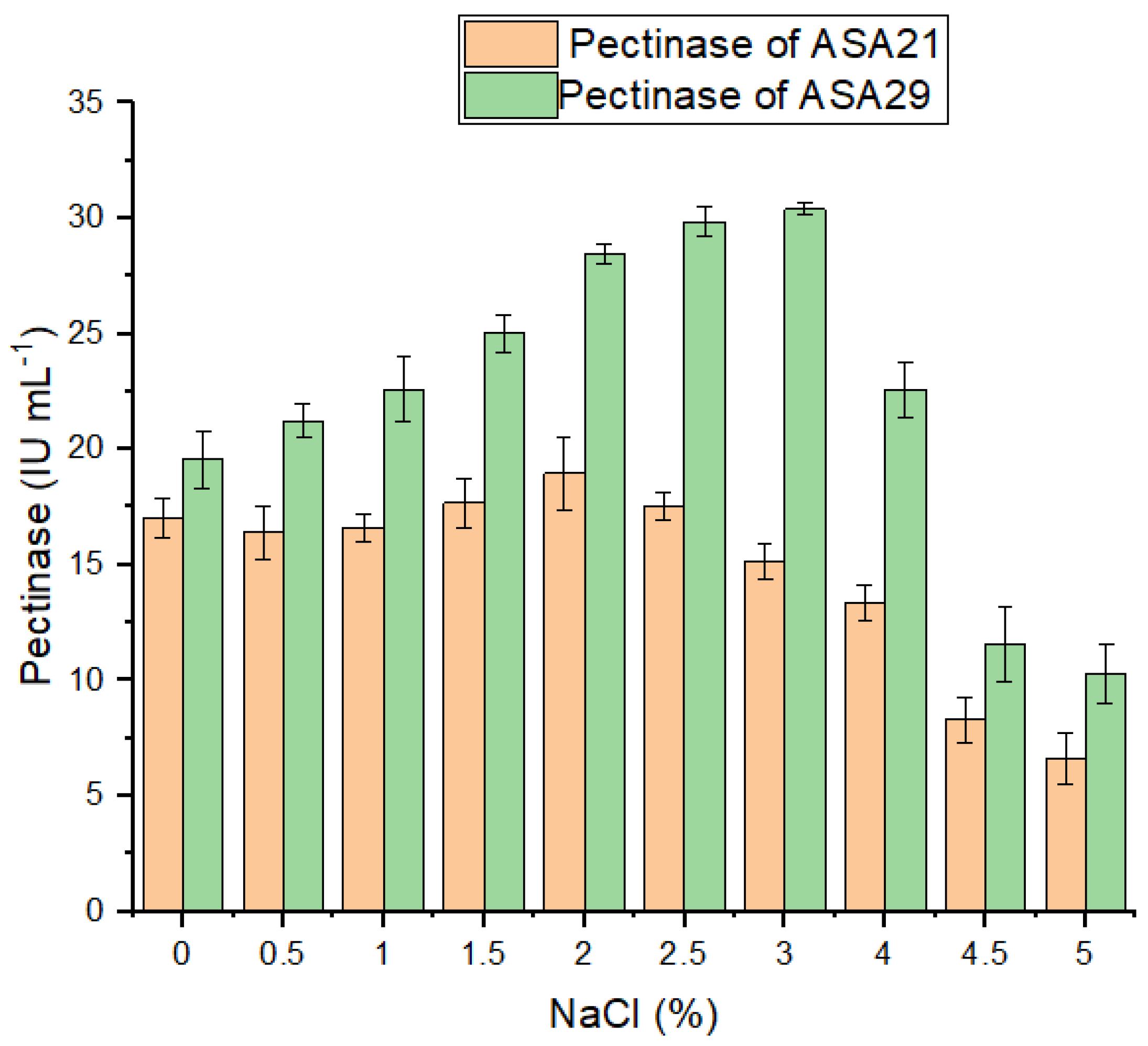
3.4. Effect of Salt Concentration on Pectinase Activity Caused by Strains ASA21 and ASA29
3.5. Clarification of Beetroot, Carrot and Apple Mix Juice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Schulze-Lefert, P. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Huang, X.F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.; Shen, Q.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2014, 8, e55731. [Google Scholar]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Dangl, J.L. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Dangl, J.L. Defining the core Arabidopsis thaliana root microbiome. Nature 2015, 488, 86–90. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2015, 115, E1157–E1165. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Subramaniyan, S.; Prema, P. Biotechnology of microbial xylanases: Enzymology, molecular biology, and application. Crit. Rev. Biotechnol. 2002, 22, 33–64. [Google Scholar] [CrossRef]
- Liu, X.; Yu, L.; Han, J.; Wang, J.; Wang, J. Complete genome sequence of Bacillus paralicheniformis MDJK30, a plant growth-promoting Rhizobacterium with multiple beneficial traits. J. Biotechnol. 2020, 310, 23–27. [Google Scholar]
- Sharma, H.P.; Patel, H.; Sugandha. Enzymatic added extraction and clarification of fruit juices—A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 1215–1227. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Chattopadhyay, P.; Pan, D.; Patra, S.; Bhattacharyya, P.; Paul, A.K.; Mitra, S. Enhancement of plant growth and suppression of Phytophthora capsici by the rhizosphere bacterium Bacillus paralicheniformis in vitro and in field conditions. Microbiol. Res. 2020, 234, 126422. [Google Scholar]
- Wang, C.; Dong, D.; Wang, H.; Müller, K.; Qin, Y.; Wang, H.; Wu, W. Xylan degradation by a consortium of Cellulomonas, Bacillus, and Aspergillus: Impact on the fate of polychlorinated biphenyls in soil. J. Hazard. Mater. 2020, 388, 121775. [Google Scholar]
- Rybakova, D.; Mancinelli, R.; Wikström, M.; Birch-Jensen, A.S.; Postma, J.; Ehlers, R.U.; Berg, G. The role of Paenibacillus sp. in rhizosphere microbiome functioning. Microb. Ecol. 2017, 74, 947–960. [Google Scholar]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef]
- Raza, W.; Yuan, J.; Ling, N.; Huang, Q.; Shen, Q. Production of volatile organic compounds by an antagonistic strain Paenibacillus polymyxa WR-2 in the presence of root exudates and their antifungal activity against Fusarium oxysporum. Appl. Soil Ecol. 2015, 96, 52–59. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Wang, X.; Fan, L.; Ye, X.; Li, Z.; Cui, Z.; Huang, Y. Characterization of an acidic pectin methylesterase from Paenibacillus xylanexedens and its application in fruit processing. Protein Expr. Purif. 2021, 179, 105798. [Google Scholar] [CrossRef]
- Sheladiya, P.; Kapadia, C.; Prajapati, V.; Ali El Enshasy, H.; Abd Malek, R.; Marraiki, N.; Zaghloul, N.S.S.; Sayyed, R.Z. Production, statistical optimization, and functional characterization of alkali stable pectate lyase of Paenibacillus lactis PKC5 for use in juice clarification. Sci. Rep 2022, 12, 7564. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, L.; Gao, B.; Wei, W.; Wang, H.; Xie, J. Protopectinase production by Paenibacillus polymyxa Z6 and its application in pectin extraction from apple pomace. Food Process. Preserv. 2017, 42, e13367. [Google Scholar] [CrossRef]
- García, N.M.; Cely, N.M.; Méndez, P.A. Study of the Pectinase Production from Soursop and Cherimoya Pulp for Agro-Industrial Waste Reduction in Colombia. Waste Biomass Valor. 2024, 15, 6357–6365. [Google Scholar] [CrossRef]
- Mahto, R.B.; Yadav, M.; Muthuraj, M.; Sharma, A.K.; Bhunia, B. Biochemical properties and application of a novel pectinase from a mutant strain of Bacillus subtilis. Biomass Conv. Bioref. 2023, 13, 10463–10474. [Google Scholar] [CrossRef]
- Battisti, J.A.; Rocha, G.B.; Rasbold, L.M.; Delai, V.M.; Costa, M.S.S.M.; Kadowaki, M.K.; da Conceição Silva, J.L.; Simão, R.C.G.; Bifano, T.D.; Maller, A. Purification, biochemical characterization, and biotechnological applications of a multifunctional enzyme from the Thermoascus aurantiacus PI3S3 strain. Sci. Rep. 2024, 29, 5037. [Google Scholar] [CrossRef]
- Haile, S.; Ayele, A. Pectinase from Microorganisms and Its Industrial Applications. Sci. World J. 2022, 2022, 1881305. [Google Scholar] [CrossRef]
- Hassan, M.; Ejaz, U.; Rashid, R.; Moin, S.F.; Gulzar, S.; Sohail, M.; Hassan, K.A.; Alswat, A.S.; El-Bahy, Z.M. Utilization of wild Cressa cretica biomass for pectinase production from a halo-thermotolerant bacterium. Biotechnol. J. 2023, 18, 2200477. [Google Scholar] [CrossRef] [PubMed]
- Aslam, F.; Ansari, A.; Aman, A.; Baloch, G.; Nisar, G.; Baloch, A.H.; Rehman, H.U. Production of commercially important enzymes from Bacillus licheniformis KIBGE-IB3 using date fruit wastes as substrate. J. Genet. Eng. Biotechnol. 2020, 18, 46. [Google Scholar] [CrossRef]
- Tapre, A.R.; Jain, R.K. Pectinases: Enzymes for fruit processing industry. Int. Food Res. J. 2014, 21, 447–453. [Google Scholar]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Nguyen, C.L.; Nguyen, H.V.H. The quality of mulberry juice as affected by enzyme treatments. Beverages 2018, 4, 41. [Google Scholar] [CrossRef]
- Tirry, N.; Kouchou, A.; Laghmari, G.; Lemjereb, M.; Hnadi, H.; Amrani, K.; Bahafid, W.; El Ghachtouli, N. Improved salinity tolerance of Medicago sativa and soil enzyme activities by PGPR. Biocatal. Agric. Biotechnol. 2021, 31, 101914. [Google Scholar] [CrossRef]
- Du, Y.; Ma, J.; Yin, Z.; Liu, K.; Yao, G.; Xu, W.; Fan, L.; Du, B.; Ding, Y.; Wang, C. Comparative genomic analysis of Bacillus paralicheniformis MDJK30 with its closely related species reveals an evolutionary relationship between B. paralicheniformis and B. licheniformis. BMC Genom. 2019, 20, 283. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.; Li-Qun, Z.; Zeng, Q.; Atiq, M.; Ahmad, K.; Tariq, A.; Al-Ansari, N.; Blom, J.; Fenske, L.; Alodaini, H.A.; et al. Comprehensive genomic analysis of Bacillus paralicheniformis strain BP9, pan-genomic and genetic basis of biocontrol mechanism. Comput. Struct. Biotechnol. J. 2023, 3, 4647–4662. [Google Scholar] [CrossRef]
- Świątczak, J.; Kalwasińska, A.; Szabó, A.; Brzezinska, M.S. The effect of seed bacterization with Bacillus paralicheniformis 2R5 on bacterial and fungal communities in the canola rhizosphere. Microbiol. Res. 2023, 275, 127448. [Google Scholar] [CrossRef]
- Ngom, S.I.; Maski, S.; Rached, B.; Chouati, T.; Oliveira Correia, L.; Juste, C.; Meylheuc, T.; Henrissat, B.; El Fahime, E.; Amar, M.; et al. Exploring the hemicellulolytic properties and safety of Bacillus paralicheniformis as stepping stone in the use of new fibrolytic beneficial microbes. Sci. Rep. 2023, 13, 22785. [Google Scholar] [CrossRef]
- Rahman, M.S.; Choi, Y.S.; Kim, Y.K.; Park, C.; Yoo, J.C. Production of Novel Polygalacturonase from Bacillus paralicheniformis CBS32 and Application to Depolymerization of Ramie Fiber. Polymers 2019, 11, 1525. [Google Scholar] [CrossRef]
- da Silva, K.R.A.; Salles, J.F.; Seldin, L.; van Elsas, J.D. Application of a novel Paenibacillus-specific PCR-DGGE method and sequence analysis to assess the diversity of Paenibacillus spp. in the maize rhizosphere. J. Microbiol. Methods 2003, 54, 213–231. [Google Scholar] [CrossRef]
- Wang, L.Y.; Li, J.; Li, Q.X.; Chen, S.F. Paenibacillus beijingensis sp. nov., a nitrogen-fixing species isolated from wheat rhizosphere soil. Antonie Leeuwenhoek 2013, 104, 675–683. [Google Scholar] [CrossRef] [PubMed]
- von der Weid, I.; Duarte, G.F.; van Elsas, J.D.; Seldin, L. Paenibacillus brasilensis sp. nov., a novel nitrogen-fixing species isolated from the maize rhizosphere in Brazil. Int. J. Syst. Evol. Microbiol. 2002, 52, 2147–2153. [Google Scholar] [CrossRef]
- Giacobbe, S.; Olimpia, P.; Ventorino, V.; Birolo, L.; Vinciguerra, R.; Faraco, V. Identification and characterization of a pectinolytic enzyme from Paenibacillus xylanolyticus. Bioresources 2014, 9, 4873–4887. [Google Scholar] [CrossRef]
- El-Sayed, M.H.; Elsehemy, I.A.E. Paenibacillus sp. strain NBR–10, A Thermophilic Soil-Isolated Bacterium with Thermo-Alkali Stable Pectinase Activity. J. Appl. Environ. Biol. Sci. 2017, 7, 9–19. [Google Scholar]
- Abdelgalil, S.A.; Soliman, N.A.; Abo-Zaid, G.A.; Abdel-Fattah, Y.R. Dynamic consolidated bioprocessing for innovative lab-scale production of bacterial alkaline phosphatase from Bacillus paralicheniformis strain APSO. Sci. Rep. 2021, 11, 6071. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Duan, Y.; Wang, Q.; Cong, X.; He, Y.; Gao, C.; Hafeez, M.; Jan, S.; Rasheed, S.M.; et al. Enhancing the Activity of Carboxymethyl Cellulase Enzyme Using Highly Stable Selenium Nanoparticles Biosynthesized by Bacillus paralicheniformis Y4. Molecules 2022, 27, 4585. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, H.; Wu, Y.; Zeng, J.; Guo, Z.; Wang, L.; Shen, C.; Qiao, D.; Cao, Y. A new cold-adapted, alkali-stable and highly salt-tolerant esterase from Bacillus licheniformis. Int. J. Biol. Macromol. 2018, 111, 1183–1193. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, L.; Jespersen, L.; Nielsen, D.S.; Lametsch, R. The role of Bacillus strains and growth medium in shaping γ-glutamyl peptide production. Food Biosci. 2024, 62, 105103. [Google Scholar] [CrossRef]
- Gummadi, S.N.; Kumar, S.; Aneesh, C.N.A. Effect of Salts on Growth and Pectinase Production by Halotolerant Yeast, Debaryomyces nepalensis NCYC 3413. Curr. Microbiol. 2007, 54, 472–476. [Google Scholar] [CrossRef]
- de Paula, N.M.; da Silva, K.; Brugnari, T.; Haminiuk, C.W.I.; Maciel, G.M. Biotechnological potential of fungi from a mangrove ecosystem: Enzymes, salt tolerance and decolorization of a real textile effluent. Microbiol. Res. 2022, 254, 126899. [Google Scholar] [CrossRef]
- John, J.; Surendranathan, K.K.K.; Smith, M.L.; Rahman, P.K.S.M.; Chellam, P.V. Advances in upstream and downstream strategies of pectinase bioprocessing: A review. Int. J. Biol. Macromol. 2020, 162, 1086–1099. [Google Scholar] [CrossRef]
- Gundala, P.B.; Chinthala, P. Extremophilic Pectinases. In Extremophilic Enzymatic Processing of Lignocellulosic Feedstocks to Bioenergy; Sani, R., Krishnaraj, R., Eds.; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Vasconcellos, J.; Conte-Junior, C.; Silva, D.; Pierucci, A.P.; Paschoalin, V.; Alvares, T.S. Comparison of Total Antioxidant Potential, and Total Phenolic, Nitrate, Sugar, and Organic Acid Contents in Beetroot Juice, Chips, Powder, and Cooked Beetroot. Food Sci. Biotechnol. 2016, 25, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Vanga, S.K.; Raghavan, V. High-intensity ultrasound processing of kiwifruit juice: Effects on the ascorbic acid, total phenolics, flavonoids and antioxidant capacity. LWT 2019, 107, 299–307. [Google Scholar] [CrossRef]
- Yasaminshirazi, K.; Hartung, J.; Fleck, M.; Graeff-Hoenninger, S. Bioactive Compounds and Total Sugar Contents of Different Open-Pollinated Beetroot Genotypes Grown Organically. Molecules 2020, 25, 4884. [Google Scholar] [CrossRef] [PubMed]
- Kalaycıoğlu, Z.; Erim, F.B. Total phenolic contents, antioxidant activities, and bioactive ingredients of juices from pomegranate cultivars worldwide. Food Chem. 2017, 221, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Lachowicz, S.; Oszmiański, J. The influence of addition of cranberrybush juice to pear juice on chemical composition and antioxidant properties. J. Food Sci. Technol. 2018, 55, 3399–3407. [Google Scholar] [CrossRef]
| Isolate | Pectinase Production (IU mL−1) in Mineral Salt Medium Containing NaCl (%) | |||||
|---|---|---|---|---|---|---|
| 0 | 1.0 | 1.5 | 2.0 | 2.5 | 3.0 | |
| ASA3 | 3.7 (±0.21) | 4.1 (±0.18) | 1.8 (±0.11) | 0.6 (±0.47) | 0 (±0) | 0 (±0) |
| ASA8 | 7.8 (±0.42) | 6.5 (±0.09) | 7.1 (±0.68) | 2.8 (±0.54) | 1.2 (±0.21) | 0 (±0) |
| ASA9 | 1.2 (±0.27) | 0.7 (±0.06) | 0.5 (±0.01) | 0 (±0) | 0 (±0) | 0 (±0) |
| ASA14 | 6.1 (±0.31) | 5.8 (±0.47) | 2.1 (±0.08) | 0.6 (±0.11) | 0 (±0) | 0 (±0) |
| ASA19 | 2.1 (±0.17) | 4.1 (±0.19) | 3.9 (±0.26) | 1.2 (±0.19) | 0.9 (±0.10) | 0.5 (±0.03) |
| ASA20 | 5.3 (±0.22) | 3.1 (±0.25) | 2.5 (±0.38) | 0.6 (±0.17) | 0 (±0) | 0 (±0) |
| ASA21 | 11.8 (±0.86) | 12.1 (±1.01) | 12.6 (±0.87) | 11.9 (±0.90) | 13.3 (±0.71) | 8.2 (±0.76) |
| ASA22 | 9.1 (±0.98) | 8.5 (±0.77) | 8.2 (±1.1) | 9.2 (±0.76) | 6.3 (±0.78) | 2.4 (±0.55) |
| ASA24 | 6.1 (±0.91) | 5.1 (±0.28) | 3.2 (±0.41) | 3.1 (±0.78) | 2.7 (±0.01) | 1.1 (±0.12) |
| ASA26 | 1.1 (±0.12) | 1.8 (±0.17) | 2.1 (±0.29) | 2.4 (±0.34) | 0.6 (±0.47) | 0 (±0) |
| ASA29 | 13.2 (±0.08) | 13.9 (±0.92) | 13.9 (±0.71) | 13.5 (±0.77) | 14.1 (±0.85) | 10.2 (±0.11) |
| ASA31 | 4.5 (±0.59) | 4.1 (±0.72) | 4.1 (±0.46) | 3.5 (±0.13) | 3.1 (±0.31) | 1.7 (±0.19) |
| Strain | μ in Mineral Salt Medium (h−1) | Qp (IU L−1 h−1) in the Mineral Salt Medium | ||
|---|---|---|---|---|
| Without NaCl | With 2.5% NaCl | Without NaCl | With 2.5% NaCl | |
| ASA21 | 0.069 (±0.001) | 0.081 (±0.001) | 560 (±35) | 640 (±51) |
| ASA29 | 0.83 (±0.002) | 0.088 (±0.001) | 625 (±45) | 925.9 (±32) |
| Pectinase Units | Time Duration (min) | Juice Yield (%) | Total Soluble Solids (°Bx) | TPC (mgGAE/100mL) | ABTS (IC50) | DPPH (IC50) |
|---|---|---|---|---|---|---|
| 10 | 120 | 43.8 (±1.5) | 12 (±1.01) | 31.6 (±2.7) | 40 (±3.1) | 50 (±2.8) |
| 10 | 180 | 81.4 (±7.1) | 18 (±1.0) | 42.3 (±2.7) | 60 (±3.1) | 55 (±4.1) |
| 20 | 120 | 82.3 (±7.3) | 20 (±1.9) | 43.9 (±1.9) | 70 (±3.5) | 60 (±2.8) |
| 20 | 180 | 81.1 (±6.0) | 19 (±1.7) | 44.8 (±3.7) | 80 (±5.0) | 60 (±3.5) |
| 0 | 120 | 42.5 (±2.7) | 8 (±0.9) | 21.3 (±1.1) | 35 (±1.8) | 38 (±1.4) |
| 0 | 180 | 43.8 (±3.5) | 9 (±0.8) | 22.7 (±1.8) | 37 (±2.6) | 38 (±1.9) |
| Pectinase Units | Time Duration (min) | Juice Yield (%) | Total Soluble Solids (°Bx) | TPC (mgGAE/100mL) | ABTS (IC50) | DPPH (IC50) |
|---|---|---|---|---|---|---|
| 10 | 120 | 65.7 (±4.8) | 15 (±0.6) | 45.7 (±2.2) | 70 (±4.8) | 70 (±3.4) |
| 10 | 180 | 81.3 (±7.2) | 24 (±3.5) | 52.9 (±5.8) | 80 (±7.4) | 75 (±4.9) |
| 20 | 120 | 96.4 (±6.1) | 26 (±1.9) | 54.2 (±5.6) | 100 (±0.5) | 100 (±0) |
| 20 | 180 | 97.8 (±3.1) | 27 (±0.8) | 52.9 (±7.1) | 100 (±0) | 90 (±0.2) |
| 0 | 120 | 42.5 (±2.7) | 8 (±0.9) | 21.3 (±1.1) | 35 (±1.8) | 38 (±1.4) |
| 0 | 180 | 43.8 (±3.5) | 9 (±0.8) | 22.7 (±1.8) | 37 (±2.6) | 38 (±1.9) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alswat, A.S.; Alharthy, O.M.; Alzahrani, S.S.; Alhelaify, S.S. Halophilic Pectinase-Producing Bacteria from Arthrocnemum macrostachyum Rhizosphere: Potential for Fruit–Vegetable Juice Processing. Microorganisms 2024, 12, 2162. https://doi.org/10.3390/microorganisms12112162
Alswat AS, Alharthy OM, Alzahrani SS, Alhelaify SS. Halophilic Pectinase-Producing Bacteria from Arthrocnemum macrostachyum Rhizosphere: Potential for Fruit–Vegetable Juice Processing. Microorganisms. 2024; 12(11):2162. https://doi.org/10.3390/microorganisms12112162
Chicago/Turabian StyleAlswat, Amal S., Ohud Muslat Alharthy, Seham Saeed Alzahrani, and Seham Sater Alhelaify. 2024. "Halophilic Pectinase-Producing Bacteria from Arthrocnemum macrostachyum Rhizosphere: Potential for Fruit–Vegetable Juice Processing" Microorganisms 12, no. 11: 2162. https://doi.org/10.3390/microorganisms12112162
APA StyleAlswat, A. S., Alharthy, O. M., Alzahrani, S. S., & Alhelaify, S. S. (2024). Halophilic Pectinase-Producing Bacteria from Arthrocnemum macrostachyum Rhizosphere: Potential for Fruit–Vegetable Juice Processing. Microorganisms, 12(11), 2162. https://doi.org/10.3390/microorganisms12112162






