An Overview of Lsr2 Repressor Effect in Streptomyces spp. Secondary Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Design and Plasmid Construction
2.3. Metabolite Production and Chromatographic Analysis
2.4. Dereplication Analysis
2.5. Bioinformatic Tools
3. Results
3.1. Streptomyces sp. CS014
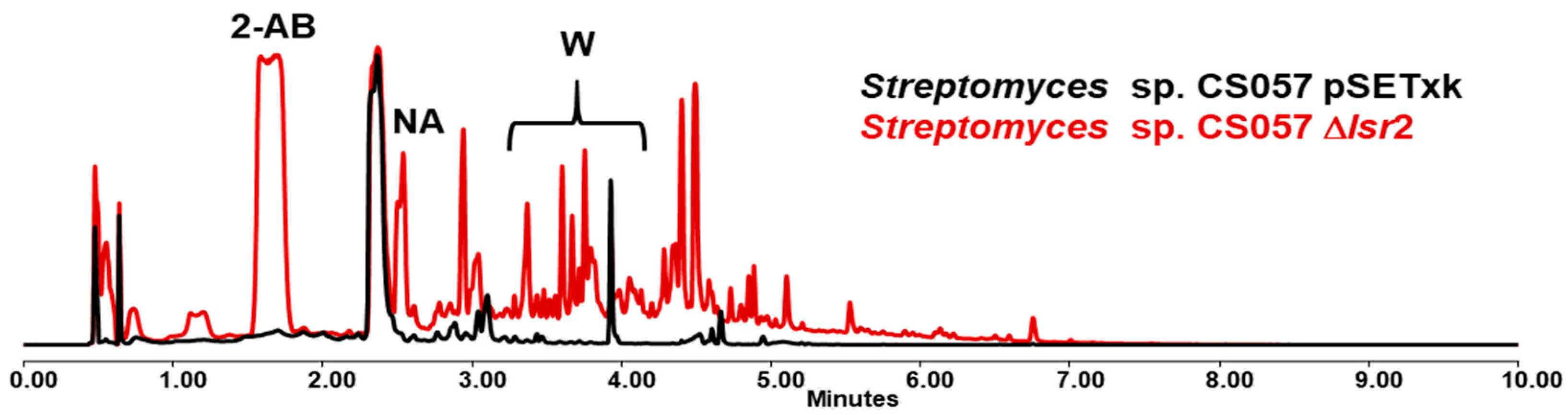
3.2. Streptomyces sp. CS057
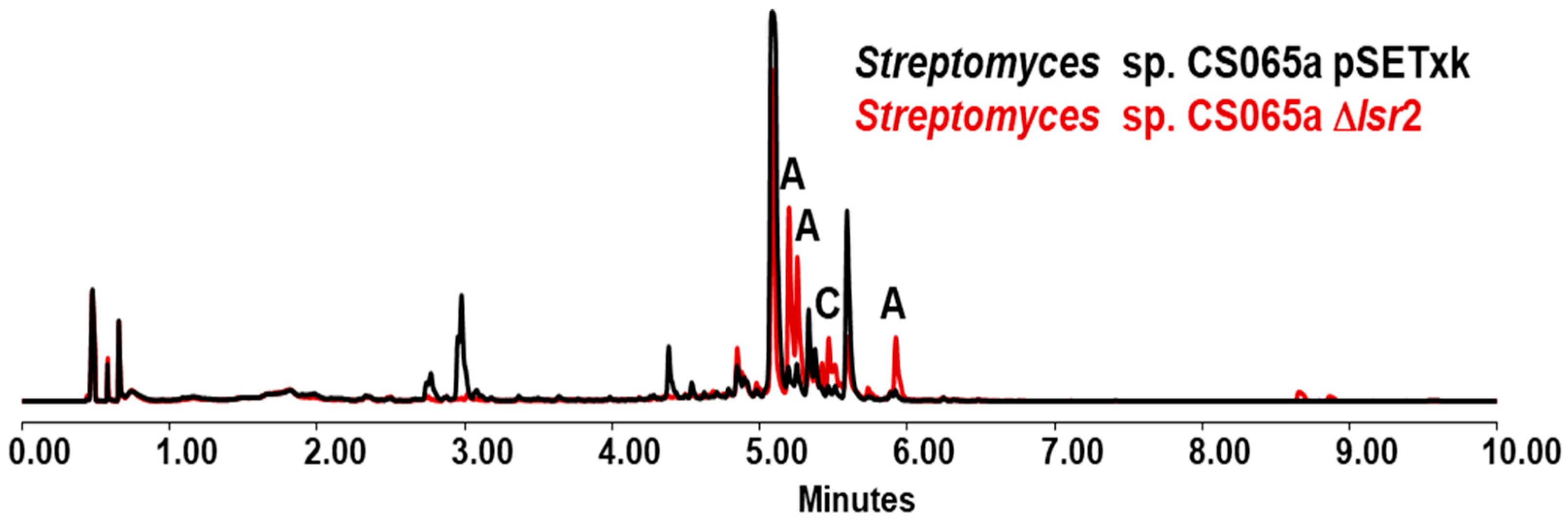
3.3. Streptomyces sp. CS065a
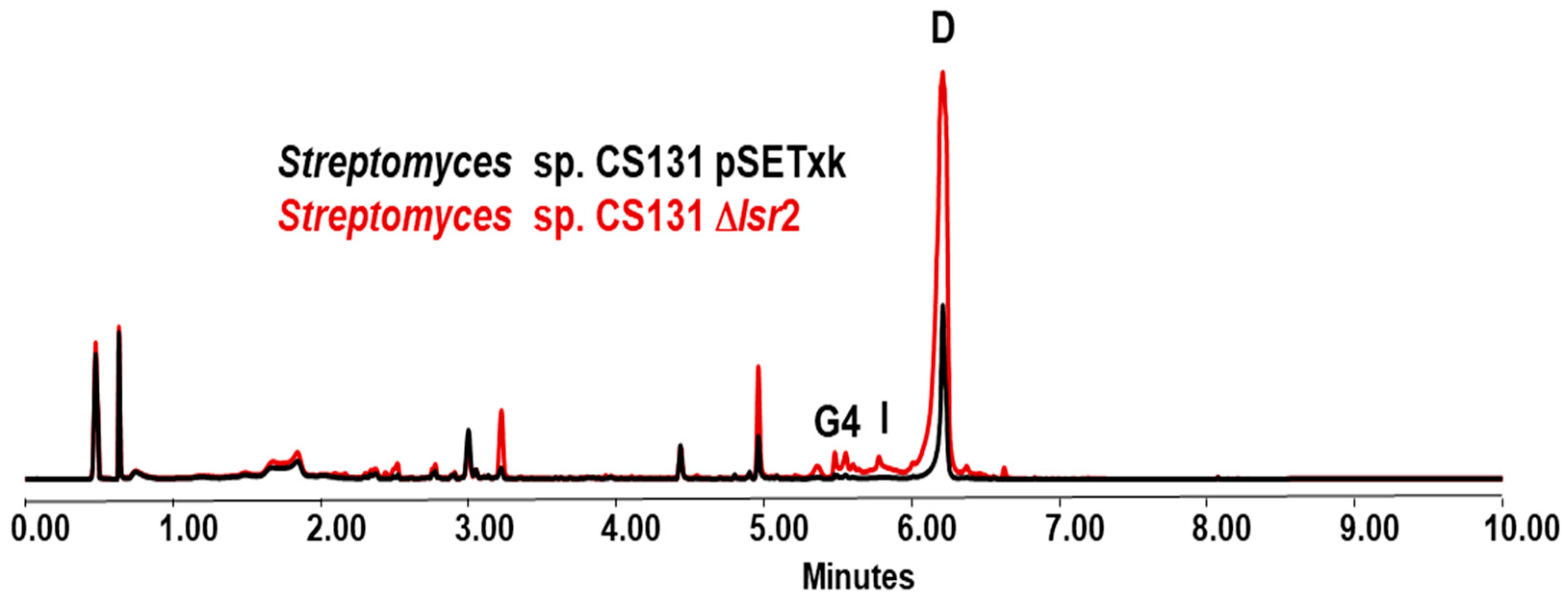
3.4. Streptomyces sp. CS131
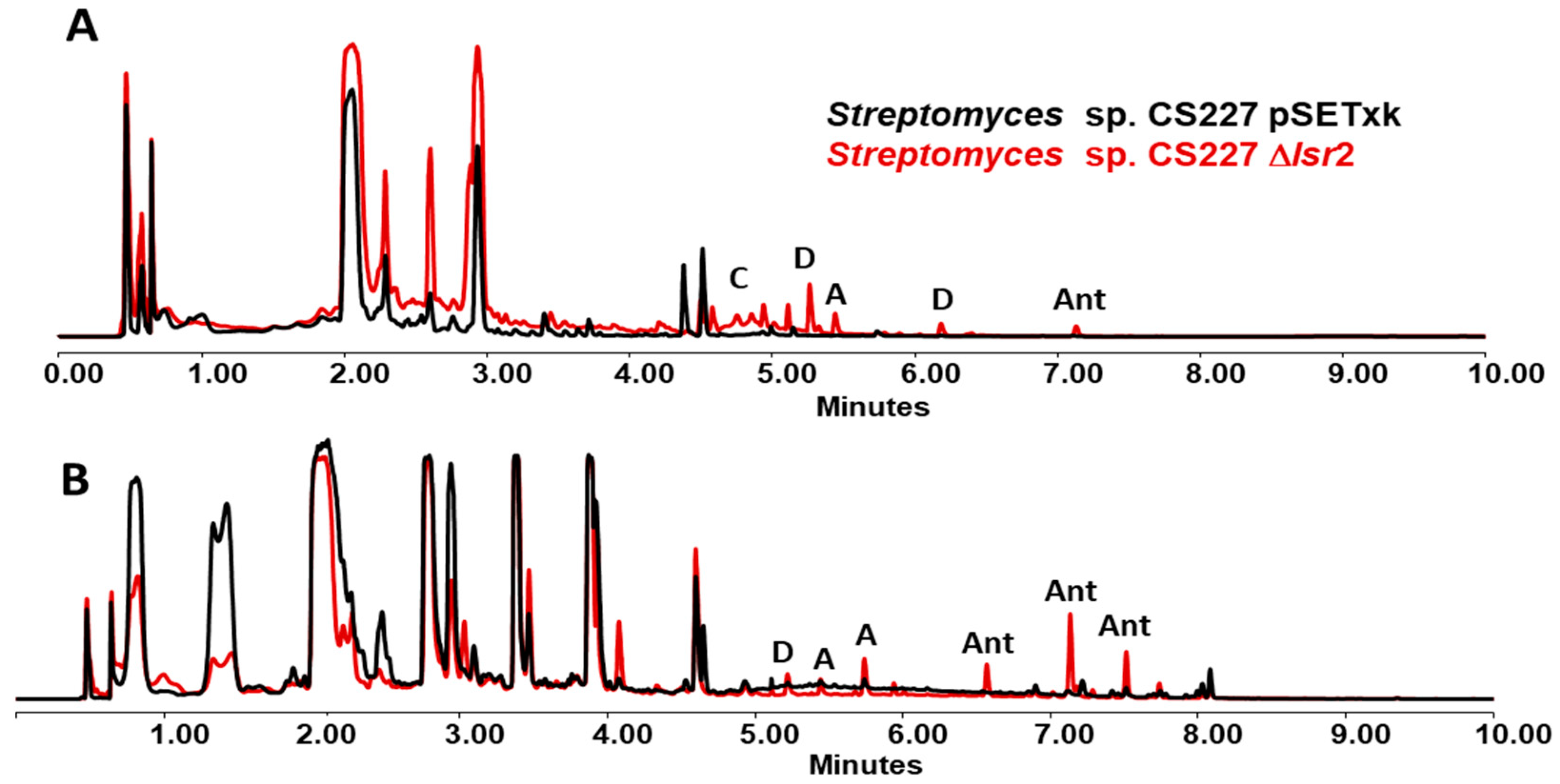
3.5. Streptomyces sp. CS227
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jaishankar, J.; Srivastava, P. Molecular Basis of Stationary Phase Survival and Applications. Front. Microbiol. 2017, 8, 2000. [Google Scholar] [CrossRef] [PubMed]
- Haruta, S.; Kanno, N. Survivability of Microbes in Natural Environments and Their Ecological Impacts. Microbes Environ. 2015, 30, 123–125. [Google Scholar] [CrossRef] [PubMed]
- Prakash, B.; Veeregowda, B.M.; Krishnappa, G. Biofilms: A survival strategy of bacteria. Curr. Sci. 2003, 478, 1299–1307. [Google Scholar]
- Rani, A.; Saini, K.C.; Bast, F.; Varjani, S.; Mehariya, S.; Bhatia, S.K.; Sharma, N.; Funk, C. A Review on Microbial Products and Their Perspective Application as Antimicrobial Agents. Biomolecules 2021, 11, 1860. [Google Scholar] [CrossRef] [PubMed]
- Ostrem Loss, E.M.; Yu, J.H. Bioremediation and Microbial Metabolism of Benzo(a)Pyrene. Mol. Microbiol. 2018, 109, 433–444. [Google Scholar] [CrossRef]
- Cibichakravarthy, B.; Jose, P.A. Biosynthetic Potential of Streptomyces Rationalizes Genome-Based Bioprospecting. Antibiotics 2021, 10, 873. [Google Scholar] [CrossRef]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral Gene Transfer and the Nature of Bacterial Innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef]
- Hołówka, J.; Zakrzewska-Czerwińska, J. Nucleoid Associated Proteins: The Small Organizers That Help to Cope with Stress. Front. Microbiol. 2020, 11, 590. [Google Scholar] [CrossRef]
- Wiechert, J.; Filipchyk, A.; Hünnefeld, M.; Gätgens, C.; Brehm, J.; Heermann, R.; Frunzke, J. Deciphering the Rules Underlying Xenogeneic Silencing and Counter-Silencing of Lsr2-like Proteins Using CgpS of Corynebacterium Glutamicum as a Model. mBio 2020, 11, e02273-19. [Google Scholar] [CrossRef]
- Arnold, B.J.; Huang, I.T.; Hanage, W.P. Horizontal Gene Transfer and Adaptive Evolution in Bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef]
- Daubin, V.; Szöllősi, G.J. Horizontal Gene Transfer and the History of Life. Cold Spring Harb Perspect Biol. 2016, 8, a018036. [Google Scholar] [CrossRef] [PubMed]
- Emamalipour, M.; Seidi, K.; Zununi Vahed, S.; Jahanban-Esfahlan, A.; Jaymand, M.; Majdi, H.; Amoozgar, Z.; Chitkushev, L.T.; Javaheri, T.; Jahanban-Esfahlan, R.; et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front Cell Dev. Biol. 2020, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; Méndez, C.; Salas, J.A. Molecular Insights on the Biosynthesis of Antitumour Compounds by Actinomycetes. Microb. Biotechnol. 2011, 4, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhao, Y.; Huang, C.; Luo, Y. Recent Advances in Silent Gene Cluster Activation in Streptomyces. Front. Bioeng. Biotechnol. 2021, 9, 632230. [Google Scholar] [CrossRef]
- Dorman, C.J. Function of Nucleoid-Associated Proteins in Chromosome Structuring and Transcriptional Regulation. J. Mol. Microbiol. Biotechnol. 2014, 24, 316–331. [Google Scholar] [CrossRef]
- Dillon, S.C.; Dorman, C.J. Bacterial Nucleoid-Associated Proteins, Nucleoid Structure and Gene Expression. Nat. Rev. Microbiol. 2010, 8, 185–195. [Google Scholar] [CrossRef]
- Gordon, B.R.G.; Li, Y.; Wang, L.; Sintsova, A.; Van Bakel, H.; Tian, S.; Navarre, W.W.; Xia, B.; Liu, J. Lsr2 Is a Nucleoid-Associated Protein That Targets AT-Rich Sequences and Virulence Genes in Mycobacterium Tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 5154–5159. [Google Scholar] [CrossRef]
- Landick, R.; Wade, J.T.; Grainger, D.C. H-NS and RNA Polymerase: A Love-Hate Relationship? Curr. Opin. Microbiol. 2015, 24, 53–59. [Google Scholar] [CrossRef]
- Lim, C.J.; Lee, S.Y.; Kenney, L.J.; Yan, J. Nucleoprotein Filament Formation Is the Structural Basis for Bacterial Protein H-NS Gene Silencing. Sci. Rep. 2012, 2, 509. [Google Scholar] [CrossRef]
- Alam, K.; Mazumder, A.; Sikdar, S.; Zhao, Y.M.; Hao, J.; Song, C.; Wang, Y.; Sarkar, R.; Islam, S.; Zhang, Y.; et al. Streptomyces: The Biofactory of Secondary Metabolites. Front. Microbiol. 2022, 13, 968053. [Google Scholar] [CrossRef]
- Dhandapani, R.; Thangavelu, S.; Ragunathan, L.; Paramasivam, R.; Velmurugan, P.; Muthupandian, S. Potential Bioactive Compounds from Marine Streptomyces sp. and Their In Vitro Antibiofilm and Antibacterial Activities Against Antimicrobial-Resistant Clinical Pathogens. Appl. Biochem. Biotechnol. 2022, 194, 4702–4723. [Google Scholar] [CrossRef] [PubMed]
- Quach, N.T.; Nguyen, Q.H.; Vu, T.H.N.; Le, T.T.H.; Ta, T.T.T.; Nguyen, T.D.; Van Doan, T.; Van Nguyen, T.; Dang, T.T.; Nguyen, X.C.; et al. Plant-Derived Bioactive Compounds Produced by Streptomyces Variabilis LCP18 Associated with Litsea Cubeba (Lour.) Pers as Potential Target to Combat Human Pathogenic Bacteria and Human Cancer Cell Lines. Braz. J. Microbiol. 2021, 52, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Malmierca, M.G.; González-Montes, L.; Pérez-Victoria, I.; Sialer, C.; Braña, A.F.; Salcedo, R.G.; Martín, J.; Reyes, F.; Méndez, C.; Olano, C.; et al. Searching for Glycosylated Natural Products in Actinomycetes and Identification of Novel Macrolactams and Angucyclines. Front. Microbiol. 2018, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Prado-Alonso, L.; Pérez-Victoria, I.; Malmierca, M.G.; Montero, I.; Rioja-Blanco, E.; Martín, J.; Reyes, F.; Méndez, C.; Salas, J.A.; Olano, C. Colibrimycins, Novel Halogenated Hybrid Polyketide Synthase-Nonribosomal Peptide Synthetase (PKS-NRPS) Compounds Produced by Streptomyces sp. Strain CS147. Appl. Environ. Microbiol. 2022, 88, e01839-21. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, L.; Braña, A.F. Cell Density Influences Antibiotic Biosynthesis in Streptomyces Clavuligerus. Microbiology 1996, 142, 1209–1220. [Google Scholar] [CrossRef][Green Version]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Fernandez, E.; Fernandez, F.; Weißbach, U.; Sar, C.É.; Sa’, S.; Reillo, S.; Braña, A.F.; Braña, B.; Mendez, C.; Ju¨, J.; et al. Identification of Two Genes from Streptomyces Argillaceus Encoding Glycosyltransferases Involved in Transfer of a Disaccharide during Biosynthesis of the Antitumor Drug Mithramycin. J. Bacteriol. 1998, 180, 4929–4937. [Google Scholar] [CrossRef]
- Hobbs, G.; Frazer, C.M.; Gardner, D.C.J.; Cullum, J.A.; Oliver, S.G. Dispersed Growth of Streptomyces in Liquid Culture. Appl. Microbiol. Biotechnol. 1989, 31, 272–277. [Google Scholar] [CrossRef]
- Cuervo, L.; Méndez, C.; Salas, J.A.; Olano, C.; Malmierca, M.G. Volatile Compounds in Actinomycete Communities: A New Tool for Biosynthetic Gene Cluster Activation, Cooperative Growth Promotion, and Drug Discovery. Cells 2022, 11, 3510. [Google Scholar] [CrossRef]
- Peláez, A.; Ribas-Aparicio, R.; Gómez, A.; Rodicio, M. Structural and Functional Characterization of the RecR Gene of Streptomyces. Mol. Genet. Genom. 2001, 265, 663–672. [Google Scholar] [CrossRef]
- Sun, Y.; He, X.; Liang, J.; Zhou, X.; Deng, Z. Analysis of Functions in Plasmid PHZ1358 Influencing Its Genetic and Structural Stability in Streptomyces Lividans 1326. Appl. Microbiol. Biotechnol. 2009, 82, 303–310. [Google Scholar] [CrossRef]
- Cuervo, L.; Malmierca, M.G.; García-Salcedo, R.; Méndez, C.; Salas, J.A.; Olano, C.; Ceniceros, A. Co-Expression of Transcriptional Regulators and Housekeeping Genes in Streptomyces Spp.: A Strategy to Optimize Metabolite Production. Microorganisms 2023, 11, 1585. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for Occurrences of a given Motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Gordon, B.R.G.; Li, Y.; Cote, A.; Weirauch, M.T.; Ding, P.; Hughes, T.R.; Navarre, W.W.; Xia, B.; Liu, J. Structural Basis for Recognition of AT-Rich DNA by Unrelated Xenogeneic Silencing Proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 10690–10695. [Google Scholar] [CrossRef] [PubMed]
- Vior, N.M.; Olano, C.; García, I.; Méndez, C.; Salas, J.A. Collismycin a Biosynthesis in Streptomyces sp. CS40 Is Regulated by Iron Levels through Two Pathway-Specific Regulators. Microbiology 2014, 160, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Gomi, S.; Amano, S.; Sato, E.; Miyadoh, S.; Kodama, Y. Novel antibiotics SF2738A, B and C, and their analogs produced by Streptomyces sp. J. Antibiot. 1994, 47, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Toriya, M.; Yaginuma, S.; Murofushi, S.; Ogawa, K.; Muto, N.; Hayashi, M.; Matsumoto, K. Zincphyrin, a novel coproporphyrin III with zinc from Streptomyces sp. J. Antibiot. 1993, 46, 196–200. [Google Scholar] [CrossRef]
- Wattana-Amorn, P.; Charoenwongsa, W.; Williams, C.; Crump, M.P.; Apichaisataienchote, B. Antibacterial Activity of Cyclo(L-Pro-L-Tyr) and Cyclo(D-Pro-L-Tyr) from Streptomyces sp. Strain 22–4 against Phytopathogenic Bacteria. Nat. Prod. Res. 2016, 30, 1980–1983. [Google Scholar] [CrossRef]
- Cuervo, L.; Méndez, C.; Salas, J.A.; Olano, C.; Malmierca, M.G. Volatile Communication in Actinobacteria: A Language for Secondary Metabolism Regulation. Microb. Cell Fact. 2024, 23, 181. [Google Scholar] [CrossRef]
- Helaly, S.E.; Goodfellow, M.; Zinecker, H.; Imhoff, J.F.; Süssmuth, R.D.; Fiedler, H.P. Warkmycin, a Novel Angucycline Antibiotic Produced by Streptomyces sp. Acta 2930 *. J. Antibiot. 2013, 66, 669–674. [Google Scholar] [CrossRef]
- Gil, J.A.; Naharro, G.; Villanueva, J.R.; Martín, J.F. Characterization and Regulation of P-Aminobenzoic Acid Synthase from Stveptomyces Gviseus. J. Gen. Microbiol. 1989, 131, 1279–1287. [Google Scholar]
- Nirwati, H.; Damayanti, E.; Sholikhah, E.N.; Mutofa, M.; Widada, J. Soil-Derived Streptomyces sp. GMR22 Producing Antibiofilm Activity against Candida Albicans: Bioassay, Untargeted LC-HRMS, and Gene Cluster Analysis. Heliyon 2022, 8, e09333. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; García, I.; González, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sánchez-Hidalgo, M.; Braña, A.F.; Méndez, C.; Salas, J.A. Activation and Identification of Five Clusters for Secondary Metabolites in Streptomyces AlbusJ1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zeng, J.; Cui, P.; Wang, W.; Yu, D.; Zhan, J. Manipulation of Two Regulatory Genes for Efficient Production of Chromomycins in Streptomyces Reseiscleroticus. J. Biol. Eng. 2018, 12, 9. [Google Scholar] [CrossRef]
- Machushynets, N.V.; Elsayed, S.S.; Du, C.; Siegler, M.A.; de la Cruz, M.; Genilloud, O.; Hankemeier, T.; van Wezel, G.P. Discovery of Actinomycin L, a New Member of the Actinomycin Family of Antibiotics. Sci. Rep. 2022, 12, 2813. [Google Scholar] [CrossRef]
- Shiomi, K.; Hatae, K.; Hatano, H.; Matsumoto, A.; Takahashi, Y.; Jiang, C.-L.; Tomoda, H.; Kobayashi, S.; Tanaka, H.; Mura, S.O.; et al. A New Antibiotic, Antimycin A 9, Produced by Streptomyces sp. K01-0031. J. Antibiot. 2005, 58, 74–78. [Google Scholar] [CrossRef]
- Haeder, S.; Wirth, R.; Herz, H.; Spiteller, D. Candicidin-Producing Streptomyces Support Leaf-Cutting Ants to Protect Their Fungus Garden against the Pathogenic Fungus Escovopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 4742–4746. [Google Scholar] [CrossRef]
- Jakobi, M.; Winkelmann, G.; Kaiser, D.; Kempter, C.; Jung, G.; Berg, G.; Bahl, H. Maltophilin: A New Antifimgal Compound Produced by Stenotvophomonas Maltophilia R3089. J. Antibiot. 1996, 49, 1101–1104. [Google Scholar] [CrossRef]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The Sound of Silence: Activating Silent Biosynthetic Gene Clusters in Marine Microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef]
- Piña-Iturbe, A.; Suazo, I.D.; Hoppe-Elsholz, G.; Ulloa-Allendes, D.; González, P.A.; Kalergis, A.M.; Bueno, S.M. Horizontally Acquired Homologs of Xenogeneic Silencers: Modulators of Gene Expression Encoded by Plasmids, Phages and Genomic Islands. Genes 2020, 11, 142. [Google Scholar] [CrossRef]
- Gehrke, E.J.; Zhang, X.; Pimentel-Elardo, S.M.; Johnson, A.R.; Rees, C.A.; Jones, S.E.; Gehrke, S.S.; Turvey, S.; Boursalie, S.; Hill, J.E.; et al. Silencing Cryptic Specialized Metabolism in Streptomyces by the Nucleoid-Associated Protein Lsr2. Microbiol. Infect. Dis. 2019, 8, e47691. [Google Scholar] [CrossRef] [PubMed]
- Cendra, M.d.M.; Juárez, A.; Madrid, C.; Torrents, E. H-NS Is a Novel Transcriptional Modulator of the Ribonucleotide Reductase Genes in Escherichia Coli. J. Bacteriol. 2013, 195, 4255–4263. [Google Scholar] [CrossRef] [PubMed]
- Rico, S.; Santamaría, R.I.; Yepes, A.; Rodríguez, H.; Laing, E.; Bucca, G.; Smith, C.P.; Díaz, M. Deciphering the Regulon of Streptomyces Coelicolor AbrC3, a Positive Response Regulator of Antibiotic Production. Appl. Envrion. Microbiol. 2014, 80, 2417–2428. [Google Scholar] [CrossRef] [PubMed]
- Prudence, S.M.M.; Addington, E.; Castaño-Espriu, L.; Mark, D.R.; Pintor-Escobar, L.; Russell, A.H.; McLean, T.C. Advances in Actinomycete Research: An Actinobase Review of 2019. Microbiology 2020, 166, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Kołodziej, M.; Trojanowski, D.; Bury, K.; Hołówka, J.; Matysik, W.; Kąkolewska, H.; Feddersen, H.; Giacomelli, G.; Konieczny, I.; Bramkamp, M.; et al. Lsr2, a Nucleoid-Associated Protein Influencing Mycobacterial Cell Cycle. Sci. Rep. 2021, 11, 2910. [Google Scholar] [CrossRef]
- Kołodziej, M.; Łebkowski, T.; Płociński, P.; Hołówka, J.; Paściak, M.; Wojtaś, B.; Bury, K.; Konieczny, I.; Dziadek, J.; Zakrzewska-Czerwińska, J. Lsr2 and Its Novel Paralogue Mediate the Adjustment of Mycobacterium Smegmatis to Unfavorable Environmental Conditions. mSphere 2021, 6, 1110–1128. [Google Scholar] [CrossRef]
- Bartek, I.L.; Woolhiser, L.K.; Baughn, A.D.; Basaraba, R.J.; Jacobs, W.R.; Lenaerts, A.J.; Voskuil, M.I. Mycobacterium Tuberculosis Lsr2 Is a Global Transcriptional Regulator Required for Adaptation to Changing Oxygen Levels and Virulence. mBio 2014, 5, 3. [Google Scholar] [CrossRef]
- Chen, J.M.; German, G.J.; Alexander, D.C.; Ren, H.; Tan, T.; Liu, J. Roles of Lsr2 in Colony Morphology and Biofilm Formation of Mycobacterium Smegmatis. J. Bacteriol. 2006, 188, 633–641. [Google Scholar] [CrossRef]
- Bradshaw, E.; Saalbach, G.; McArthur, M. Proteomic Survey of the Streptomyces Coelicolor Nucleoid. J. Proteom. 2013, 83, 37–46. [Google Scholar] [CrossRef]
- Summers, E.L.; Meindl, K.; Usón, I.; Mitra, A.K.; Radjainia, M.; Colangeli, R.; Alland, D.; Arcus, V.L. The Structure of the Oligomerization Domain of Lsr2 from Mycobacterium Tuberculosis Reveals a Mechanism for Chromosome Organization and Protection. PLoS ONE 2012, 7, e38542. [Google Scholar] [CrossRef]
- Olano, C.; Rodríguez, M. Actinomycetes Associated with Arthropods as a Source of New Bioactive Compounds. Curr. Issues Mol. Biol. 2024, 46, 3822–3838. [Google Scholar] [CrossRef] [PubMed]
- Ceniceros, A.; Cuervo, L.; Méndez, C.; Salas, J.A.; Olano, C.; Malmierca, M.G. A Multidisciplinary Approach to Unraveling the Natural Product Biosynthetic Potential of a Streptomyces Strain Collection Isolated from Leaf-Cutting Ants. Microorganisms 2021, 9, 2225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Andres, S.N.; Elliot, M.A. Interplay between Nucleoid-Associated Proteins and Transcription Factors in Controlling Specialized Metabolism in Streptomyces. Mbio 2021, 12, 1077–1098. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Burgo, Y.; Santos-Aberturas, J.; Rodríguez-García, A.; Barreales, E.G.; Tormo, J.R.; Truman, A.W.; Reyes, F.; Aparicio, J.F.; Liras, P. Activation of Secondary Metabolite Gene Clusters in Streptomyces Clavuligerusby the Pimm Regulator of Streptomyces Natalensis. Front. Microbiol. 2019, 10, 580. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Álvarez, R.; Botas, A.; Albillos, S.M.; Rumbero, A.; Martín, J.F.; Liras, P. Molecular Genetics of Naringenin Biosynthesis, a Typical Plant Secondary Metabolite Produced by Streptomyces clavuligerus. Microb. Cell Fact. 2015, 14, 178. [Google Scholar] [CrossRef]
- Luo, Y.; Huang, H.; Liang, J.; Wang, M.; Lu, L.; Shao, Z.; Cobb, R.E.; Zhao, H. Activation and Characterization of a Cryptic Polycyclic Tetramate Macrolactam Biosynthetic Gene Cluster. Nat. Commun. 2013, 4, 2894. [Google Scholar] [CrossRef]
- Xia, H.; Li, X.; Li, Z.; Zhan, X.; Mao, X.; Li, Y. The Application of Regulatory Cascades in Streptomyces: Yield Enhancement and Metabolite Mining. Front. Microbiol. 2020, 11, 406. [Google Scholar] [CrossRef]
- Álvarez-Álvarez, R.; Rodríguez-García, A.; Santamarta, I.; Pérez-Redondo, R.; Prieto-Domínguez, A.; Martínez-Burgo, Y.; Liras, P. Transcriptomic Analysis of Streptomyces Clavuligerus ΔccaR: Tsr: Effects of the Cephamycin C-Clavulanic Acid Cluster Regulator CcaR on Global Regulation. Microb. Biotechnol. 2014, 7, 221–231. [Google Scholar] [CrossRef]
- Hwang, S.; Lee, N.; Jeong, Y.; Lee, Y.; Kim, W.; Cho, S.; Palsson, B.O.; Cho, B.K. Primary Transcriptome and Translatome Analysis Determines Transcriptional and Translational Regulatory Elements Encoded in the Streptomyces Clavuligerus Genome. Nucleic Acids Res. 2019, 47, 6114–6129. [Google Scholar] [CrossRef]
- Zorro-Aranda, A.; Escorcia-Rodríguez, J.M.; González-Kise, J.K.; Freyre-González, J.A. Curation, Inference, and Assessment of a Globally Reconstructed Gene Regulatory Network for Streptomyces Coelicolor. Sci. Rep. 2022, 12, 2840. [Google Scholar] [CrossRef]
- Šmídová, K.; Ziková, A.; Pospíšil, J.; Schwarz, M.; Bobek, J.; Vohradsky, J. DNA Mapping and Kinetic Modeling of the HrdB Regulon in Streptomyces Coelicolor. Nucleic Acids Res. 2019, 47, 621–633. [Google Scholar] [CrossRef] [PubMed]


| Motif 1: ATTTAAAT | Motif 7: AATTAAAT | Motif 13: TATATTAA |
| Motif 2: AATTTAAT | Motif 8: AAATAATT | Motif 14: AAATATTT |
| Motif 3: TAATTTAA | Motif 9: AATAAATT | Motif 15: ATATAATA |
| Motif 4: AATTTAAA | Motif 10: ATTATAAT | Motif 16: TAAAATAA |
| Motif 5: ATATTAAT | Motif 11: AATAAATA | Motif 17: TTAATTAA |
| Motif 6: AAATAAAT | Motif 12: AATTATAA | Motif 18: AAATTATA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuervo, L.; Malmierca, M.G.; Olano, C. An Overview of Lsr2 Repressor Effect in Streptomyces spp. Secondary Metabolism. Microorganisms 2024, 12, 2317. https://doi.org/10.3390/microorganisms12112317
Cuervo L, Malmierca MG, Olano C. An Overview of Lsr2 Repressor Effect in Streptomyces spp. Secondary Metabolism. Microorganisms. 2024; 12(11):2317. https://doi.org/10.3390/microorganisms12112317
Chicago/Turabian StyleCuervo, Lorena, Mónica G. Malmierca, and Carlos Olano. 2024. "An Overview of Lsr2 Repressor Effect in Streptomyces spp. Secondary Metabolism" Microorganisms 12, no. 11: 2317. https://doi.org/10.3390/microorganisms12112317
APA StyleCuervo, L., Malmierca, M. G., & Olano, C. (2024). An Overview of Lsr2 Repressor Effect in Streptomyces spp. Secondary Metabolism. Microorganisms, 12(11), 2317. https://doi.org/10.3390/microorganisms12112317






