Gut Microbiome Alteration in HIV/AIDS and the Role of Antiretroviral Therapy—A Scoping Review
Abstract
1. Introduction
1.1. Effects of HIV Infection on the Immune System and Intestinal Barrier
1.2. Effects of HIV Infection on the Gut Microbiota
1.3. Effects of HIV Infection on the Gut Metabolome
1.4. Effect of HIV Progression and the Effect of AIDS on the Gut Microbiota
1.5. Effects of HIV Infection on Systematic Elevation Markers
1.6. Antiretroviral Therapy
1.7. Aims of the Scoping Review
- Mapping the gut microbiome alterations in PLWH who are receiving INSTI-based therapy compared to ART-naïve PLWH.
- Mapping the gut microbiome alterations in PLWH who are receiving an NNRTI-based regime compared to ART-naïve PLWH.
- Mapping the gut microbiome alterations in PLWH who are receiving INSTI-based therapy compared to those on an NNRTI-based regime.
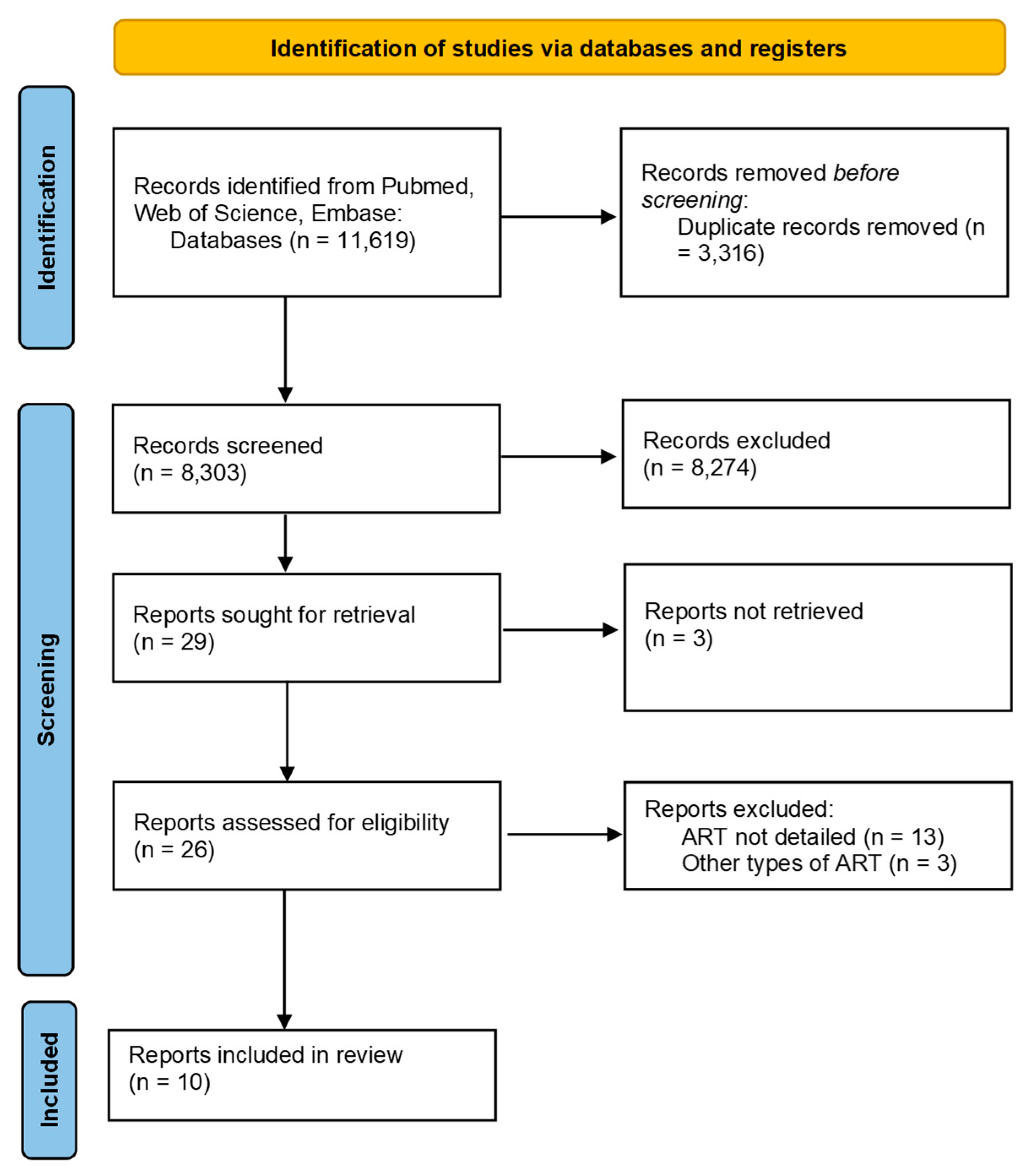
2. Materials and Methods
3. Results
3.1. Overview
3.2. Comparison of Gut Microbiota Composition in INSTI-Treated Patients and ART-Naïve Individuals
3.3. Comparison of Gut Microbiota Composition in NNRTI-Treated Patients and ART-Naïve Individuals
3.4. INSTI-Based Treatment Regime Compared to NNRTI-Based Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nganou-Makamdop, K.; Douek, D.C. The Gut and the Translocated Microbiomes in HIV Infection: Current Concepts and Future Avenues. Pathog. Immun. 2024, 9, 168–194. [Google Scholar] [CrossRef] [PubMed]
- Onabajo, O.O.; Mattapallil, J.J. Gut Microbiome Homeostasis and the CD4 T- Follicular Helper Cell IgA Axis in Human Immunodeficiency Virus Infection. Front. Immunol. 2021, 12, 657679. [Google Scholar] [CrossRef] [PubMed]
- Hummelen, R.; Vos, A.P.; Land, B.V.T.; Norren, K.V.; Reid, G. Altered host-microbe interaction in HIV: A target for intervention with pro- and prebiotics. Int. Rev. Immunol. 2010, 29, 485–513. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F.; Überla, K.; Ng, S.C. Gut as viral reservoir: Lessons from gut viromes, HIV and COVID-19. Gut 2021, 70, 1605–1608. [Google Scholar] [CrossRef] [PubMed]
- Baltazar-Díaz, T.A.; Andrade-Villanueva, J.F.; Sánchez-Álvarez, P.; Amador-Lara, F.; Holguín-Aguirre, T.; Sánchez-Reyes, K.; González-Hernández, L.A. A Two-Faced Gut Microbiome: Butyrogenic and Proinflammatory Bacteria Predominate in the Intestinal Milieu of People Living with HIV from Western Mexico. Int. J. Mol. Sci. 2024, 25, 4830. [Google Scholar] [CrossRef]
- Rocafort, M.; Gootenberg, D.B.; Luévano, J.M., Jr.; Paer, J.M.; Hayward, M.R.; Bramante, J.T.; Kwon, D.S. HIV-associated gut microbial alterations are dependent on host and geographic context. Nat. Commun. 2024, 15, 1055. [Google Scholar] [CrossRef]
- Lazzaro, A.; Innocenti, G.P.; Santinelli, L.; Pinacchio, C.; Girolamo, G.D.; Vassalini, P.; d’Ettorre, G. Antiretroviral Therapy Dampens Mucosal CD4+ T Lamina Propria Lymphocytes Immune Activation in Long-Term Treated People Living with HIV-1. Microorganisms 2021, 9, 1624. [Google Scholar] [CrossRef]
- Khan, S.; Telwatte, S.; Trapecar, M.; Yukl, S.; Sanjabi, S. Differentiating Immune Cell Targets in Gut-Associated Lymphoid Tissue for HIV Cure. AIDS Res. Hum. Retroviruses 2017, 33, S40–S58. [Google Scholar] [CrossRef]
- Pan, Z.; Wu, N.; Jin, C. Intestinal Microbiota Dysbiosis Promotes Mucosal Barrier Damage and Immune Injury in HIV-Infected Patients. Can. J. Infect. Dis. Med. Microbiol. 2023, 2023, 3080969. [Google Scholar] [CrossRef]
- Yang, X.; Su, B.; Zhang, X.; Liu, Y.; Wu, H.; Zhang, T. Incomplete immune reconstitution in HIV/AIDS patients on antiretroviral therapy: Challenges of immunological non-responders. J. Leukoc. Biol. 2020, 107, 597–612. [Google Scholar] [CrossRef]
- Routy, J.P.; Royston, L.; Isnard, S. Aging with Grace for People Living With HIV: Strategies to Overcome Leaky Gut and Cytomegalovirus Coinfection. J. Acquir. Immune Defic. Syndr. 2022, 89, S29–S33. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Yu, F.; Yan, L.; Zhao, H.; Zhang, F. Alterations in circulating markers in HIV/AIDS patients with poor immune reconstitution: Novel insights from microbial translocation and innate immunity. Front. Immunol. 2022, 17, 1026070. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Qi, Q. Gut microbial metabolites associated with HIV infection. Future Virol. 2019, 14, 335–347. [Google Scholar] [CrossRef]
- Ortiz, A.M.; Brenchley, J.M. Untangling the role of the microbiome across the stages of HIV disease. Curr. Opin. HIV AIDS 2024, 19, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Kantamala, D.; Praparattanapan, J.; Taejaroenkul, S.; Srithep, S.; Yoosupap, R.; Supparatpinyo, K. High microbial translocation limits gut immune recovery during short-term HAART in the area with high prevalence of foodborne infection. Cytokine 2020, 136, 155257. [Google Scholar] [CrossRef] [PubMed]
- MacCann, R.; Landay, A.L.; Mallon PW, G. HIV and comorbidities—The importance of gut inflammation and the kynurenine pathway. Curr. Opin. HIV AIDS 2023, 18, 102–110. [Google Scholar] [CrossRef]
- Pereira, L.L.; Amorim DV, S.; Sampaio, W.B.; Azevêdo TA, C.; Cardoso VB, P.; Lemos, F.B.; Chang, A.S.; Machado, F.; Lima, F.P.; Neves, F.S.; et al. Factors Associated with Periodontitis in Patients with and without HIV. Int. J. Dent. 2023, 2023, 9929835. [Google Scholar] [CrossRef]
- Ng, Q.X.; Yau, C.E.; Yaow CY, L.; Chong RI, H.; Chong, N.Z.; Teoh, S.E.; Lim, Y.L.; Soh AY, S.; Ng, W.K.; Thumboo, J. What Has Longitudinal ‘Omics’ Studies Taught Us about Irritable Bowel Syndrome? A Systematic Review. Metabolites 2023, 13, 484. [Google Scholar] [CrossRef]
- Wensel, C.R.; Pluznick, J.L.; Salzberg, S.L.; Sears, C.L. Next-generation sequencing: Insights to advance clinical investigations of the microbiome. J. Clin. Investig. 2022, 132, e154944. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef]
- Bai, X.; Narayanan, A.; Nowak, P.; Ray, S.; Neogi, U.; Sönnerborg, A. Whole-Genome Metagenomic Analysis of the Gut Microbiome in HIV-1-Infected Individuals on Antiretroviral Therapy. Front. Microbiol. 2021, 12, 667718. [Google Scholar] [CrossRef] [PubMed]
- Tuddenham, S.A.; Koay, W.L.; Zhao, N.; White, J.R.; Ghanem, K.G.; Sears, C.L. The Impact of Human Immunodeficiency Virus Infection on Gut Microbiota α-Diversity: An Individual-level Meta-analysis. Clin. Infect. Dis. 2020, 70, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Tao, J.; Abu, Y.; Sussman, D.A.; Girotra, M.; Franceschi, D.; Roy, S. HIV-Positive Patients on Antiretroviral Therapy Have an Altered Mucosal Intestinal but Not Oral Microbiome. Microbiol. Spectr. 2023, 11, e02472-22. [Google Scholar] [CrossRef]
- Ray, S.; Narayanan, A.; Giske, C.G.; Neogi, U.; Sönnerborg, A.; Nowak, P. Altered Gut Microbiome Under Antiretroviral Therapy: Impact of Efavirenz and Zidovudine. ACS Infect. Dis. 2021, 7, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, A.; Kieri, O.; Vesterbacka, J.; Manoharan, L.; Chen, P.; Ghorbani, M.; Nowak, P. Exploring the interplay between antiretroviral therapy and the gut-oral microbiome axis in people living with HIV. Sci. Rep. 2024, 14, 17820. [Google Scholar] [CrossRef]
- Salvador, P.B.; Altavas, P.J.; del Rosario, M.A.; Ornos, E.D.; Dalmacio, L.M. Alterations in the Gut Microbiome Composition of People Living with HIV in the Asia–Pacific Region: A Systematic Review. Clin. Pract. 2024, 14, 846–861. [Google Scholar] [CrossRef]
- Gootenberg, D.B.; Paer, J.M.; Luevano, J.M.; Kwon, D.S. HIV-associated changes in the enteric microbial community: Potential role in loss of homeostasis and development of systemic inflammation. Curr. Opin. Infect. Dis. 2017, 30, 31–43. [Google Scholar] [CrossRef]
- Pinto-Cardoso, S.; Klatt, N.R.; Reyes-Terán, G. Impact of antiretroviral drugs on the microbiome: Unknown answers to important questions. Curr. Opin. HIV AIDS 2018, 13, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Rocafort, M.; Noguera-Julian, M.; Rivera, J.; Pastor, L.; Guillén, Y.; Langhorst, J.; Paredes, R. Evolution of the gut microbiome following acute HIV-1 infection. Microbiome 2019, 7, 73. [Google Scholar] [CrossRef]
- Zhang, Y.; Andreu-Sánchez, S.; Vadaq, N.; Wang, D.; Matzaraki, V.; van der Heijden, W.A.; Fu, J. Gut dysbiosis associates with cytokine production capacity in viral-suppressed people living with HIV. Front. Cell Infect. Microbiol. 2023, 13, 1202035. [Google Scholar] [CrossRef]
- Vujkovic-Cvijin, I.; Sortino, O.; Verheij, E.; Sklar, J.; Wit, F.W.; Kootstra, N.A.; Sereti, I. HIV-associated gut dysbiosis is independent of sexual practice and correlates with noncommunicable diseases. Nat. Commun. 2020, 11, 2448. [Google Scholar] [CrossRef]
- Parbie, P.K.; Mizutani, T.; Ishizaka, A.; Kawana-Tachikawa, A.; Runtuwene, L.R.; Seki, S.; Matano, T. Dysbiotic Fecal Microbiome in HIV-1 Infected Individuals in Ghana. Front. Cell Infect. Microbiol. 2021, 11, 646467. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, Y.; Cui, P.; Luo, L.; Chen, H.; Liang, B.; Jiang, J.; Ning, C.; Tian, L.; Zhong, X.; et al. Gut Microbiome Changes Associated with HIV Infection and Sexual Orientation. Front. Cell Infect. Microbiol. 2020, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Sönnerborg, A.; Nowak, P. Elite controllers microbiome: Unraveling the mystery of association and causation. Curr. Opin. HIV AIDS 2024, 19, 261–267. [Google Scholar] [CrossRef] [PubMed]
- do Nascimento, W.M.; Machiavelli, A.; Ferreira, L.G.; Silveira, L.C.; de Azevedo, S.S.; Bello, G.; Pinto, A.R. Gut Microbiome Profiles and Associated Metabolic Pathways in HIV-Infected Treatment-Naïve Patients. Cells 2021, 10, 385. [Google Scholar] [CrossRef] [PubMed]
- Sperk, M.; Ambikan, A.T.; Ray, S.; Singh, K.; Mikaeloff, F.; Diez, R.C.; Narayanan, A.; Vesterbacka, J.; Nowak, P.; Sönnerborg, A.; et al. Fecal Metabolome Signature in the HIV-1 Elite Control Phenotype: Enrichment of Dipeptides Acts as an HIV-1 Antagonist but a Prevotella Agonist. J. Virol. 2021, 95, e0047921. [Google Scholar] [CrossRef]
- Zaongo, S.D.; Ouyang, J.; Isnard, S.; Zhou, X.; Harypursat, V.; Cui, H.; Chen, Y. Candida albicans can foster gut dysbiosis and systemic inflammation during HIV infection. Gut Microbes 2023, 15, 2167171. [Google Scholar] [CrossRef]
- Li, S.; Yang, X.; Moog, C.; Wu, H.; Su, B.; Zhang, T. Neglected mycobiome in HIV infection: Alterations, common fungal diseases and antifungal immunity. Front. Immunol. 2022, 13, 1015775. [Google Scholar] [CrossRef]
- Gosalbes, M.J.; Jimenéz-Hernandéz, N.; Moreno, E.; Artacho, A.; Pons, X.; Ruíz-Pérez, S.; Serrano-Villar, S. Interactions among the mycobiome, bacteriome, inflammation, and diet in people living with HIV. Gut Microbes 2022, 14, 2089002. [Google Scholar] [CrossRef]
- Ma, J.; Wen, S.; Dong, A.; Fan, W.; Kang, Y. Gut Microbiome (Bacteria, Fungi, and Viruses) and HIV Infection: Revealing Novel Treatment Strategies. Mol. Nutr. Food Res. 2023, 67, e2300566. [Google Scholar] [CrossRef]
- Monaco, C.L.; Gootenberg, D.B.; Zhao, G.; Handley, S.A.; Ghebremichael, M.S.; Lim, E.S.; Virgin, H.W. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe 2016, 19, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lin, H.; Cole, M.; Morris, A.; Martinson, J.; Mckay, H.; Rinaldo, C.R. Signature changes in gut microbiome are associated with increased susceptibility to HIV-1 infection in MSM. Microbiome 2021, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Deng, J.; Zhang, C.; Lai, G.; Xie, B.; Zhong, X. Gut microbiome dysbiosis in men who have sex with men increases HIV infection risk through immunity homeostasis alteration. Front. Cell Infect. Microbiol. 2023, 13, 1260068. [Google Scholar] [CrossRef] [PubMed]
- Villanueva-Millán, M.J.; Pérez-Matute, P.; Recio-Fernández, E.; Rosales, J.M.; Oteo, J.A. Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients. J. Int. AIDS Soc. 2017, 20, 21526. [Google Scholar] [CrossRef]
- Nowak, R.G.; Bentzen, S.M.; Ravel, J.; Crowell, T.A.; Dauda, W.; Ma, B.; Liu, H.; Blattner, W.A.; Baral, S.D.; Charurat, M.E.; et al. Rectal microbiota among HIV-uninfected, untreated HIV, and treated HIV-infected in Nigeria. AIDS 2017, 31, 857–862. [Google Scholar] [CrossRef]
- Tuddenham, S.; Koay, W.L.; Sears, C. HIV, Sexual Orientation, and Gut Microbiome Interactions. Dig. Dis. Sci. 2020, 65, 800–817. [Google Scholar] [CrossRef]
- Sánchez-Conde, M.; Alba, C.; Castro, I.; Dronda, F.; Ramírez, M.; Arroyo, R.; Brañas, F. Comparison of the Fecal Bacteriome of HIV-Positive and HIV-Negative Older Adults. Biomedicines 2023, 11, 2305. [Google Scholar] [CrossRef]
- de la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems 2019, 4, e00261-19. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef]
- Ashuro, A.A.; Lobie, T.A.; Ye, D.Q.; Leng, R.X.; Li, B.Z.; Pan, H.F.; Fan, Y.G. Review on the Alteration of Gut Microbiota: The Role of HIV Infection and Old Age. AIDS Res. Hum. Retroviruses 2020, 36, 556–565. [Google Scholar] [CrossRef]
- Kim, Y.S.; Unno, T.; Kim, B.Y.; Park, M.S. Sex Differences in Gut Microbiota. World J. Mens Health 2020, 38, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Ng, Q.X.; Lim, Y.L.; Yaow, C.Y.L.; Ng, W.K.; Thumboo, J.; Liew, T.M. Effect of Probiotic Supplementation on Gut Microbiota in Patients with Major Depressive Disorders: A Systematic Review. Nutrients 2023, 15, 1351. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.S.; Chu, Q.S.; Ashuro, A.A.; Di, D.S.; Zhang, Q.; Liu, X.M.; Fan, Y.G. The Effect of Probiotics, Prebiotics, and Synbiotics on CD4 Counts in HIV-Infected Patients: A Systematic Review and Meta-Analysis. Biomed. Res. Int. 2020, 2020, 7947342. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Wu, C. Modulation of Gut Microbiota and Immune System by Probiotics, Pre-biotics, and Post-biotics. Front. Nutr. 2022, 8, 634897. [Google Scholar] [CrossRef]
- Blázquez-Bondia, C.; Parera, M.; Català-Moll, F.; Casadellà, M.; Elizalde-Torrent, A.; Aguiló, M.; Espadaler-Mazo, J.; Santos, J.R.; Paredes, R.; Noguera-Julian, M. Probiotic effects on immunity and microbiome in HIV-1 discordant patients. Front. Immunol. 2022, 13, 1066036. [Google Scholar] [CrossRef] [PubMed]
- Tenore, S.B.; Avelino-Silva, V.I.; Costa, P.R.; Franco, L.M.; Sabino, E.C.; Kalil, J.; Cerqueira, N.B.; Nakagawa, Z.; Kallas, E.G. Immune effects of Lactobacillus casei Shirota in treated HIV-infected patients with poor CD4+ T-cell recovery. AIDS 2020, 34, 381–389. [Google Scholar] [CrossRef]
- Herrera, S.; Martínez-Sanz, J.; Serrano-Villar, S. HIV. Cancer, and the Microbiota: Common Pathways Influencing Different Diseases. Front. Immunol. 2019, 10, 1466. [Google Scholar] [CrossRef]
- Enriquez, A.B.; Caten, F.T.; Ghneim, K.; Sekaly, R.P. Regulation of Immune Homeostasis, Inflammation, and HIV Persistence by the Microbiome, Short-Chain Fatty Acids, and Bile Acids. Annu. Rev. Virol. 2023, 10, 397–422. [Google Scholar] [CrossRef]
- Sereti, I.; Verburgh, M.L.; Gifford, J.; Lo, A.; Boyd, A.; Verheij, E.; Vujkovic-Cvijin, I. Impaired gut microbiota-mediated short-chain fatty acid production precedes morbidity and mortality in people with HIV. Cell Rep. 2023, 42, 113336. [Google Scholar] [CrossRef]
- Hishiya, N.; Uno, K.; Nakano, A.; Konishi, M.; Higashi, S.; Eguchi, S.; Yano, H. Association between the gut microbiome and organic acid profiles in a Japanese population with HIV infection. J. Infect. Chemother. 2024, 30, 58–66. [Google Scholar] [CrossRef]
- Koay, W.; Siems, L.; Persaud, D. The Microbiome and HIV Persistence: Implications for Viral Remission and Cure. Curr. Opin. HIV AIDS 2018, 13, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Ghare, S.; Singhal, R.; Bryant, V.; Gautam, S.; Tirumala, C.C.; Srisailam, P.K.; Barve, S. Age-Associated Gut Dysbiosis, Marked by Loss of Butyrogenic Potential, Correlates with Altered Plasma Tryptophan Metabolites in Older People Living with HIV. J. Acquir. Immune Defic. Syndr. 2022, 89, S56–S64. [Google Scholar] [CrossRef] [PubMed]
- Swinkels, H.M.; Vaillant, A.A.; Nguyen, A.D.; Gulick, P.G. HIV and AIDS; StatPearls Publishing: St. Petersburg, FL, USA, 2024. [Google Scholar]
- Geng, S.T.; Zhang, Z.Y.; Wang, Y.X.; Lu, D.; Yu, J.; Zhang, J.B.; Wang, K.H. Regulation of Gut Microbiota on Immune Reconstitution in Patients with Acquired Immunodeficiency Syndrome. Front. Microbiol. 2020, 11, 594820. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Rozas, G.; Lopez, M.N.; Soto-Rifo, R.; Vidal, R.; Cortes, C.P. Enteropathy and gut dysbiosis as obstacles to achieve immune recovery in undetectable people with HIV: A clinical view of evidence, successes, and projections. AIDS 2023, 37, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, Z.; Zhou, J.; Li, Y.; Ning, C.; Su, Q.; Huang, J. The altered metabolites contributed by dysbiosis of gut microbiota are associated with microbial translocation and immune activation during HIV infection. Front. Immunol. 2022, 13, 1020822. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Zhang, J.B.; Wang, Y.X.; Geng, S.T.; Zhang, Z.; Xu, Y.; Kuang, Y.Q. Association between CD4+ T cell counts and gut microbiota and serum cytokines levels in HIV-infected immunological non-responders. BMC Infect. Dis. 2021, 21, 742. [Google Scholar] [CrossRef]
- Lu, J.; Ma, S.S.; Zhang, W.Y.; Duan, J.P. Changes in peripheral blood inflammatory factors (TNF-α and IL-6) and intestinal flora in AIDS and HIV-positive individuals. J. Zhejiang Univ. Sci. 2019, 20, 793–802. [Google Scholar] [CrossRef]
- Xie, Y.; Sun, J.; Wei, L.; Jiang, H.; Hu, C.; Yang, J.; Huang, Y.; Ruan, B.; Zhu, B. Altered gut microbiota correlate with different immune responses to HAART in HIV-infected individuals. BMC Microbiol. 2021, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Ishizaka, A.; Koga, M.; Mizutani, T.; Parbie, P.K.; Prawisuda, D.; Yusa, N.; Yotsuyanagi, H. Unique Gut Microbiome in HIV Patients on Antiretroviral Therapy (ART) Suggests Association with Chronic Inflammation. Microbiol. Spectr. 2021, 9, 10. [Google Scholar] [CrossRef]
- Isnard, S.; Lin, J.; Bu, S.; Fombuena, B.; Royston, L.; Routy, J.P. Gut Leakage of Fungal-Related Products: Turning Up the Heat for HIV Infection. Front. Immunol. 2021, 12, 656414. [Google Scholar] [CrossRef]
- Fulcher, J.A.; Li, F.; Tobin, N.H.; Zabih, S.; Elliott, J.; Clark, J.L.; Aldrovandi, G.M. Gut dysbiosis and inflammatory blood markers precede HIV with limited changes after early seroconversion. EBioMedicine 2022, 84, 104286. [Google Scholar] [CrossRef] [PubMed]
- Mehraj, V.; Ramendra, R.; Isnard, S.; Dupuy, F.P.; Ponte, R.; Chen, J.; Kema, I.; Jenabian, M.A.; Costinuik, C.T.; Lebouché, B.; et al. Montreal Primary HIV Infection Study and Canadian HIV and Aging Cohort Study Groups. Circulating (1 → 3)-β-D-glucan Is Associated with Immune Activation During Human Immunodeficiency Virus Infection. Clin. Infect. Dis. 2020, 70, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Neff, C.P.; Krueger, O.; Xiong, K.; Arif, S.; Nusbacher, N.; Schneider, J.M.; Cunningham, A.W.; Armstrong, A.; Li, S.; McCarter, M.D.; et al. Fecal Microbiota Composition Drives Immune Activation in HIV-infected Individuals. EBioMedicine 2018, 30, 192–202. [Google Scholar] [CrossRef]
- Ellis, R.J.; Iudicello, J.E.; Heaton, R.K.; Isnard, S.; Lin, J.; Routy, J.P.; Knight, R. Markers of Gut Barrier Function and Microbial Translocation Associate with Lower Gut Microbial Diversity in People with HIV. Viruses 2021, 13, 1891. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, L. Development of Anti-HIV Therapeutics: From Conventional Drug Discovery to Cutting-Edge Technology. Pharmaceuticals 2024, 17, 887. [Google Scholar] [CrossRef]
- Azzman, N.; Gill MS, A.; Hassan, S.S.; Christ, F.; Debyser, Z.; Mohamed WA, S.; Ahemad, N. Pharmacological advances in anti-retroviral therapy for human immunodeficiency virus-1 infection: A comprehensive review. Rev. Med. Virol. 2024, 34, e2529. [Google Scholar] [CrossRef]
- Zhao, A.V.; Crutchley, R.D.; Guduru, R.C.; Ton, K.; Lam, T.; Min, A.C. A clinical review of HIV integrase strand transfer inhibitors (INSTIs) for the prevention and treatment of HIV-1 infection. Retrovirology 2022, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. Available online: https://clinicalinfo.hiv.gov/en/guidelines/hiv-clinical-guidelines-adult-and-adolescent-arv/whats-new (accessed on 15 October 2024).
- EACS Guidelines v12.0. Available online: https://www.eacsociety.org/media/guidelines-12.0.pdf (accessed on 18 October 2024).
- Waters, L.; Winston, A.; Reeves, I.; Boffito, M.; Churchill, D.; Cromarty, B.; Dunn, D.; Fink, D.; Fidler, S.; Foster, C.; et al. BHIVA guidelines on antiretroviral treatment for adults living with HIV-1 2022. HIV Med. 2022, 23, 3–115. [Google Scholar] [CrossRef]
- Sayyed, S.K.; Quraishi, M.; Jobby, R.; Rameshkumar, N.; Kayalvizhi, N.; Krishnan, M.; Sonawane, T. A computational overview of integrase strand transfer inhibitors (INSTIs) against emerging and evolving drug-resistant HIV-1 integrase mutants. Arch. Microbiol. 2023, 205, 142. [Google Scholar] [CrossRef]
- Kolakowska, A.; Maresca, A.F.; Collins, I.J.; Cailhol, J. Update on Adverse Effects of HIV Integrase Inhibitors. Curr. Treat. Options Infect. Dis. 2019, 11, 372–387. [Google Scholar] [CrossRef]
- Baltazar-Díaz, T.A.; Amador-Lara, F.; Andrade-Villanueva, J.F.; González-Hernández, L.A.; Cabrera-Silva, R.I.; Sánchez-Reyes, K.; Bueno-Topete, M.R. Gut Bacterial Communities in HIV-Infected Individuals with Metabolic Syndrome: Effects of the Therapy with Integrase Strand Transfer Inhibitor-Based and Protease Inhibitor-Based Regimens. Microorganisms 2023, 11, 951. [Google Scholar] [CrossRef] [PubMed]
- Namasivayam, V.; Vanangamudi, M.; Kramer, V.G.; Kurup, S.; Zhan, P.; Liu, X.; Byrareddy, S.N. The Journey of HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs) from Lab to Clinic. J. Med. Chem. 2019, 62, 4851–4883. [Google Scholar] [CrossRef]
- Ma, S.; Xie, X.; Fu, Y.; Gan, L.; Yang, X.; Kong, L.; Li, J.; Long, H. Clinical benefits of novel non-nucleoside reverse transcriptase inhibitors: A prospective cohort study. Immun. Inflamm. Dis. 2024, 12, e1217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, Y.J.; Li, M.; Pannecouque, C.; De Clercq, E.; Wang, S.; Chen, F.E. Deciphering the enigmas of non-nucleoside reverse transcriptase inhibitors (NNRTIs): A medicinal chemistry expedition towards combating HIV drug resistance. Med. Res. Rev. 2024; ahead of print. [Google Scholar]
- Ishizaka, A.; Koga, M.; Mizutani, T.; Suzuki, Y.; Matano, T.; Yotsuyanagi, H. Sustained gut dysbiosis and intestinal inflammation show correlation with weight gain in person with chronic HIV infection on antiretroviral therapy. BMC Microbiol. 2024, 24, 274. [Google Scholar] [CrossRef]
- Imahashi, M.; Ode, H.; Kobayashi, A.; Nemoto, M.; Matsuda, M.; Hashiba, C.; Iwatani, Y. Impact of long-term antiretroviral therapy on gut and oral microbiotas in HIV-1-infected patients. Sci. Rep. 2021, 11, 960. [Google Scholar] [CrossRef]
- Russo, E.; Nannini, G.; Sterrantino, G.; Kiros, S.T.; Di Pilato, V.; Coppi, M.; Amedei, A. Effects of viremia and CD4 recovery on gut “microbiome-immunity” axis in treatment-naïve HIV-1-infected patients undergoing antiretroviral therapy. World J. Gastroenterol. 2022, 28, 635–652. [Google Scholar] [CrossRef]
- Churchill, M.J.; Deeks, S.G.; Margolis, D.M.; Siliciano, R.F.; Swanstrom, R. HIV reservoirs: What, where and how to target them. Nat. Rev. Microbiol. 2016, 14, 55–60. [Google Scholar] [CrossRef]
- Estes, J.D.; Kityo, C.; Ssali, F.; Swainson, L.; Makamdop, K.N.; Del Prete, G.Q.; Deeks, S.G.; Luciw, P.A.; Chipman, J.G.; Beilman, G.J.; et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat. Med. 2017, 23, 1271–1276. [Google Scholar] [CrossRef] [PubMed]
- Moretti, S.; Schietroma, I.; Sberna, G.; Maggiorella, M.T.; Sernicola, L.; Farcomeni, S.; Borsetti, A. HIV-1-Host Interaction in Gut-Associated Lymphoid Tissue (GALT): Effects on Local Environment and Comorbidities. Int. J. Mol. Sci. 2023, 24, 12193. [Google Scholar] [CrossRef]
- Yan, L.; Xu, K.; Xiao, Q.; Tuo, L.; Luo, T.; Wang, S.; Yang, R.; Zhang, F.; Yang, X. Cellular and molecular insights into incomplete immune recovery in HIV/AIDS patients. Front. Immunol. 2023, 14, 1152951. [Google Scholar] [CrossRef]
- Patterson, K.B.; Prince, H.A.; Stevens, T.; Shaheen, N.J.; Dellon, E.S.; Madanick, R.D.; Kashuba, A.D. Differential Penetration of Raltegravir throughout Gastrointestinal Tissue: Implications for Eradication and Cure. AIDS 2013, 27, 1413–1419. [Google Scholar] [CrossRef] [PubMed]
- Hladik, F.; Burgener, A.; Ballweber, L.; Gottardo, R.; Vojtech, L.; Fourati, S.; McGowan, I. Mucosal effects of tenofovir 1% gel. eLife 2015, 4, e04525. [Google Scholar] [CrossRef] [PubMed]
- Villoslada-Blanco, P.; Pérez-Matute, P.; Íñiguez, M.; Recio-Fernández, E.; Jansen, D.; De Coninck, L.; Oteo, J.A. Impact of HIV infection and integrase strand transfer inhibitors-based treatment on the gut virome. Sci. Rep. 2022, 12, 21658. [Google Scholar] [CrossRef]
- Townsend, E.M.; Kelly, L.; Muscatt, G.; Box, J.D.; Hargraves, N.; Lilley, D.; Jameson, E. The Human Gut Phageome: Origins and Roles in the Human Gut Microbiome. Front. Cell Infect. Microbiol. 2021, 11, 643214. [Google Scholar] [CrossRef]
- Dillon, S.M.; Kibbie, J.; Lee, E.J.; Santiago, M.L.; Austin, G.L.; Guo, K.; Wilson, C.C. Low abundance of colonic butyrate-producing bacteria in HIV infection is associated with microbial translocation and immune activation. AIDS 2017, 31, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Villoslada-Blanco, P.; Pérez-Matute, P.; Íñiguez, M.; Recio-Fernández, E.; Blanco-Navarrete, P.; Metola, L.; Oteo, J.A. Integrase Inhibitors Partially Restore Bacterial Translocation, Inflammation and Gut Permeability Induced by HIV Infection: Impact on Gut Microbiota. Infect. Dis. Ther. 2022, 11, 1541–1557. [Google Scholar] [CrossRef]
- Sortino, O.; Phanuphak, N.; Schuetz, A.; Ortiz, A.M.; Chomchey, N.; Belkaid, Y.; Sereti, I. Impact of Acute HIV Infection and Early Antiretroviral Therapy on the Human Gut Microbiome. Open Forum Infect. Dis. 2020, 7, ofz367. [Google Scholar] [CrossRef]
- Ji, Y.; Zhang, F.; Zhang, R.; Shen, Y.; Liu, L.; Wang, J.; Yang, J.; Tang, Q.; Xun, J.; Qi, T.; et al. Changes in intestinal microbiota in HIV-1-infected subjects following cART initiation: Influence of CD4+ T cell count. Emerg. Microbes Infect. 2018, 7, 113. [Google Scholar] [CrossRef]
- Hanttu, A.M.; Pekkalab, S.; Satokari, R.; Hartikainen, A.K.; Arkkila, P.; Pietiläinen, K.H.; Sutinen, J.P. Gut microbiota alterations after switching from a protease inhibitor or efavirenz to raltegravir in a randomized, controlled study. AIDS 2023, 37, 323–332. [Google Scholar] [CrossRef]
- Fu, Y.; Ke, S.; Tang, G.; Guo, Q.; Guo, Q.; Wang, Z.; Fan, Y. Characterization of the intestinal microbiota in MSM with HIV infection. BMC Microbiol. 2024, 24, 192. [Google Scholar] [CrossRef] [PubMed]
- Feranchuk, S.; Belkova, N.; Potapova, U.; Kuzmin, D.; Belikov, S. Evaluating the use of diversity indices to distinguish between microbial communities with different traits. Res. Microbiol. 2018, 169, 254–261. [Google Scholar] [CrossRef] [PubMed]
| Inclusion Criteria: | |||
| Studies written in English were considered if they fell into the following categories: reviews, randomized controlled trials, single-arm trials, cohort studies, and case-control studies. | |||
| Population | Intervention | Comparator | |
| PICO 1 | Adults diagnosed with HIV infection confirmed by serological tests | PLWH receiving INSTI-based therapy | ART-naïve PLWH |
| PICO 2 | Adults diagnosed with HIV infection confirmed by serological tests | PLWH receiving NNRTI-based therapy | ART-naïve PLWH |
| PICO 3 | Adults diagnosed with HIV infection confirmed by serological tests | PLWH receiving INSTI-based therapy | PLWH receiving NNRTI-based therapy |
| Exclusion criteria: | |||
| The following types of publications were excluded: case series, case reports, clinical guidelines, non-peer-reviewed literature, conference abstracts, letters, and editorials. | |||
| Study Title | Author | Publication Year | Participants | INSTI Mediated Alpha Diversity Changes | INSTI Mediated Beta Diversity Changes | INSTI Mediated Changes in Microbiome Composition | Stool Sample Analysis | INSTI Mediated Change on Bacterial Translocation or Systematic Inflammation Markers |
|---|---|---|---|---|---|---|---|---|
| Integrase Inhibitors Partially Restore Bacterial Translocation, Inflammation and Gut Permeability Induced by HIV Infection: Impact on Gut Microbiota | Villoslada-Blanco et al. [101] | 2022 | PLWH on INSTI treatment vs. ART-naïve PLWH vs. seronegative controls | INSTI restored alpha diversity (Chao1) | Seronegative controls differed significantly independent of ART treatment |
| 16S rRNS gene sequencing |
|
| Impact of HIV infection and integrase strand transfer inhibitors-based treatment on the gut virome | Villoslada-Blanco et al. [97] | 2022 | PLWH on INSTI treatment vs. ART-naïve PLWH vs. seronegative controls | INSTI restored alpha diversity among bacteriophages (Fisher’s alpha indexes) | Seronegative controls differed significantly independent of ART treatment regarding bacteriophage composition |
| 16S rRNS gene sequencing | - |
| Study Title | Author | Publication Year | Participants | NNRTI Mediated Alpha Diversity Changes | NNRTI Mediated Beta Diversity Changes | NNRTI Mediated Changes in Microbiome Composition | Stool Sample Analysis | NNRTI Mediated Change on Bacterial Translocation or Systematic Inflammation Markers |
|---|---|---|---|---|---|---|---|---|
| Impact of Acute HIV Infection and Early Antiretroviral Therapy on the Human Gut Microbiome | Sortino et al. [52] | 2020 | PLWH before and 6 months after NNRTI treatment vs. seronegative controls | Compared to seronegative controls, alpha diversity was still significantly lower (Chao1, Shannon index) | NNRTI PLWH showed partial restoration |
| 16S rRNS gene sequencing |
|
| Altered Gut Microbiome under Antiretroviral Therapy: Impact of Efavirenz and Zidovudine | Ray et al. [27] | 2021 | PLWH before and 10 months after NNRTI treatment vs. seronegative controls | Decreased after NNRTI treatment (Fischer, Chao1, ACE) | Only moderate differences could be observed |
| 16S rRNS gene sequencing | - |
| High microbial translocation limits gut immune recovery during short-term HAART in the area with high prevalence of foodborne infection | Kantamala et al. [15] | 2020 | PLWH before and 48 weeks after NNRTI treatment | - | - | - | 16S rRNS gene sequencing |
|
| Changes in intestinal microbiota in HIV-1-infected subjects following cART initiation: influence of CD4+ T cell count | Ji et al. [102] | 2018 | PLWH before and 12 months after NNRTI treatment | PLWH with a baseline of <300/mm3 CD4 T cells had a significant elevation after NNRTI treatment | Significant differences after NNRTI treatment |
| 16S rRNS gene sequencing |
|
| Study Title | Author | Publication Year | Participants | ART Mediated Alpha Diversity Changes | ART Mediated Beta Diversity Changes | ART Mediated Changes in Microbiome Composition | Stool Sample Analysis | ART Mediated Change on Bacterial Translocation or Systematic Inflammation Markers |
|---|---|---|---|---|---|---|---|---|
| Exploring the interplay between antiretroviral therapy and the gut-oral microbiome axis in people living with HIV | Narayanan et al. [25] | 2024 | NNRTI vs. INSTI treated PLWH vs. seronegative individuals | - | - |
| 16S rRNS gene sequencing | - |
| Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients | Villanueva-Millán et al. [44] | 2017 | NNRTI vs. INSTI treated PLWH vs. ART-naïve PLWH vs. seronegative individuals |
| - | INSTI:
| 16S rDNA pyrosequencing |
|
| Gut microbiota alterations after switching from a protease inhibitor or efavirenz to raltegravir in a randomized, controlled study | Hanttu et al. [103] | 2023 | NNRTI-based to INSTI-based vs. PI-based to INSTI-based vs. NNRT-based vs. PI-based ART vs. seronegative controls | INSTI: alpha diversity restored | - | NNRTI to INSTI:
| 16S rRNS gene sequencing | NNRTI to INSTI: I-FABP and LBP unchanged |
| Characterization of the intestinal microbiota in MSM with HIV infection | Fu et al. [104] | 2024 | NNRTI-based vs. INSTI-based vs. PI-based ART vs. ART-naïve PLWH vs. seronegative controls | NNRTI: reduced alpha diversity compared to ART-naïve PLWH | NNRTI: significantly different beta diversity compared to ART-naïve PLWH | NNRTI:
| 16S rRNS gene sequencing | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gáspár, Z.; Nagavci, B.; Szabó, B.G.; Lakatos, B. Gut Microbiome Alteration in HIV/AIDS and the Role of Antiretroviral Therapy—A Scoping Review. Microorganisms 2024, 12, 2221. https://doi.org/10.3390/microorganisms12112221
Gáspár Z, Nagavci B, Szabó BG, Lakatos B. Gut Microbiome Alteration in HIV/AIDS and the Role of Antiretroviral Therapy—A Scoping Review. Microorganisms. 2024; 12(11):2221. https://doi.org/10.3390/microorganisms12112221
Chicago/Turabian StyleGáspár, Zsófia, Blin Nagavci, Bálint Gergely Szabó, and Botond Lakatos. 2024. "Gut Microbiome Alteration in HIV/AIDS and the Role of Antiretroviral Therapy—A Scoping Review" Microorganisms 12, no. 11: 2221. https://doi.org/10.3390/microorganisms12112221
APA StyleGáspár, Z., Nagavci, B., Szabó, B. G., & Lakatos, B. (2024). Gut Microbiome Alteration in HIV/AIDS and the Role of Antiretroviral Therapy—A Scoping Review. Microorganisms, 12(11), 2221. https://doi.org/10.3390/microorganisms12112221







