Abstract
The transition towards a sustainable society involves the utilization of lignocellulosic biomass as a renewable feedstock for materials, fuel, and base chemicals. Lignocellulose consists of cellulose, hemicellulose, and lignin, forming a complex, recalcitrant matrix where efficient enzymatic saccharification is pivotal for accessing its valuable components. This study investigated microbial communities from brackish Lauwersmeer Lake, in The Netherlands, as a potential source of xylan-degrading enzymes. Environmental sediment samples were enriched with wheat arabinoxylan (WAX) and beechwood glucuronoxylan (BEX), with enrichment on WAX showing higher bacterial growth and complete xylan degradation compared to BEX. Metagenomic sequencing revealed communities consisting almost entirely of bacteria (>99%) and substantial shifts in composition during the enrichment. The first generation of seven-day enrichments on both xylans led to a high accumulation of Gammaproteobacteria (49% WAX, 84% BEX), which were largely replaced by Alphaproteobacteria (42% WAX, 69% BEX) in the fourth generation. Analysis of the protein function within the sequenced genomes showed elevated levels of genes associated with the carbohydrate catabolic process, specifically targeting arabinose, xylose, and xylan, indicating an adaptation to the primary monosaccharides present in the carbon source. The data open up the possibility of discovering novel xylan-degrading proteins from other sources aside from the thoroughly studied Bacteroidota.
1. Introduction
To move towards a more sustainable society, it is important to harness the potential of lignocellulosic biomasses as feedstock for materials and fuel production as an alternative to petrol-based sources. Lignocellulosic biomass is generated as a byproduct from agricultural and industrial processes and is the focus of second-generation biorefineries as a more ecological and non-food competitive counterpart to the sugar-rich first-generation biorefineries [1,2]. Its constituents, cellulose, hemicellulose, and lignin, are present in different proportions depending on the type of biomass and species and may even vary throughout the year [3,4].
Cellulose is the most abundant component in the plant cell wall, accounting for 25–45 w/w%. This polysaccharide is composed of a regular structure made up of (1,4)-β-D-glucosyl units that form microfibrils via hydrogen bonds and van der Waals forces. Additionally, several inter- and intra-molecular forces from the microfibrils generate a crystalline macromolecular structure [5,6], which is more resistant to enzymatic saccharification and protects the plants from bacterial degradation [7].
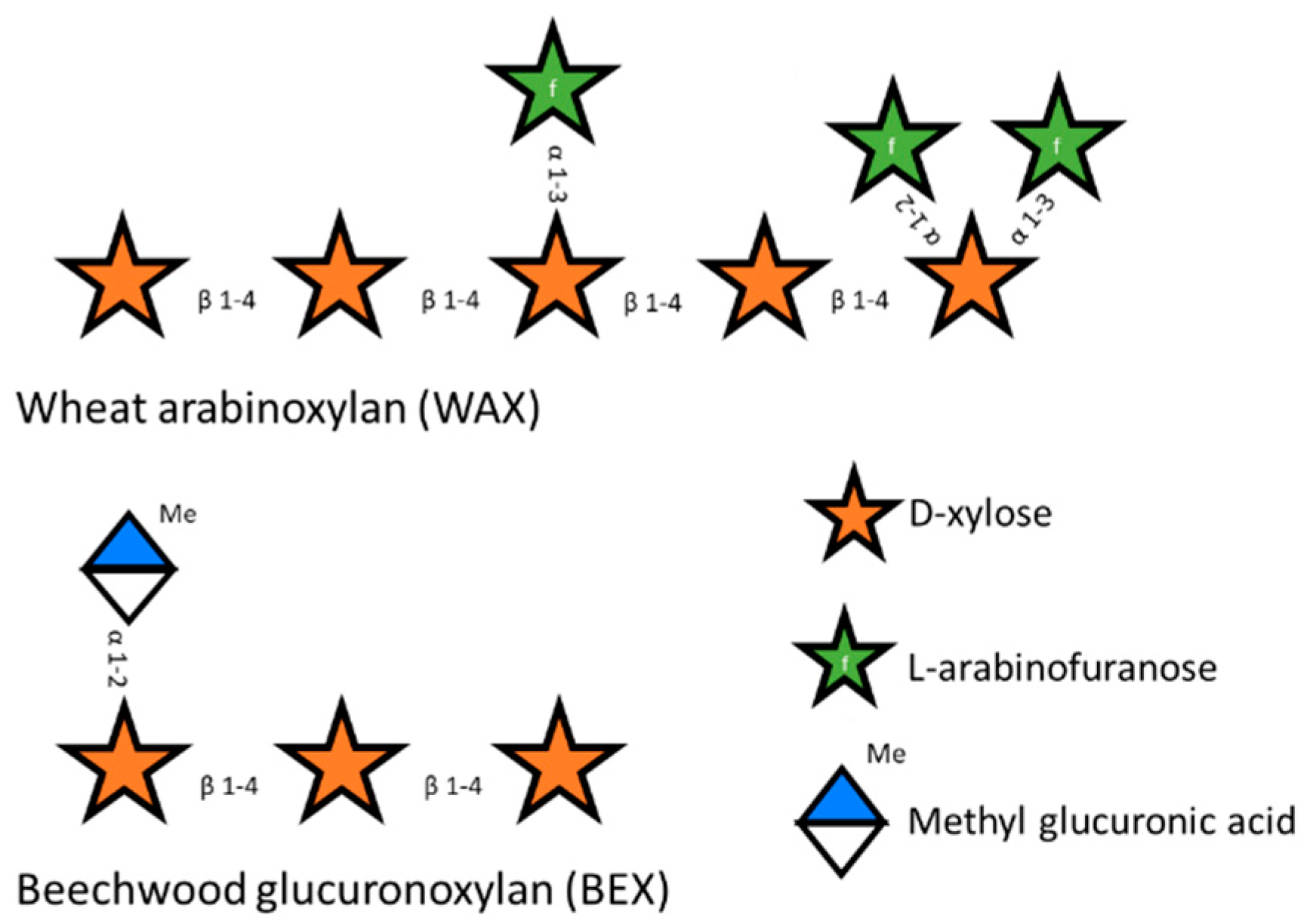
Hemicelluloses are more variable than cellulose and, depending on their plant source, can be present as mannan, xyloglucan, xylan, and glucomannan [8]. The most abundant type of hemicellulose is xylan, consisting of xylose residues as a β-1-4 linked backbone that are partly substituted with different groups through various linkages. The main substituent defines the name of the subtype of xylan. Glucoronoxylan is the simplest xylan polysaccharide. It is mostly present in softwoods like birchwood and is characterized by the substitution of the backbone with glucuronic acid and methyl glucuronic acid [9]. Arabinoxylan is mainly substituted by arabinose at the O2 position or at both the O2 and the O3 position and is common in some plants of the Poaceae family. Glucuronoarabinoxylan (Figure 1) is a combination of both glucoronoxylan and arabinoxylan and is substituted with glucuronic acid, methyl glucuronic acid, arabinose, and ferulic acid [6,10,11].
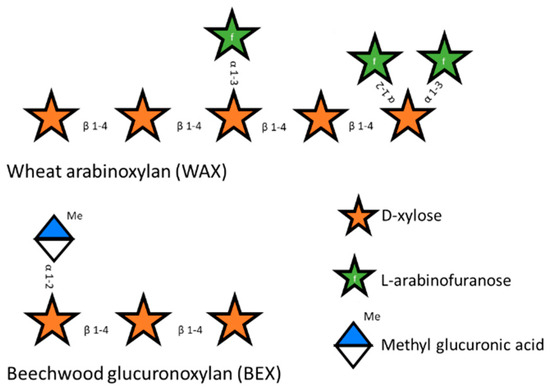
Figure 1.
Schematic representation of wheat arabinoxylan (WAX) and beechwood glucuronoxylan (BEX).
The last main polymer in plant cell walls is lignin. Its biosynthesis involves the oxidative polymerization of phenylpropanoid monomers, primarily coniferyl, sinapyl, and p-coumaryl alcohols, yielding a heterogeneous and irregular macromolecular network that protects the plant from bacterial attacks by recalcitrance against the enzymatic hydrolysis of plant cell walls [12,13,14]. All three main polymers are also inter- and intra-connected, creating a complex and recalcitrant matrix [9,15,16].
In nature, bacteria have the capacity to hydrolyze xylan, and the different substitutions employ distinct selective enzymes [17]. First, endo-xylanases (EC 3.2.1.8) cleave the main backbone to generate smaller oligosaccharides. The generated fragments are then attacked by accessory xylanolytic enzymes, such as α-l-arabinofuranosidases (EC 3.2.1.55), cleaving the arabinose substitutions, α-glucuronidases (EC 3.2.1.139), removing the linkages between glucuronic acid and xylose, and 1,4-β-xylosidases (EC 3.2.1.37), targeting the bonds between xyloses [17].
Several organisms have been described as capable of degrading plant polysaccharides, including fungi and bacterial taxa such as Actinobacteria, Alphaproteobacteria, Sphingobacteria, and Betaproteobacteria [18]. Aquatic microbial communities may contain such species, as some have been described to degrade grasses [19]. In marine studies, the predominant taxa were typically Pseudomonata [20,21]. In a long-term temperate marine study, Alphaproteobacteria was found to be the most abundant class [21], whereas a study on the communities in an artic fjord identified Gammaproteobacteria to be the predominant class in marine sediment and Flavobacteriales, Verrucomicrobia, and Actinobacteriales in water [20].
The degradation of xylan into monomeric sugars has proved to be a key challenge in using lignocellulosic biomass due to its complex structure [22]. To overcome this limitation of xylan saccharification for a more holistic biorefinery, it is important to find new enzymes capable of xylan degradation. This study aimed to elucidate a possible new source for lignocellulose-degrading bacteria that could contain suitable xylan-degrading enzymes. To date, there have been a large number of studies on microbial communities in the sediment and surface water of both fresh and marine water around the world, but brackish water remains largely understudied. Therefore, the Lauwersmeer in the north of The Netherlands was selected for sampling as it is a lake with a unique environment. Located next to the Wadden Sea and fed by several rivers and canals, it receives both salty and fresh water. The existing gradient of salinity is expected to influence the microbial communities in this environment and may provide a wealthy source for novel lignocellulose-degrading bacteria.
2. Material and Methods
2.1. Sampling and Site Description
The sampling was conducted in the Lauwersmeer lake in the nature park of Lauwersmeer (Groningen, The Netherlands 53°22′09.2″ N 6°14′17.9″ E), which is in contact with the Wadden Sea in the north (brackish water) and fed with fresh water by canals in the south. Samples were taken in the south from superficial sediment (<10 cm) using two sterile 500 mL bottles and kept at 4 °C during transport and storage.
2.2. Substrate Preparation
Beechwood glucuronoxylan (BEX) and wheat arabinoxylan (WAX) were purchased from Megazyme Ltd. (Bray, Ireland). Each substrate was obtained from a single batch. 9.7 g/L substrate solutions of WAX and BEX were prepared by solubilizing the xylan in sterile MQ water with 4.2% ethanol for 20 min at 120 °C. The solution was then added to the medium.
2.3. Bacterial Enrichment
The media used for the enrichment was non-reduced Widdel mineral media [23].
The sediment was homogenized and centrifuged at 300 rpm (3 min, 4 °C) to remove soil and vegetation. Then, the bacteria were harvested by centrifugation (10,000 rpm, 15 min, 4 °C) and the cell pellet was washed twice with media before resuspending it in 80 mL media.
For the cultures, 95 mL media (containing 10 g/L xylan) were inoculated with 5 mL cell suspension and incubated in Erlenmeyer flasks for 7 days at 30 °C and shaken at 100 rpm. Three additional generations were grown, which were inoculated with 5 mL of culture from the previous generation. In addition to the closed cultures, open cultures were grown for the first and last generation to conduct daily sampling of 5 mL aliquots.
Both aliquots and cultures were harvested by centrifugation (10,000 rpm, 15 min, 4 °C) and the cell pellet and supernatant were separately flash frozen and stored at −80 °C. For the aliquots, the supernatant was filtered through a 0.22 µm filter before freezing.
During the enrichment, bacterial growth was monitored by measuring the optical density (600 nm) of 100 µL culture in microtiter plates using a spectrophotometer (SpectraMax from Molecular Devices).
All treatments were conducted in duplicates and a negative control (no inoculation) was included in each generation.
2.4. Metagenomic Sequencing
Five samples were selected for metagenomic sequencing, being the original sample and after enrichment on either WAX or BEX (1st and 4th generation, closed cultures). The total community DNA was extracted from these samples using the ZymoBIOMICS DNA Miniprep Kit (Zymo Research; Irvine, CA, USA) following the protocol provided by the manufacturer. The purity and quantity of the extracted DNA was verified with a NanoPhotometer (Implen; Munich, Germany; N50) and 1.5% agarose gel.
Metagenomic sequencing on the purified DNA samples was conducted by Eurofins Genomics (Ebersberg, Germany) with the option of bioinformatic analysis “Advance”. The quality of the obtained raw data was first verified by FastQC (bioinformatics.babraham.ac.uk/projects/fastqc/, Version: 0.11.9). The taxonomic distribution was obtained with Kraken 2 [24] (Version 2.1.2) using the standard reference database (github.com/DerrickWood/kraken2, Standard Database, accessed on 23 June 2023) and the samples were combined using KrakenReportAnalyzer (github.com/gaenssle/Kraken_ReportAnalyzer, accessed on 8 August 2023). The functional analysis was conducted with DIAMOND [25] (Version 2.0.13) and all data were visualized using Stata16 (StataCorp; College Station, TX, USA).
2.5. Gel Permeation Chromatography
The filtered supernatant from the cultured aliquots (1st and 4th generation) were analyzed with gel permeation chromatography (Agilent Technologies 1200 Series, Santa Clara, CA, USA) with two Suprema PSS columns (100 Å and 1000 Å; 8 × 300 mm 10 µm), tempered at 40 °C and equipped with a refractive index detector. The used eluent was 0.05 M NaNO3 and the samples (injection volume 10 µL) were eluted with a flow rate of 1 mg/mL with ethylene glycol as internal standard and a series of 9 pullulan standards (1–708 kDa) as universal standard. Both replicates of the enrichment were analyzed and a representative chromatogram was selected for the graphical representation.
3. Results and Discussion
3.1. Polysaccharide Degradation and Bacterial Growth
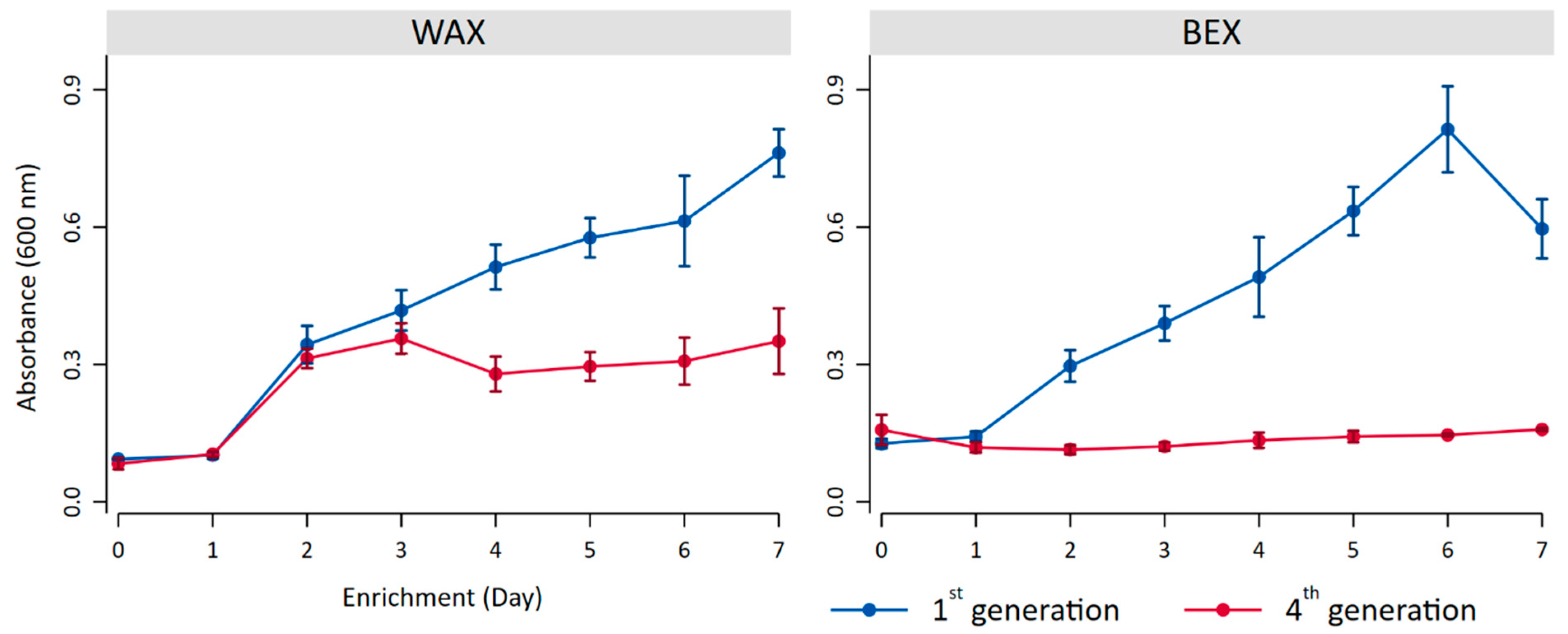
Samples of superficial sediments were collected from a lake with a salinity gradient in The Netherlands and subjected to incubation with two types of xylan: wheat arabinoxylan (WAX) and beechwood glucuronoxylan (BEX). Bacterial growth, estimated through turbidity, polysaccharide degradation, and gel permeation chromatography (GPC), was monitored daily during the first and fourth generations of seven-day enrichment.
During the first generation of enrichment, the bacterial growth exhibited similar patterns for both substrates (Figure 2), demonstrating continuous growth with only a decrease in turbidity for BEX on day 7. It is noteworthy that turbidity served merely as an indicator of growth, as the substrates themselves contribute to turbidity and may fluctuate during degradation processes throughout bacterial growth, potentially explaining the decreased turbidity observed on the final day of BEX incubation.
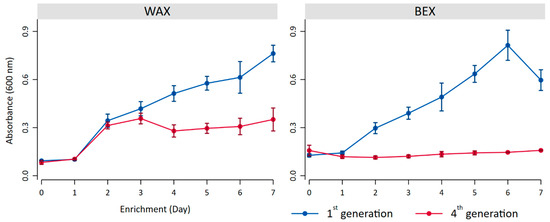
Figure 2.
Bacterial growth indicated by the turbidity at 600 nm with BEX and WEX as substrates during the first generation (blue) and the fourth generation of enrichment (red).
The fourth generation of enrichment on WAX resulted in initial bacterial growth, followed by stabilization on day 4. Incubation with BEX, on the other hand, showed only low levels of growth, indicating a potential preference of the bacterial community for WAX.
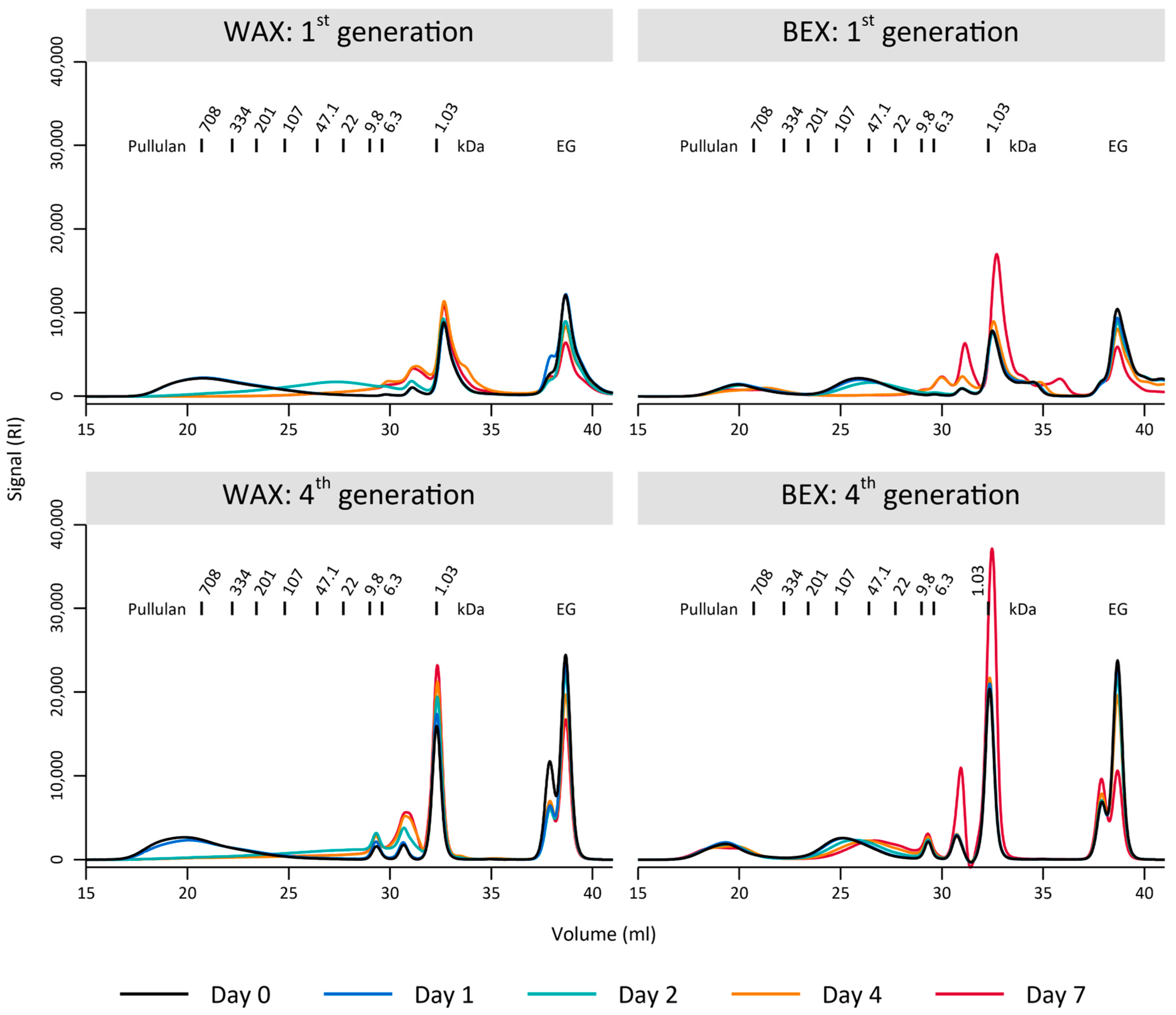
The preference of the environmental sample for the WAX substrate was further evident in the polymer degradation observed through GPC (Figure 3). During the initial generation of enrichment, the bacterial culture completely depolymerized the WAX sample, with a population of partly degraded substrate appearing temporarily at day 2 (Figure 3). In contrast, the bacterial culture was unable to fully degrade the BEX, as indicated by the remaining fraction of the initial xylan population between 708–334 kDa, even after 7 days of growth (Figure 3). These differences were even more pronounced after four-week-long generations of enrichment, where WAX was completely degraded after 7 days, with improved depolymerization compared to the first generation (complete degradation at day 2). In contrast, there was only minimal degradation of BEX, notable by the preservation of the largest substrate population between 708–334 kDa and the very limited decrease in the population around 22 kDa.
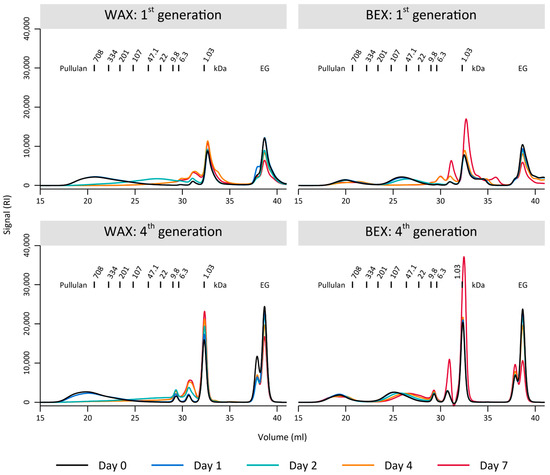
Figure 3.
Gel permeation chromatography of the substrates BEX and WAX on several days during the first and fourth generation of 7-day enrichments. EG marks the internal standard ethylene glycol.
Therefore, it is plausible that the environmental sample, extracted from an area mostly populated by grass-like plants, was better adapted to degrade WAX, which is more prevalent in such plants compared to glucuronoxylan, which is more commonly found in hardwood biomass [26]. Nevertheless, the loss of the initial BEX degradation and growth is surprising. This could indicate that the bacterial culture was negatively impacted during the different enrichment generations. Some loss of activity could be due to substrate preferences or potential errors during the enrichment period, such as substrate solubilization by adding ethanol, even though this step was conducted for both substrates.
Moreover, the bacterial culture growing with WAX as the only carbon source presented an intriguing culture morphology during the growth. The bacteria in the culture were aggregated in granules (Figure S1), showing the possible production of compounds of interest such as exopolysaccharides [23,27].
3.2. Taxonomic Profiling
To study the microbial community responsible for the degradation during the enrichment period, samples were taken after the first generation (WAX1 and BEX1) and fourth generation (WAX4 and BEX4) and subjected to metagenomic sequencing. All four samples resulted in total reads of 14–20 k (Table 1), whereas the original sample (OS) had a lower number of total reads (11 k). Notably, only less than 75% of reads were classified, with under 46% in OS and BEX4. The incomplete classification is most likely due to the reference database used for the identification. The standard database of kraken2 contained about 100 GB of reference genome sequences from Bacteria, Archaea, Viruses, Vectors, and the human genome. As neither the microbial community of the original sample (a brackish lake) nor the enrichment on the substrate (xylan) were studied previously in greater detail, it is plausible that some of the species present in the studied samples were not included in the standard database. Another reason could be the short length of the fragments (150 bp) and the sequencing of the entire genomes. Thus, many fragments could not contain a recognizable marker for the taxonomic analysis.

Table 1.
Taxonomic classification of the metagenomic sequencing data.
The determined Shannon Diversity Index ranged from 4.0 to 5.2, and the Equability Index ranged from 0.44 to 0.58. WAX4 had both the highest diversity and evenness, and BEX1 had the lowest values of all samples.
The OS had the highest number of identified species (9.1 k), which reduced to approx. 8.6 k after the first generation of enrichment and to 7.3 k and 6.2 k after the fourth generation for WAX and BEX, respectively. However, only a fraction (<40) were present, with over 20,000 reads (~1% relative abundance).
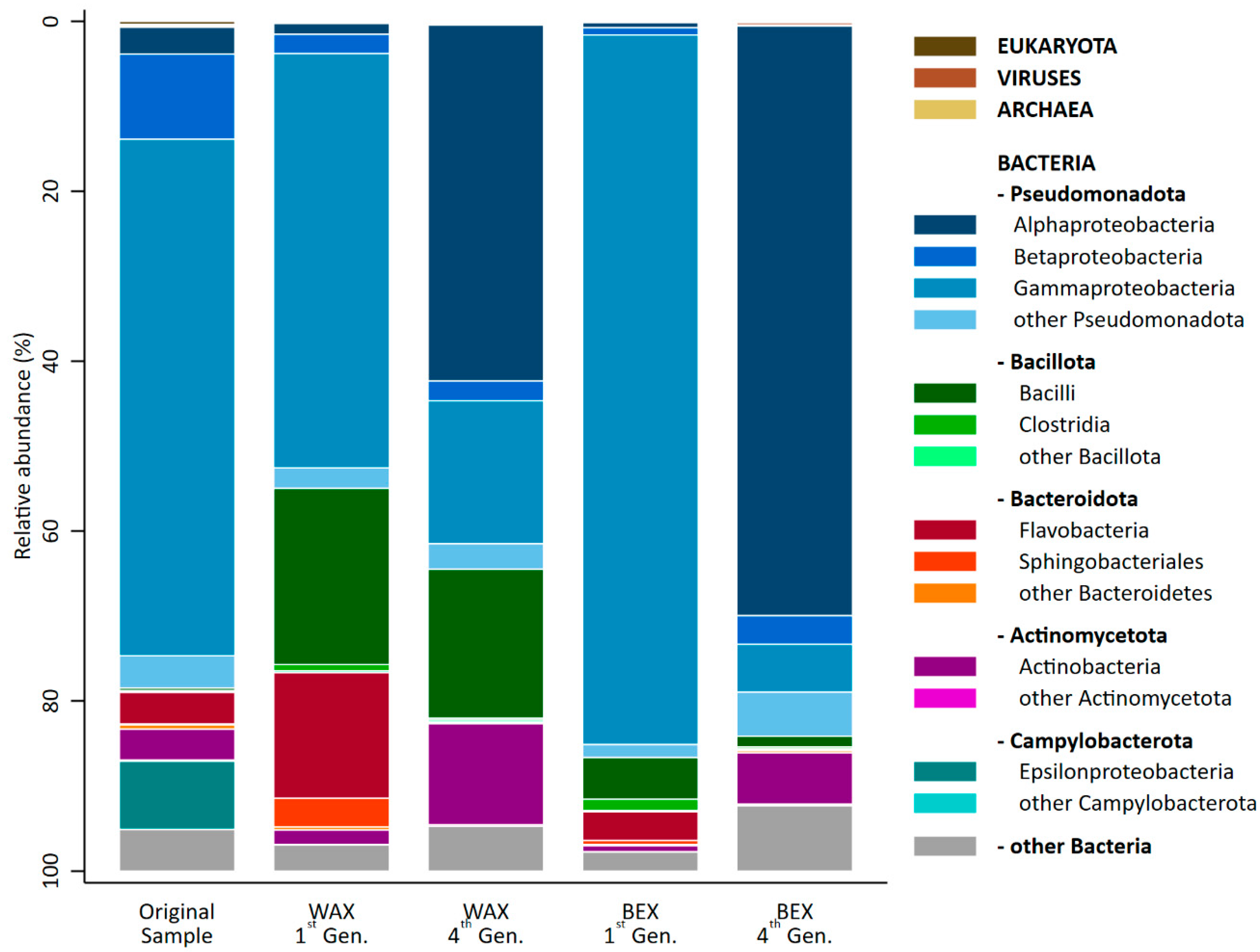
All cultures consisted almost entirely of bacteria (Figure 4), ranging from 99.32% (OS) to 99.86% (BEX1). Additionally, the samples contained traces of Eukaryota (<0.40%), Viruses (<0.30%), and Archaea (<0.20%).
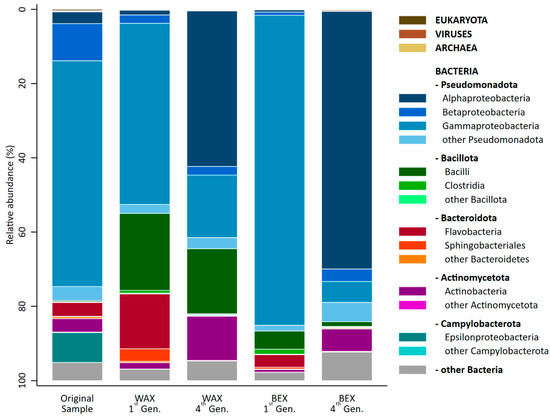
Figure 4.
Taxonomic distribution across domains, phyla, and classes of the original sample and the first and fourth generation of enrichment on WAX and BEX. Taxa with fewer than 20,000 reads in all samples were summarized in the categories labelled “other”.
The predominant phylum in all cultures was found to be Pseudomonata, contributing over half of the relative abundance of the microbial community. Notably, samples enriched on BEX contained the highest amount of Pseudomonata (84–87%), followed by the original sample (78%) and the cultures grown in WAX (55–64%). The second-most dominant phylum in the OS was Campylobacteria, which disappeared almost entirely during the xylan enrichment. Enrichment with WAX led to a substantial increase in Bacillota (22% first generation, 18% fourth generation) and a temporary increase in Bacteroidota (19%) in the first generation, which were replaced by Actinomycetota (12%) in the fourth generation. The same phyla and pattern could also be observed for enrichment on BEX, though it was to a lower degree.
All four detected phyla have been described as capable of degrading biomass [18] and are thus in agreement with previous results. Sediment samples from a lake in Groningen (The Netherlands) as well as seawater samples from the southwest coast of India predominantly consisted of Pseudomonata, Bacteroidota, Bacillota, and Actinomycetota [28]. The seawater from the Indian coast further contained Cyanobacteria and Verrucomicrobia [22]. Incubation of the Dutch lake sediment on pre-treated switch grass favored the enrichment of Pseudomonata and Bacillota, with relative abundances changing substantially over time [28]. Furthermore, enrichment studies of the microbial communities on the biomass of various chipped wood piles resulted in an overwhelming majority (>90%) of the species belonging to Actinomycoetota, Pseudomonata, Bacteroidota, and Bacillota [18].
The most apparent change in the microbial community was the dramatic shift (>40%) in both xylan enrichments from Gammaproteobacteria in the original sample and the samples from the first generation to Alphaproteobacteria in the fourth generation. BEX1 consisted almost entirely of Gammaproteobacteria (84%), which were substantially replaced by Alphaproteobacteria (69%).
Gammaproteobacteria usually thrive in anaerobic conditions, such as the sediments of tidal flooded mangrove forests [29], are involved in carbon fixation [22], and occur in response to marine phytoplankton blooms [30]. These blooms lead to the growth of heterotrophic bacteria, classified as Alpha- and Gammaproteobacteria, and Bacteroidota, which have been speculated to perform a division of labor during the degradation of algal-derived organic substrates. While Bacteroidota have been considered to target compounds of high molecular weight, such as proteins and polysaccharides, Gammaproteobacteria were proposed to primarily act as remineralizers of the same polymers, but with less specialization on polysaccharides. Alphaproteobacteria, on the other hand, were generally observed to prefer organic compounds of small molecular weight, such as ethanol, sugar monomers, and amino acids [30]. Thus, even though fresh substrate was supplied at the beginning of each generation, there might have been an accumulation of organic compounds of small molecular weight, favoring an enrichment of Alphaproteobacteria.
Table 2 lists the 20 species with the highest relative abundance. The Actinomycetota Mycobacterium canetti was present in all samples. The most abundant species in the original sample were two Arcobacter species and Pseudomona fluorescens. All other species were only present in traces. Enrichment on WAX led to the accumulation of several Bacillota, as well as two Cloacibacterium species and several Acinetobacter species. In the fourth generation, almost 8.6% could be attributed to Niallia circulans. Incubation with the substrate BEX led to a similar increase in Acinetobacter species as well as Dickeya. However, in the fourth generation, multiple Acetobater and Komagataeibacter species were detected.

Table 2.
Overall top 20 species with the highest relative abundance.
Pseudomonas has been found in multiple environmental sources, associated with lignin degradation, and observed during xylan enrichment [31]. Bacteria belonging to the genus Bacillus/Niallia have been described to produce high amounts of xylanases and possess significant xylanase activity [32]. Furthermore, Enterobacter, Acinetobacter, Pseudomonas, Flavobacterium, and Stenotrophonas are capable of degrading plant lignin, cellulose, and/or hemicellulose [33]. Therefore, the orders found were in good agreement with previous results on microbial biomass degradation. Half of the orders detected in the microbial flora of termite gut were also found in this study, including all three orders of Bacillota (Bacillales, Lactobacillales, and Clostridiales) and three out of four orders of Gammaproteobacteria (Enterobacterales, Pseudomonadales, and Xanthomonadales) [34].
The high amount of Acetobacter and other members of the Acetobacteraceae family in the fourth generation on BEX was interesting as they are obligate aerobic bacteria adapted to environments rich in sugars and ethanol, such as in the guts of insects with sugar-based diets (e.g., bees and fruitflies) [35]. Analysis of the gut microbiome of the fruitfly Drosophila melongaster resulted in a very low diversity, with just five species accounting for 97% of the relative abundance. All five species belonged to just two genera, Acetobacter and Lactobacillus [36]. Furthermore, the most prominent species (Acetobacter papayae, Acetobacter okinawensis) have been reported to grow on D-xlyose [37]. Hence, the significant reduction in microbial diversity observed in BEX enrichment, characterized by the selective enrichment of bacteria acclimated to ethanol, notably Acetobacter, alongside diminished growth and polymer degradation, suggests a potential loss of enrichment likely attributable to elevated ethanol levels within the BEX enrichment.
3.3. Enrichment of Genes Associated with Xylan Degradation and Catabolism
KEGG (kegg.jp) is a database that assigns unique IDs to all of the genomes and genes it includes. Additionally, it assigns IDs to the enzymes encoded in the genes. This enabled an estimation of the occurrence of specific enzymes in specific taxa. Table 3 summarizes the relative occurrence of xylan-specific enzymes present in the genomes of the phyla that were detected in this study (Figure 4).

Table 3.
Relative occurrence of xylan-specific enzymes in genomes (KEGG database) of relevant phyla.
The three dominant phyla in the original sample were found to only exhibit a low occurrence of the target enzymes. For Campylobacteria, there were no hits for any of the listed enzymes in any genome. This phylum disappeared entirely from the enriched samples. Betaprotebacteria and Gammaproteobacteria, which also only had genomes low on xylan enzymes, decreased in abundance during the enrichment as well. These phyla were replaced by others generally rich in xylan-specific genes. Almost half the genomes of Bacillota were found to contain a xylanase and/or an arabinofurosidase. Genomes classified as Bacteroidota included, on average, one xylosidase, one glycosylceramidase, and several others to a lesser degree. Actinomycetota, found in both enrichments in the fourth generation, had the highest general occurrence of xylanases.
Although the data in Table 3 were not specific for the species found in the samples, the trend towards phyla generally rich in xylan specific genes supported the experimentally obtained results.
A significant portion of carbohydrate-active enzymes (CAZYmes) were identified in bacteria associated with the gut microbiota of various animals, as highlighted by Wardman et al., 2022 [38], with a particular focus on research and identification within the bacterial phylum Bacteroidota [39]. The enrichment of xylan-degrading bacteria from other phyla, such as Pseudomonata, presents a promising opportunity to uncover novel xylan-degrading enzymes.
The hypothesis that the enriched cultures have expressed a sophisticated enzymatic toolkit for the degradation of WAX was supported by the observed increase in genes dedicated to carbohydrate transport and metabolism during the enrichment process (Figure S2). Table 4 highlights the most significant GO (gene orthology) term assignments [40,41], which have been enhanced by over 2000 reads during the enrichment (Table S1). These data demonstrate how the bacterial community, enriched with WAX, has accumulated a higher number of genes associated with the carbohydrate catabolic process (GO:0016052), specifically targeting arabinose (GO:0019568) and xylose/xylan (GO:0045493) catabolic processes. The increased prevalence of these specific catabolic pathways indicates an adaptation to the primary monosaccharides present in the carbon source.

Table 4.
Most relevant GO terms of which gene occurrence has been increased by at least 2000 reads during the enrichment of the environmental sample with WAX as substrate.
Despite the prominence of genes related to arabinose and xylose catabolism, other carbohydrate catabolic processes have also been enhanced, including those for cellulose (GO:0030245) and arabinan (GO:0031222). Furthermore, the enrichment has led to a significant accumulation of genes associated with specific enzymatic activities, such as xylan 1,4-beta-xylosidase (GO:0009044) and endo-1,4-beta-xylanase (GO:0031176). In addition to hydrolytic activities, there has also been an increase in genes related to carbohydrate binding (GO:0030246).
The analysis of GO terms provides further insights into the predominant degradation and transport mechanisms within the environmental sample. As shown in Table 4, several GO terms are associated with carbohydrate transport (GO:0008643) via ATP-binding cassette (ABC) transporters, including ABC-type D-ribose transporter activity (GO:0015611) and ABC-type L-arabinose transporter activity (GO:0015612). This observation is consistent with previous reports indicating that, in non-Bacteroidota species, ABC transporters are the most common transport mechanism [42,43].
Overall the increased occurrence of xylan-degrading genes associated with the enriched phyla (Table 3), as well as the high enhancement of those in our specific enrichments (Table 4), showcases the adaptation of the enriched culture to use WAX as a carbon source, showing a promising source of non-bacteroidetes carbohydrate-active enzymes.
4. Conclusions
In this study, we presented an enrichment of a bacterial community derived from the superficial sediments of a lake characterized by a salinity gradient and an influx of plant material. The environmental bacterial community showed a pronounced and swift adaptation to the substrate, wheat arabinoxylan (WAX), from the onset of the enrichment process. In contrast, beechwood glucuronoxylan (BEX) degradation was nearly lost after four enrichment generations, while WAX degradation was notably enhanced. This trend was further supported by the observed loss of microbial diversity, with the enrichment predominantly favoring Alphaproteobacteria in BEX, while a more diverse and robust culture, including Gammaproteobacteria and Bacillota, was enriched with WAX. Despite the evident bacterial degradation of WAX, supported by optical density (OD), gel permeation chromatography (GPC), and metagenomics analysis, the enriched phyla exhibited only a limited number of known xylan-degrading enzymes, underscoring the potential of the enriched culture as a promising source of novel xylan-degrading enzymes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12081715/s1, Figure S1: Image of the 7th day of the 4th generation of the culture with WAX as the only carbon source; Figure S2. COG assignment of proteins classified with DIAMOND Table S1: All the GO terms that have been overrepresented more than 2000 reads during the 4 weeks of enrichment with WAX.
Author Contributions
A.L.O.G. was involved in conceptualization and writing the original draft and conducted the metagenomic sequencing, as well as subsequent and related analysis. S.B.-L. initiated the research, gathered the environmental samples, conducted the enrichment, and was involved in the conceptualization, writing the original draft, and analyzing the data. P.J.D. and E.J. were both involved in the conceptualization process, the critical reviewing of the manuscript, funding acquisition, and supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
All described data is presented in the manuscript and supplementary materials.
Acknowledgments
We thank Anne de Jong for his introduction and guidance for analyzing the metagenomic sequencing data.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- de Jong, E.; Higson, A.; Walsh, P.; Wellisch, M. Bio-Based Chemicals: Value Added Products from Biorefinerie. IEA Bioenergy—Task 42 Biorefinery. 2012. Available online: https://www.nnfcc.co.uk/publications/biorefinery-biobased-chemicals (accessed on 1 July 2024).
- Ginni, G.; Kavitha, S.; Yukesh Kannah, R.; Bhatia, S.K.; Adish Kumar, S.; Rajkumar, M.; Kumar, G.; Pugazhendhi, A.; Chi, N.T.L.; Rajesh Banu, J. Valorization of Agricultural Residues: Different Biorefinery Routes. J. Environ. Chem. Eng. 2021, 9, 105435. [Google Scholar] [CrossRef]
- Himmel, M.E.; Ding, S.-Y.; Johnson, D.K.; Adney, W.S.; Nimlos, M.R.; Brady, J.W.; Foust, T.D. Biomass Recalcitrance: Engineering Plants and Enzymes for Biofuels Production. Science 2007, 315, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Okolie, J.A.; Nanda, S.; Dalai, A.K.; Kozinski, J.A. Chemistry and Specialty Industrial Applications of Lignocellulosic Biomass. Waste Biomass Valorization 2021, 12, 2145–2169. [Google Scholar] [CrossRef]
- Arantes, V.; Saddler, J.N. Cellulose Accessibility Limits the Effectiveness of Minimum Cellulase Loading on the Efficient Hydrolysis of Pretreated Lignocellulosic Substrates. Biotechnol. Biofuels 2011, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Bichot, A.; Delgenès, J.P.; Méchin, V.; Carrère, H.; Bernet, N.; García-Bernet, D. Understanding Biomass Recalcitrance in Grasses for Their Efficient Utilization as Biorefinery Feedstock. Rev. Environ. Sci. Bio/Technol. 2018, 17, 707–748. [Google Scholar] [CrossRef]
- Cortes-Tolalpa, L.; Salles, J.F.; van Elsas, J.D. Bacterial Synergism in Lignocellulose Biomass Degradation-Complementary Roles of Degraders as Influenced by Complexity of the Carbon Source. Front. Microbiol. 2017, 8, 1628. [Google Scholar] [CrossRef] [PubMed]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Mosbech, C.; Holck, J.; Meyer, A.S.; Agger, J.W. The Natural Catalytic Function of CuGE Glucuronoyl Esterase in Hydrolysis of Genuine Lignin–Carbohydrate Complexes from Birch. Biotechnol. Biofuels 2018, 11, 71. [Google Scholar] [CrossRef]
- Vismeh, R.; Lu, F.; Chundawat, S.P.S.; Humpula, J.F.; Azarpira, A.; Balan, V.; Dale, B.E.; Ralph, J.; Jones, A.D. Profiling of Diferulates (Plant Cell Wall Cross-Linkers) Using Ultrahigh-Performance Liquid Chromatography-Tandem Mass Spectrometry. Analyst 2013, 138, 6683. [Google Scholar] [CrossRef]
- de Oliveira, D.M.; Finger-Teixeira, A.; Rodrigues Mota, T.; Salvador, V.H.; Moreira-Vilar, F.C.; Correa Molinari, H.B.; Craig Mitchell, R.A.; Marchiosi, R.; Ferrarese-Filho, O.; Dantas dos Santos, W. Ferulic Acid: A Key Component in Grass Lignocellulose Recalcitrance to Hydrolysis. Plant Biotechnol. J. 2015, 13, 1224–1232. [Google Scholar] [CrossRef]
- Wang, Z.; Deuss, P.J. The Isolation of Lignin with Native-like Structure. Biotechnol. Adv. 2023, 68, 108230. [Google Scholar] [CrossRef]
- Raikwar, D.; Van Aelst, K.; Vangeel, T.; Corderi, S.; Van Aelst, J.; Van den Bosch, S.; Servaes, K.; Vanbroekhoven, K.; Elst, K.; Sels, B.F. Elucidating the Effect of the Physicochemical Properties of Organosolv Lignins on Its Solubility and Reductive Catalytic Depolymerization. Chem. Eng. J. 2023, 461, 141999. [Google Scholar] [CrossRef]
- Abu-Omar, M.M.; Barta, K.; Beckham, G.T.; Luterbacher, J.S.; Ralph, J.; Rinaldi, R.; Román-Leshkov, Y.; Samec, J.S.M.; Sels, B.F.; Wang, F. Guidelines for Performing Lignin-First Biorefining. Energy Environ. Sci. 2021, 14, 262–292. [Google Scholar] [CrossRef]
- Tarasov, D.; Leitch, M.; Fatehi, P. Lignin–Carbohydrate Complexes: Properties, Applications, Analyses, and Methods of Extraction: A Review. Biotechnol. Biofuels 2018, 11, 269. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D.M.D.; Lahtinen, M.H.; Lawoko, M.; Mikkonen, K.S. Enrichment and Identification of Lignin-Carbohydrate Complexes in Softwood Extract. ACS Sustain. Chem. Eng. 2020, 8, 11795–11804. [Google Scholar] [CrossRef]
- Rogowski, A.; Briggs, J.A.; Mortimer, J.C.; Tryfona, T.; Terrapon, N.; Lowe, E.C.; Baslé, A.; Morland, C.; Day, A.M.; Zheng, H.; et al. Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine. Nat. Commun. 2015, 6, 7481. [Google Scholar] [CrossRef]
- Ventorino, V.; Aliberti, A.; Faraco, V.; Robertiello, A.; Giacobbe, S.; Ercolini, D.; Amore, A.; Fagnano, M.; Pepe, O. Exploring the Microbiota Dynamics Related to Vegetable Biomasses Degradation and Study of Lignocellulose-Degrading Bacteria for Industrial Biotechnological Application. Sci. Rep. 2015, 5, 8161. [Google Scholar] [CrossRef] [PubMed]
- Bout, A.E.; Pfau, S.F.; van der Krabben, E.; Dankbaar, B. Residual Biomass from Dutch Riverine Areas-from Waste to Ecosystem Service. Sustainability 2019, 11, 509. [Google Scholar] [CrossRef]
- Cardman, Z.; Arnosti, C.; Durbin, A.; Ziervogel, K.; Cox, C.; Steen, A.D.; Teske, A. Verrucomicrobia Are Candidates for Polysaccharide-Degrading Bacterioplankton in an Arctic Fjord of Svalbard. Appl. Environ. Microbiol. 2014, 80, 3749–3756. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbrück, L.; Reeder, J.; Temperton, B.; Huse, S.; McHardy, A.C.; Knight, R.; Joint, I.; et al. Defining Seasonal Marine Microbial Community Dynamics. ISME J. 2012, 6, 298–308. [Google Scholar] [CrossRef]
- Parvathi, A.; Jasna, V.; Aswathy, V.K.; Nathan, V.K.; Aparna, S.; Balachandran, K.K. Microbial Diversity in a Coastal Environment with Co-Existing Upwelling and Mud-Banks along the South West Coast of India. Mol. Biol. Rep. 2019, 46, 3113–3127. [Google Scholar] [CrossRef] [PubMed]
- Marín, P.; Martirani-Von Abercron, S.M.; Urbina, L.; Pacheco-Sánchez, D.; Castañeda-Cataña, M.A.; Retegi, A.; Eceiza, A.; Marqués, S. Bacterial Nanocellulose Production from Naphthalene. Microb. Biotechnol. 2019, 12, 662–676. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Ebringerová, A. Structural Diversity and Application Potential of Hemicelluloses. Macromol. Symp. 2006, 232, 1–12. [Google Scholar] [CrossRef]
- Netrusov, A.I.; Liyaskina, E.V.; Kurgaeva, I.V.; Liyaskina, A.U.; Yang, G.; Revin, V.V. Exopolysaccharides Producing Bacteria: A Review. Microorganisms 2023, 11, 1541. [Google Scholar] [CrossRef]
- Korenblum, E.; Jiménez, D.J.; van Elsas, J.D. Succession of Lignocellulolytic Bacterial Consortia Bred Anaerobically from Lake Sediment. Microb. Biotechnol. 2016, 9, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Behera, P.; Mohapatra, M.; Kim, J.Y.; Adhya, T.K.; Pattnaik, A.K.; Rastogi, G. Spatial and Temporal Heterogeneity in the Structure and Function of Sediment Bacterial Communities of a Tropical Mangrove Forest. Environ. Sci. Pollut. Res. 2019, 26, 3893–3908. [Google Scholar] [CrossRef]
- Francis, B.; Urich, T.; Mikolasch, A.; Teeling, H.; Amann, R. North Sea Spring Bloom-Associated Gammaproteobacteria Fill Diverse Heterotrophic Niches. Environ. Microbiome 2021, 16, 15. [Google Scholar] [CrossRef]
- Carlos, C.; Fan, H.; Currie, C.R. Substrate Shift Reveals Roles for Members of Bacterial Consortia in Degradation of Plant Cell Wall Polymers. Front. Microbiol. 2018, 9, 364. [Google Scholar] [CrossRef]
- Parab, P.D.; Khandeparker, R.D.; Shenoy, B.D.; Sharma, J. Phylogenetic Diversity of Culturable Marine Bacteria from Mangrove Sediments of Goa, India: A Potential Source of Xylanases Belonging to Glycosyl Hydrolase Family 10. Appl. Biochem. Microbiol. 2020, 56, 718–728. [Google Scholar] [CrossRef]
- Jiménez, D.J.; Dini-Andreote, F.; Van Elsas, J.D. Metataxonomic Profiling and Prediction of Functional Behaviour of Wheat Straw Degrading Microbial Consortia. Biotechnol. Biofuels 2014, 7, 92. [Google Scholar] [CrossRef] [PubMed]
- Do, T.H.; Nguyen, T.T.; Nguyen, T.N.; Le, Q.G.; Nguyen, C.; Kimura, K.; Truong, N.H. Mining Biomass-Degrading Genes through Illumina-Based de Novo Sequencing and Metagenomic Analysis of Free-Living Bacteria in the Gut of the Lower Termite Coptotermes Gestroi Harvested in Vietnam. J. Biosci. Bioeng. 2014, 118, 665–671. [Google Scholar] [CrossRef]
- Crotti, E.; Rizzi, A.; Chouaia, B.; Ricci, I.; Favia, G.; Alma, A.; Sacchi, L.; Bourtzis, K.; Mandrioli, M.; Cherif, A.; et al. Acetic Acid Bacteria, Newly Emerging Symbionts of Insects. Appl. Environ. Microbiol. 2010, 76, 6963–6970. [Google Scholar] [CrossRef]
- Wong, C.N.A.; Ng, P.; Douglas, A.E. Low-Diversity Bacterial Community in the Gut of the Fruitfly Drosophila Melanogaster. Environ. Microbiol. 2011, 13, 1889–1900. [Google Scholar] [CrossRef] [PubMed]
- Iino, T.; Suzuki, R.; Kosako, Y.; Ohkuma, M.; Komagata, K.; Uchimura, T. Acetobacter Okinawensis Sp. Nov., Acetobacter Papayae Sp. Nov., and Acetobacter Persicus Sp. Nov.; Novel Acetic Acid Bacteria Isolated from Stems of Sugarcane, Fruits, and a Flower in Japan. J. Gen. Appl. Microbiol. 2012, 58, 235–243. [Google Scholar] [CrossRef][Green Version]
- Wardman, J.F.; Bains, R.K.; Rahfeld, P.; Withers, S.G. Carbohydrate-Active Enzymes (CAZymes) in the Gut Microbiome. Nat. Rev. Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and Gut Bacteroidetes: The Food Connection. Front. Microbiol. 2011, 2, 9588. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology Knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Chandravanshi, M.; Sharma, A.; Dasgupta, P.; Mandal, S.K.; Kanaujia, S.P. Identification and Characterization of ABC Transporters for Carbohydrate Uptake in Thermus Thermophilus HB8. Gene 2019, 696, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Schneider, E. ABC Transporters Catalyzing Carbohydrate Uptake. Res. Microbiol. 2001, 152, 303–310. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).