Roles and Regulation of Quorum Sensing of Acidophiles in Bioleaching: A Review
Abstract
:1. Introduction
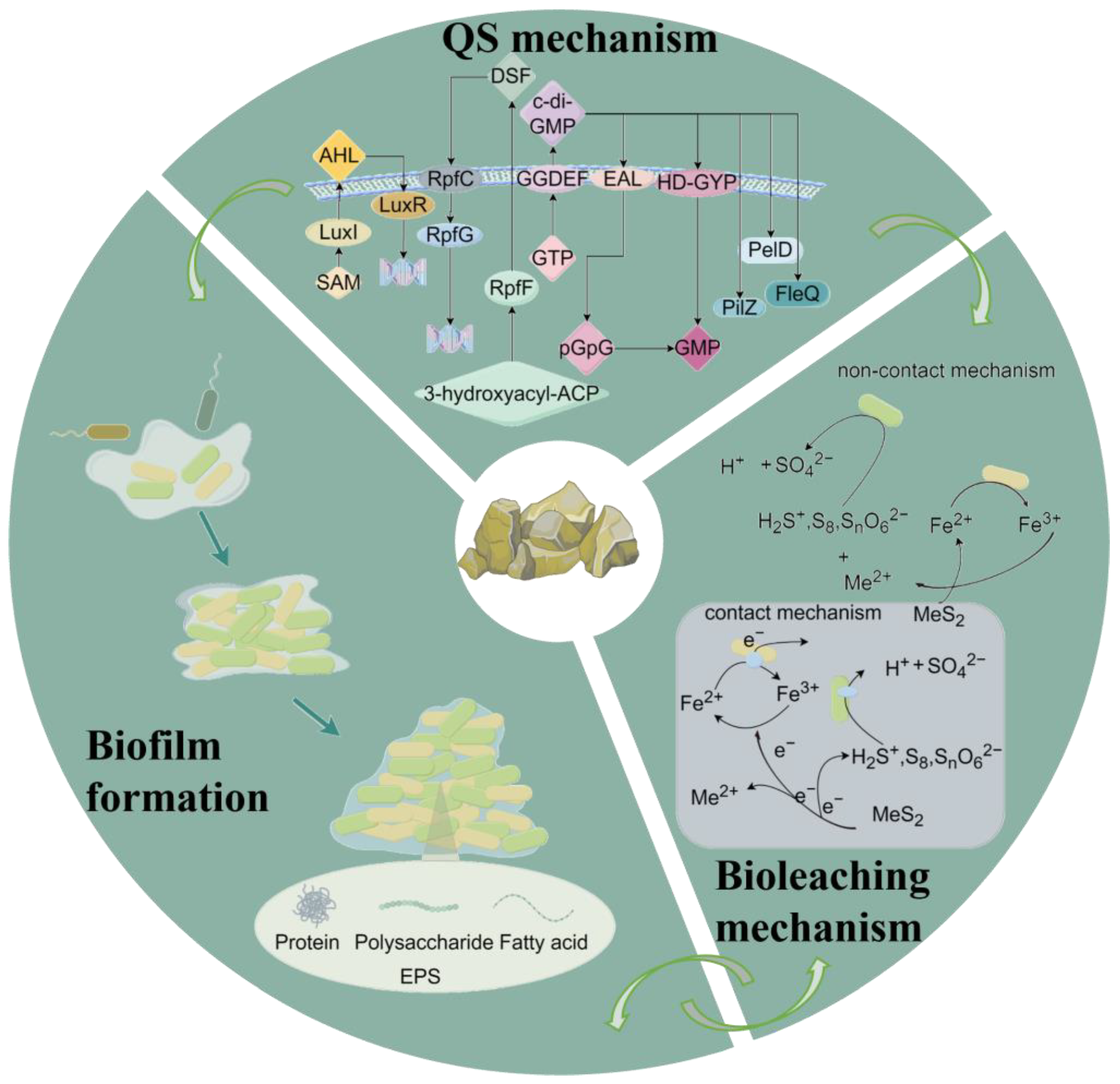
2. QS and Second Messenger in Acidophiles
2.1. N-Acyl Homoserine Lactones System
2.2. Diffusible Signal Factor System
2.3. Cyclic Dimeric Guanosine Monophosphate System
3. Interactions between Different Systems
4. The Role of QS in Bioleaching
4.1. Cell Morphology
4.2. Community Structure
4.3. Biofilm Formation
4.4. Microbial Metabolism
5. Regulation of QS in Bioleaching
5.1. Endogenous Regulation
5.2. Exogenous Regulation
5.2.1. N-Acyl Homoserine Lactones
5.2.2. N-Acyl Homoserine Lactones Analogues
5.2.3. Diffusible Signal Factor
6. Research Needs and Future Direction
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Full Name |
| QS | quorum sensing |
| EPS | extracellular polymers |
| AIs | autoinducers |
| AHL | N-acyl homoserine lactones |
| c-di-GMP | cyclic di-guanosine |
| DGCs | diguanylate cyclases |
| PDEs | phosphodiesterases |
| pGpG | 5′-phosphoguanylyl-(3′-5′)-guanosine |
| DSF | diffusible signal factor |
| BDSF | Burkholderia diffusible signal factor |
| AHL-QS | AfeI/R-type QS system |
| SAM | S-adenosyl-L-methionine |
| acyl-ACP | acyl–acyl carrier protein |
| HSL | homoserine lactone |
| OH-HSL | hydroxy-homoserine lactone |
| oxo-HSL | oxygen-homoserine lactone |
| 3-oxo-C8-HSL | N-(3-Oxooctanoyl)-L-homoserine lactone |
| 3-OH-C14-HSL | N-(3-Hydroxytetradecanoyl)-DL-homoserine lactone |
References
- Wang, X.; Ma, L.; Wu, J.; Xiao, Y.; Tao, J.; Liu, X.D. Effective bioleaching of low-grade copper ores: Insights from microbial cross experiments. Bioresour. Technol. 2020, 308, 123273. [Google Scholar] [CrossRef]
- Gu, T.; Rastegar, S.O.; Mousavi, S.M.; Li, M.; Zhou, M. Advances in bioleaching for recovery of metals and bioremediation of fuel ash and sewage sludge. Bioresour. Technol. 2018, 261, 428–440. [Google Scholar] [CrossRef]
- Roberto, F.F.; Schippers, A. Progress in bioleaching: Part B, applications of microbial processes by the minerals industries. Appl. Microbiol. Biotechnol. 2022, 106, 5913–5928. [Google Scholar] [CrossRef]
- Zhang, R.; Bellenberg, S.; Neu, T.; Sand, W.; Vera, M. The biofilm lifestyle of acidophilic metal/sulfur-oxidizing microorganisms. In Biotechnology of Extremophiles: Grand Challenges in Biology and Biotechnology; Rampelotto, P., Ed.; Springer: Cham, Switzerland, 2016; pp. 177–213. [Google Scholar]
- Li, L.; Liu, Z.; Zhang, M.; Meng, D.; Liu, X.; Wang, P.; Li, X.; Jiang, Z.; Zhong, S.; Jiang, C.; et al. Insights into the metabolism and evolution of the genus Acidiphilium, a typical acidophile in acid mine drainage. mSystems 2020, 5, e00867-20. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Liang, Y.; Xiao, Y.; Ma, L.; Guo, X.; Miao, B.; Liu, H.; Peng, D.; Huang, W.; et al. Comparative genomics unravels the functional roles of co-occurring acidophilic bacteria in bioleaching heaps. Front. Microbiol. 2017, 8, 790. [Google Scholar] [CrossRef]
- Rawlings, D.E. Heavy metal mining using microbes. Annu. Rev. Microbiol. 2002, 56, 65–91. [Google Scholar] [CrossRef]
- Sand, W.; Gehrke, T.; Jozsa, P.; Schippers, A. (Bio)chemistry of bacterial leaching—Direct vs. indirect bioleaching. Hydrometallurgy 2001, 59, 159–175. [Google Scholar] [CrossRef]
- Quatrini, R.; Johnson, D.B. Microbiomes in extremely acidic environments: Functionalities and interactions that allow survival and growth of prokaryotes at low pH. Curr. Opin. Microbiol. 2018, 43, 139–147. [Google Scholar] [CrossRef]
- Ma, L.; Huang, S.; Wu, P.; Xiong, J.; Wang, H.; Liao, H.; Liu, X. The interaction of acidophiles driving community functional responses to the re-inoculated chalcopyrite bioleaching process. Sci. Total Environ. 2021, 798, 149186. [Google Scholar] [CrossRef]
- Vardanyan, A.; Vardanyan, N.; Khachatryan, A.; Zhang, R.; Sand, W. Adhesion to mineral surfaces by cells of Leptospirillum, Acidithiobacillus and Sulfobacillus from Armenian sulfide ores. Minerals 2019, 9, 69. [Google Scholar] [CrossRef]
- Moreno-Paz, M.; Gómez, M.J.; Arcas, A.; Parro, V. Environmental transcriptome analysis reveals physiological differences between biofilm and planktonic modes of life of the iron oxidizing bacteria Leptospirillum spp. in their natural microbial community. BMC Genom. 2010, 11, 404. [Google Scholar] [CrossRef]
- Ye, M.; Liang, J.; Liao, X.; Li, L.; Feng, X.; Qian, W.; Zhou, S.; Sun, S. Bioleaching for detoxification of waste flotation tailings: Relationship between EPS substances and bioleaching behavior. J. Environ. Manag. 2020, 279, 111795. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Sand, W. Mechanical and chemical studies on EPS from Sulfobacillus thermosulfidooxidans: From planktonic to biofilm cells. Colloids Surf. B 2017, 153, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, X.; Wang, J.; Yuan, X.; Jiang, X.; Wang, Y.; Zhong, C.; Xu, D.; Gu, T.; Wang, F. Bacterial biofilms as platforms engineered for diverse applications. Biotechnol. Adv. 2022, 57, 107932. [Google Scholar] [CrossRef] [PubMed]
- Rohwerder, T.; Gehrke, T.; Kinzler, K.; Sand, W. Bioleaching review part A: Progress in bioleaching: Fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 2003, 63, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Bellenberg, S.; Díaz, M.; Noël, N.; Sand, W.; Poetsch, A.; Guiliani, N.; Vera, M. Biofilm formation, communication and interactions of leaching bacteria during colonization of pyrite and sulfur surfaces. Res. Microbiol. 2014, 165, 773–781. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, X.; Liang, Y.; Fan, F.; Zhang, X.; Yin, H. Metabolic diversity and adaptive mechanisms of iron- and/or sulfur-oxidizing autotrophic acidophiles in extremely acidic environments. Environ. Microbiol. Rep. 2016, 8, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-X.; Huang, L.-N.; Méndez-García, C.; Kuang, J.; Hua, Z.; Liu, J.; Shu, W. Microbial communities, processes and functions in acid mine drainage ecosystems. Curr. Opin. Biotechnol. 2016, 38, 150–158. [Google Scholar] [CrossRef]
- Amouric, A.; Brochier-Armanet, C.; Johnson, D.B.; Bonnefoy, V.; Hallberg, K.B. Phylogenetic and genetic variation among Fe(II)-oxidizing acidithiobacilli supports the view that these comprise multiple species with different ferrous iron oxidation pathways. Microbiology 2011, 157, 111–122. [Google Scholar] [CrossRef]
- Weiner, J.H.; Ullrich, S.R.; Poehlein, A.; Tischler, J.S.; González, C.; Ossandon, F.J.; Daniel, R.; Holmes, D.S.; Schlömann, M.; Mühling, M. Genome analysis of the biotechnologically relevant acidophilic iron oxidising strain JA12 indicates phylogenetic and metabolic diversity within the novel genus “Ferrovum”. PLoS ONE 2016, 11, e0146832. [Google Scholar] [CrossRef]
- Méndez-García, C.; Mesa, V.; Sprenger, R.; Richter, M.; Diez, M.; Solano, J.; Bargiela, R.; Golyshina, O.; Manteca, Á.; Ramos, J.; et al. Microbial stratification in low pH oxic and suboxic macroscopic growths along an acid mine drainage. ISME J. 2014, 8, 1259–1274. [Google Scholar] [CrossRef] [PubMed]
- Urbieta, M.S.; Rascovan, N.; Vázquez, M.P.; Donati, E. Genome analysis of the thermoacidophilic archaeon Acidianus copahuensis focusing on the metabolisms associated to biomining activities. BMC Genom. 2017, 18, 445. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Wang, H.; Zhang, Z.; Li, K.; Zhang, X.; Mora-López, M.; Jiang, C.; Liu, C.; Wang, L.; Zhu, Y.; et al. Genome sequencing of sulfolobus sp. A20 from Costa Rica and comparative analyses of the putative pathways of carbon, nitrogen, and sulfur metabolism in Various Sulfolobus Strains. Front. Microbiol. 2016, 7, 1902. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Winzer, K.; Chan, W.C.; Camara, M. Look who’s talking: Communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. B 2007, 362, 1119–1134. [Google Scholar] [CrossRef]
- Sharma, A.; Singh, P.; Sarmah, B.K.; Nandi, S.P. Quorum sensing: Its role in microbial social networking. Res. Microbiol. 2020, 171, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C.; Greenberg, E.P. Listening in on bacteria: Acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 2002, 3, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Bassler, B.L.; Losick, R. Bacterially speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef]
- Williams, P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 2007, 153, 3923–3938. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, J.; Liu, C.; Yang, A.; Qiao, J. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 2020, 77, 1319–1343. [Google Scholar] [CrossRef]
- Kuo, A.; Blough, N.V.; Dunlap, P.V. Multiple N-acyl-L-homoserine lactone autoinducers of Luminescence in the marine symbiotic bacterium Vibrio fischerit. J. Bacteriol. 1994, 176, 7558–7565. [Google Scholar] [CrossRef]
- Higgins, D.A.; Pomianek, M.E.; Kraml, C.M.; Taylor, R.K.; Semmelhack, M.F.; Bassler, B.L. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 2007, 450, 883–886. [Google Scholar] [CrossRef]
- Pesci, E.C.; Milbank, J.B.; Pearson, J.P.; McKnight, S.; Kende, A.S.; Greenberg, E.P.; Iglewski, B.H. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 11229–11234. [Google Scholar] [CrossRef]
- Sperandio, V.; Torres, A.G.; Jarvis, B.; Nataro, J.P.; Kaper, J.B. Bacteria–host communication: The language of hormones. Proc. Natl. Acad. Sci. USA 2003, 100, 8951–8956. [Google Scholar] [CrossRef] [PubMed]
- Barber, C.E.; Tang, J.L.; Feng, J.X.; Pan, M.Q.; Wilson, T.J.G.; Slater, H.; Dow, J.M.; Williams, P.; Daniels, M.J. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 2003, 24, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Vera, M.; Schippers, A.; Hedrich, S.; Sand, W. Progress in bioleaching: Fundamentals and mechanisms of microbial metal sulfide oxidation—Part A. Appl. Microbiol. Biotechnol. 2022, 106, 6933–6952. [Google Scholar] [CrossRef] [PubMed]
- Farah, C.; Vera, M.; Morin, D.; Haras, D.; Jerez, C.; Guiliani, N. Evidence for a functional quorum-sensing type AI-1 system in the extremophilic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2005, 71, 7033–7040. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.; Seeger, M.; Jedlicki, E.; Holmes, D. Second acyl homoserine lactone production system in the extreme acidophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2007, 73, 3225–3231. [Google Scholar] [CrossRef]
- Bellenberg, S.; Salas, B.; Ganji, S.; Jorquera-Román, C.; Valenzuela, M.; Buetti-Dinh, A.; Unelius, C.R.; Dopson, M.; Vera, M. Diffusible signal factor signaling controls bioleaching activity and niche protection in the acidophilic, mineral-oxidizing leptospirilli. Sci. Rep. 2021, 11, 16275. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; Castro, M.; Barriga, A.; Jerez, C.; Guiliani, N. The extremophile Acidithiobacillus ferrooxidans possesses a c-di-GMP signalling pathway that could play a significant role during bioleaching of minerals. Lett. Appl. Microbiol. 2012, 54, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liu, X.; Yang, W.; Ma, L.; Li, H.; Liu, R.; Qiu, J.; Li, Y. Insights into adaptive mechanisms of extreme acidophiles based on quorum sensing/quenching-related proteins. mSystems 2022, 7, e0149121. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.; Seeger, M.; Holmes, D.; Jedlicki, E. A Lux-like quorum sensing system in the extreme acidophile Acidithiobacillus ferrooxidans. Biol. Res. 2005, 38, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Díaz, M.; Martin, D.S.; Castro, M.; Vera, M.; Guiliani, N. Quorum sensing signaling molecules positively regulate c-di-GMP effector PelD encoding gene and PEL exopolysaccharide biosynthesis in extremophile bacterium Acidithiobacillus thiooxidans. Genes 2021, 12, 69. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; Valenzuela, S.; Castro, M.; Gonzalez, A.R.; Frezza, M.; Soulère, L.; Rohwerder, T.; Queneau, Y.; Doutheau, A.; Sand, W.; et al. AHL communication is a widespread phenomenon in biomining bacteria and seems to be involved in mineral-adhesion efficiency. Hydrometallurgy 2008, 94, 133–137. [Google Scholar] [CrossRef]
- Castro, M.; Ruiz, L.; Barriga, A.; Jerez, C.; Holmes, D.; Guiliani, N. C-di-GMP pathway in biomining bacteria. Adv. Mater. Res. 2009, 71–73, 223–226. [Google Scholar] [CrossRef]
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef]
- Li, Z.; Nair, S.K. Quorum sensing: How bacteria can coordinate activity and synchronize their response to external signals? Protein Sci. 2012, 21, 1403–1417. [Google Scholar] [CrossRef]
- Sanders, J.G.; Akl, H.; Hagen, S.J.; Xue, B. Crosstalk enables mutual activation of coupled quorum sensing pathways through “jump-start” and “push-start” mechanisms. Sci. Rep. 2023, 13, 19230. [Google Scholar] [CrossRef]
- Mamani, S.; Moinier, D.; Denis, Y.; Soulère, L.; Queneau, Y.; Talla, E.; Bonnefoy, V.; Guiliani, N. Insights into the quorum sensing regulon of the acidophilic Acidithiobacillus ferrooxidans revealed by transcriptomic in the presence of an acyl homoserine lactone superagonist analog. Front. Microbiol. 2016, 7, 1365. [Google Scholar] [CrossRef]
- Banderas, A.; Guiliani, N. Bioinformatic prediction of gene functions regulated by quorum sensing in the bioleaching bacterium Acidithiobacillus ferrooxidans. Int. J. Mol. Sci. 2013, 14, 16901–16916. [Google Scholar] [CrossRef]
- Prescott, R.D.; Decho, A.W. Flexibility and Adaptability of Quorum Sensing in Nature. Trends Microbiol. 2020, 28, 436–444. [Google Scholar] [CrossRef]
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.E.; Greenberg, E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 1999, 96, 4360–4365. [Google Scholar] [CrossRef]
- Ortori, C.A.; Atkinson, S.; Chhabra, S.R.; Cámara, M.; Williams, P.; Barrett, D.A. Comprehensive profiling of N-acylhomoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadrupole–linear ion trap mass spectrometry. Anal. Bioanal. Chem. 2006, 387, 497–511. [Google Scholar] [CrossRef]
- Ruiz, L.; Gonzalez, A.R.; Frezza, M.; Soulère, L.; Queneau, Y.; Doutheau, A.; Rohwerder, T.; Sand, W.; Jerez, C.; Guiliani, N. Is the quorum sensing type AI-1 system of Acidithiobacillus ferrooxidans involved in its attachment to mineral surfaces? Adv. Mater. Res. 2007, 20–21, 345–349. [Google Scholar] [CrossRef]
- Churchill, M.E.A.; Chen, L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem. Rev. 2011, 111, 68–85. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-W.; Deng, Y.; Miao, Y.; Chatterjee, S.; Tran, T.M.; Tian, J.; Lindow, S. DSF-family quorum sensing signal-mediated intraspecies, interspecies, and inter-kingdom communication. Trends Microbiol. 2023, 31, 36–50. [Google Scholar] [CrossRef]
- Deng, Y.; Wu, J.; Tao, F.; Zhang, L.-H. Listening to a new language: DSF-based quorum sensing in gram-negative bacteria. Chem. Rev. 2011, 111, 160–173. [Google Scholar] [CrossRef] [PubMed]
- He, Y.W.; Wang, C.; Zhou, L.; Song, H.; Dow, J.M.; Zhang, L.H. Dual signaling functions of the hybrid sensor kinase RpfC of Xanthomonas campestris involve either phosphorelay or receiver domain-protein Interaction. J. Biol. Chem. 2006, 281, 33414–33421. [Google Scholar] [CrossRef] [PubMed]
- Lian, Z.; Chao, Y.L.; Yu, W.X.; Le, J.B.; Shuang, S.; Wen, H.Y. Identification and characterization of naturally occurring DSF-family quorum sensing signal turnover system in the phytopathogen Xanthomonas. Environ. Microbiol. 2015, 17, 4646–4658. [Google Scholar] [CrossRef]
- Bellenberg, S.; Buetti-Dinh, A.; Galli, V.; Ilie, O.; Herold, M.; Christel, S.; Boretska, M.; Pivkin, I.; Wilmes, P.; Sand, W.; et al. Automated microscopic analysis of metal sulfide colonization by acidophilic microorganisms. Appl. Environ. Microbiol. 2018, 84, e01835-18. [Google Scholar] [CrossRef] [PubMed]
- Christel, S.; Herold, M.; Bellenberg, S.; El Hajjami, M.; Buetti-Dinh, A.; Pivkin, I.V.; Sand, W.; Wilmes, P.; Poetsch, A.; Dopson, M. Multi-omics reveals the lifestyle of the acidophilic, mineral-oxidizing model species Leptospirillum ferriphilum(T). Appl. Environ. Microbiol. 2018, 84, e02091-17. [Google Scholar] [CrossRef]
- Moya-Beltrán, A.; Rojas-Villalobos, C.; Díaz, M.; Guiliani, N.; Quatrini, R.; Castro, M. Nucleotide second messenger-based signaling in extreme acidophiles of the Acidithiobacillus species complex: Partition between the core and variable gene complements. Front. Microbiol. 2019, 10, 381. [Google Scholar] [CrossRef]
- Díaz, M.; Guiliani, N. Molecular regulatory network involved in biofilm structure development by Acidithiobacillus thiooxidans includes Pel exopolysaccharide machinery. Solid State Phenom. 2017, 262, 330–333. [Google Scholar] [CrossRef]
- Chan, C.; Paul, R.; Samoray, D.; Amiot, N.C.; Giese, B.; Jenal, U.; Schirmer, T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 2004, 101, 17084–17089. [Google Scholar] [CrossRef]
- Ryan, R.P. Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiology 2013, 159, 1286–1297. [Google Scholar] [CrossRef]
- Díaz, M.; Castro, M.; Copaja, S.; Guiliani, N. Biofilm formation by the acidophile bacterium Acidithiobacillus thiooxidans involves c-di-GMP pathway and Pel exopolysaccharide. Genes 2018, 9, 113. [Google Scholar] [CrossRef]
- Prentice, J.A.; Bridges, A.A.; Bassler, B.L. Synergy between c-di-GMP and quorum-sensing signaling in Vibrio cholerae biofilm morphogenesis. J. Bacteriol. 2022, 204, e0024922. [Google Scholar] [CrossRef]
- Lin, Y.; Mi, D.; Hou, Y.; Lin, M.; Xie, Q.; Niu, X.; Chen, Y.; He, C.; Tao, J.; Li, C. Systematic analysis of the roles of c-di-GMP signaling in Xanthomonas oryzae pv. oryzae virulence. FEMS Microbiol. Lett. 2022, 369, fnac068. [Google Scholar] [CrossRef]
- Ryan, R.P.; Fouhy, Y.; Lucey, J.F.; Jiang, B.L.; He, Y.Q.; Feng, J.X.; Tang, J.L.; Dow, J.M. Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris. Mol. Microbiol. 2006, 63, 429–442. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Choub, S.-H. Sequence conservation, domain architectures, and phylogenetic distribution of the HD-GYP type c-di-GMP phosphodiesterases. J. Bacteriol. 2022, 204, e0056121. [Google Scholar] [CrossRef]
- Valdés, J.; Pedroso, I.; Quatrini, R.; Holmes, D. Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: Insights into their metabolism and ecophysiology. Hydrometallurgy 2008, 94, 180–184. [Google Scholar] [CrossRef]
- Yang, C.L.; Chen, X.K.; Wang, R.; Lin, J.Q.; Liu, X.M.; Pang, X.; Zhang, C.J.; Lin, J.Q.; Chen, L. Essential role of σ Factor RpoF in flagellar biosynthesis and flagella-mediated motility of Acidithiobacillus caldus. Front. Microbiol. 2019, 10, 1130. [Google Scholar] [CrossRef]
- Feng, S.; Yang, H.; Xin, Y.; Zhang, L.; Kang, W.; Wang, W. Isolation of an extremely acidophilic and highly efficient strain Acidithiobacillus sp for chalcopyrite bioleaching. J. Ind. Microbiol. Biotechnol. 2012, 39, 1625–1635. [Google Scholar] [CrossRef]
- Xia, J.; Peng, A.; He, H.; Yang, Y.; Liu, X.; Qiu, G. A new strain Acidithiobacillus albertensis BY-05 for bioleaching of metal sulfides ores. Trans. Nonferrous Met. Soc. China 2007, 17, 168–175. [Google Scholar] [CrossRef]
- Yin, X.; Li, X.; Li, Q.; Wang, B.; Zheng, L. Complete genome analysis reveals environmental adaptability of sulfur-oxidizing bacterium Thioclava nitratireducens M1-LQ-LJL-11 and symbiotic relationship with deep-sea hydrothermal vent Chrysomallon squamiferum. Mar. Genom. 2023, 71, 101058. [Google Scholar] [CrossRef]
- Zeng, X.; Zou, Y.; Zheng, J.; Qiu, S.; Liu, L.; Wei, C. Quorum sensing-mediated microbial interactions: Mechanisms, applications, challenges and perspectives. Microbiol. Res. 2023, 273, 127414. [Google Scholar] [CrossRef]
- Lv, L.; Chen, J.; Liu, X.; Gao, W.; Sun, L.; Wang, P.; Ren, Z.; Zhang, G.; Li, W. Roles and regulation of quorum sensing in anaerobic granular sludge: Research status, challenges, and perspectives. Bioresour. Technol. 2023, 387, 129644. [Google Scholar] [CrossRef]
- Urvoy, M.; Lami, R.; Dreanno, C.; Delmas, D.; L’Helguen, S.; Labry, C. Quorum sensing regulates the hydrolytic enzyme production and community composition of heterotrophic bacteria in coastal waters. Front. Microbiol. 2021, 12, 780759. [Google Scholar] [CrossRef]
- Smith, P.; Schuster, M. Public goods and cheating in microbes. Curr. Biol. 2019, 29, R442–R447. [Google Scholar] [CrossRef]
- Zhao, K.; Liu, L.; Chen, X.; Huang, T.; Du, L.; Lin, J.; Yuan, Y.; Zhou, Y.; Yue, B.; Wei, K.; et al. Behavioral heterogeneity in quorum sensing can stabilize social cooperation in microbial populations. BMC Biol. 2019, 17, 20. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Rolland, J.L.; Stien, D.; Sanchez-Ferandin, S.; Lami, R. Quorum sensing and Quorum quenching in the phycosphere of phytoplankton: A case of chemical interactions in ecology. J. Chem. Ecol. 2016, 42, 1201–1211. [Google Scholar] [CrossRef]
- Mion, S.; Carriot, N.; Lopez, J.; Plener, L.; Ortalo-Magné, A.; Chabrière, E.; Culioli, G.; Daudé, D. Disrupting quorum sensing alters social interactions in Chromobacterium violaceum. npj Biofilms Microbiomes 2021, 7, 40. [Google Scholar] [CrossRef]
- Kaufmann, G.F.; Sartorio, R.; Lee, S.-H.; Rogers, C.J.; Meijler, M.M.; Moss, J.A.; Clapham, B.; Brogan, A.P.; Dickerson, T.J.; Janda, K.D. Revisiting quorum sensing: Discovery of additional chemical and biological functions for 3-oxo-N-acyl homoserine lactones. Proc. Natl. Acad. Sci. USA 2004, 102, 309–314. [Google Scholar] [CrossRef]
- Schertzer, J.W.; Boulette, M.L.; Whiteley, M. More than a signal: Non-signaling properties of quorum sensing molecules. Trends Microbiol. 2009, 17, 189–195. [Google Scholar] [CrossRef]
- Parsons, J.B.; Yao, J.; Frank, M.W.; Jackson, P.; Rock, C.O. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J. Bacteriol. 2012, 194, 5294–5304. [Google Scholar] [CrossRef]
- Shen, L.; Cheng, J.; Wang, J.; Zhang, Y.; Zhou, H.; Wu, X.; Li, J.; Zeng, W. Biofilm formation and development during the pyrite bioleaching of moderately thermophilic microorganisms. Hydrometallurgy 2023, 222, 106183. [Google Scholar] [CrossRef]
- Recalde, A.; Gonzalez-Madrid, G.; Acevedo-Lopez, J.; Jerez, C.A. Sessile lifestyle offers protection against copper stress in Saccharolobus solfataricus. Microorganisms 2023, 11, 1421. [Google Scholar] [CrossRef]
- Zhang, R.; Neu, T.; Blanchard, V.; Vera, M.; Sand, W. Biofilm dynamics and EPS production of a thermoacidophilic bioleaching archaeon. New Biotechnol. 2019, 51, 21–30. [Google Scholar] [CrossRef] [PubMed]
- van Alin, A.; Corbett, M.K.; Fathollahzadeh, H.; Tjiam, M.C.; Rickard, W.D.A.; Sun, X.; Putnis, A.; Eksteen, J.; Kaksonen, A.H.; Watkin, E. Biofilm formation on the surface of monazite and xenotime during bioleaching. Microb. Biotechnol. 2023, 16, 1790–1802. [Google Scholar] [CrossRef] [PubMed]
- Salinas, J.I.; Nunez-Ramirez, D.M.; Manero, O.; Bautista, F.; Medina-Torres, L. Modeling of the bioleaching of process silver pulp. Chem. Eng. Technol. 2023, 47, 396–402. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.C.; Stout, R.; Mitchell, R. Mechanism of the initial events in the sorption of marine bacteria to surfaces. J. Gen. Microbiol. 1971, 68, 337–348. [Google Scholar] [CrossRef]
- Toyofuku, M.; Inaba, T.; Kiyokawa, T.; Obana, N.; Yawata, Y.; Nomura, N. Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 2016, 80, 7–12. [Google Scholar] [CrossRef]
- Hu, W.; Feng, S.; Tong, Y.; Zhang, H.; Yang, H. Adaptive defensive mechanism of bioleaching microorganisms under extremely environmental acid stress: Advances and perspectives. Biotechnol. Adv. 2020, 42, 107580. [Google Scholar] [CrossRef]
- Gonzalez, A.R.; Bellenberg, S.; Mamani, S.; Ruiz, L.; Echeverría, A.; Soulère, L.; Doutheau, A.; Demergasso, C.; Sand, W.; Queneau, Y.; et al. AHL signaling molecules with a large acyl chain enhance biofilm formation on sulfur and metal sulfides by the bioleaching bacterium Acidithiobacillus ferrooxidans. Appl. Microbiol. Biotechnol. 2013, 97, 3729–3737. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.; Díaz, M.; Beltrán, A.M.; Guiliani, N. Cyclic di-GMP signaling in extreme acidophilic bacteria. In Microbial Cyclic Di-Nucleotide Signaling; Chou, S., Guiliani, N., Lee, V., Römling, U., Eds.; Springer: Cham, Switzerland, 2020; pp. 337–353. [Google Scholar]
- Castro, M.; Deane, S.; Ruiz, L.; Rawlings, D.; Guiliani, N. Diguanylate cyclase null mutant reveals that c-di-GMP pathway regulates the motility and adherence of the extremophile bacterium Acidithiobacillus caldus. PLoS ONE 2015, 10, e0116399. [Google Scholar] [CrossRef]
- Ranava, D.; Backes, C.; Karthikeyan, G.; Ouari, O.; Soric, A.; Guiral, M.; Cardenas, M.L.; Giudici-Orticoni, M.T. Metabolic exchange and energetic coupling between nutritionally stressed bacterial species: Role of quorum-sensing molecules. mBio 2021, 12, e02758-20. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, J.; Wang, N.; Li, X.; Fang, N.; Zhuang, X. Diffusible signal factor enhances the saline-alkaline resistance and rhizosphere colonization of Stenotrophomonas rhizophila by coordinating optimal metabolism. Sci. Total Environ. 2022, 834, 155403. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.; Zhao, C.; Zhang, J.; He, W. Autoinducer-2 quorum sensing regulates biofilm formation and chain elongation metabolic pathways to enhance caproate synthesis in microbial electrochemical system. Chemosphere 2023, 344, 140384. [Google Scholar] [CrossRef]
- Gao, X.Y.; Fu, C.A.; Hao, L.; Gu, X.; Wang, R.; Lin, J.Q.; Liu, X.M.; Pang, X.; Zhang, C.J.; Lin, J.Q.; et al. The substrate-dependent regulatory effects of the AfeI/R system in Acidithiobacillus ferrooxidans reveals the novel regulation strategy of quorum sensing in acidophiles. Environ. Microbiol. 2020, 23, 757–773. [Google Scholar] [CrossRef]
- Li, Y.; Wang, S.; Ding, H.; Xiao, K.; Huang, X. Quorum sensing-Fe metabolism interplay affects biofouling on reverse osmosis membrane: Evidences from microbial shift and structure alteration. Desalination 2023, 551, 116416. [Google Scholar] [CrossRef]
- Gao, X.Y.; Liu, X.; Fu, C.A.; Gu, X.; Lin, J.; Liu, X.M.; Pang, X.; Lin, J.Q.; Chen, L. Novel strategy for improvement of the bioleaching efficiency of Acidithiobacillus ferrooxidans based on the AfeI/R quorum sensing system. Minerals 2020, 10, 222. [Google Scholar] [CrossRef]
- Jung, H.; Inaba, Y.; West, A.; Banta, S. Overexpression of quorum sensing genes in Acidithiobacillus ferrooxidans enhances cell attachment and covellite bioleaching. Biotechnol. Rep. 2023, 38, e00789. [Google Scholar] [CrossRef] [PubMed]
- Bellenberg, S.; Barthen, R.; Vera, M.; Guiliani, N.; Sand, W. Biofilm formation, communication and interactions of mesophilic leaching bacteria during pyrite oxidation. Adv. Mater. Res. 2013, 825, 107–110. [Google Scholar] [CrossRef]
- Mamani, S.; Denis, Y.; Moinier, D.; Sabbah, M.; Soulère, L.; Queneau, Y.; Bonnefoy, V.; Guiliani, N. Characterization of the quorum sensing regulon in Acidithiobacillus ferrooxidans. Adv. Mater. Res. 2013, 825, 129–132. [Google Scholar] [CrossRef]
- Lv, L.; Li, W.; Zheng, Z.; Li, D.; Zhang, N. Exogenous acyl-homoserine lactones adjust community structures of bacteria and methanogens to ameliorate the performance of anaerobic granular sludge. J. Hazard. Mater. 2018, 354, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.S.; Pan, X.R.; Cao, J.S.; Song, X.N.; Fang, F.; Tong, Z.H.; Li, W.W.; Yu, H.Q. Augmentation of acyl homoserine lactones-producing and -quenching bacterium into activated sludge for its granulation. Water Res. 2017, 125, 309–317. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Q.; Li, X.; Ma, S.; Hu, H.; Wu, B.; Zhang, X.X.; Ren, H. In-situ monitoring AHL-mediated quorum-sensing regulation of the initial phase of wastewater biofilm formation. Environ. Int. 2020, 135, 105326. [Google Scholar] [CrossRef]
- Xiong, F.; Zhao, X.; Wen, D.; Li, Q. Effects of N-acyl-homoserine lactones-based quorum sensing on biofilm formation, sludge characteristics, and bacterial community during the start-up of bioaugmented reactors. Sci. Total Environ. 2020, 735, 139449. [Google Scholar] [CrossRef]
- Lv, L.; Feng, C.; Li, W.; Zhang, G.; Wang, P.; Ren, Z. Exogenous N-acyl-homoserine lactones promote the degradation of refractory organics in oligotrophic anaerobic granular sludge. Sci. Total Environ. 2021, 761, 143289. [Google Scholar] [CrossRef]
- Liu, R.; Chen, J.; Zhou, W.; Cheng, H.; Zhou, H. Insight to the early-stage adsorption mechanism of moderately thermophilic consortia and intensified bioleaching of chalcopyrite. Biochem. Eng. J. 2019, 144, 40–47. [Google Scholar] [CrossRef]
- Zeng, W.; Liu, Z.; Amanze, C.; Cheng, J.; Liao, W.; Wu, X.-L.; Qiu, G.; Wang, Q.; Wu, Z.; Zou, L.; et al. In situ detection of Cu2+, Fe3+ and Fe2+ ions at the microbe-mineral interface during bioleaching of chalcopyrite by moderate thermophiles. Miner. Eng. 2023, 191, 107936. [Google Scholar] [CrossRef]
- Moncayo, E.A.; Debut, A.; Vizuete, K.; Jumbo-Flores, D.; Aguirre, P. Sticky bacteria: Combined effect of galactose and high ferric iron concentration on extracellular polymeric substances production and the attachment of Acidithiobacillus ferrooxidans on a polymetallic sulfide ore surface. Front. Microbiol. 2022, 13, 951402. [Google Scholar] [CrossRef]
- Zeng, W.M.; Cai, Y.X.; Hou, C.W.; Liu, A.j.; Peng, T.J.; Chen, M.; Qiu, G.Z.; Shen, L. Influence diversity of extracellular DNA on bioleaching chalcopyrite and pyrite by Sulfobacillus thermosulfidooxidans ST. J. Cent. South Univ. 2020, 27, 1466–1476. [Google Scholar] [CrossRef]
- Ning, X.J.; Ying, Y.H.; Lin, T.L. Adsorption behavior of glucuronic acid on pyrite surface: An electrochemical and DFT study. Arch. Metall. Mater. 2020, 65, 433–440. [Google Scholar] [CrossRef]
- Chu, H.; Wang, J.; Tian, B.; Qian, C.; Niu, T.; Qi, S.; Yang, Y.; Ge, Y.; Dai, X.; Xin, B. Generation behavior of extracellular polymeric substances and its correlation with extraction efficiency of valuable metals and change of process parameters during bioleaching of spent petroleum catalyst. Chemosphere 2021, 275, 130006. [Google Scholar] [CrossRef]
- Ma, C.; Zeng, W.; Meng, Q.; Wang, C.; Peng, Y. Identification of partial denitrification granulation enhanced by low C/N ratio in the aspect of metabolomics and quorum sensing. Chemosphere 2022, 286, 131895. [Google Scholar] [CrossRef]
- Chabert, N.; Bonnefoy, V.; Achouak, W. Quorum sensing improves current output with Acidithiobacillus ferrooxidans. Microb. Biotechnol. 2017, 11, 136–140. [Google Scholar] [CrossRef]
- Caicedo, J.C.; Villamizar, S.; Orlandoni, G. The use of synthetic agonists of quorum sensing N- acyl homoserine lactone pathway improves the bioleaching ability in Acidithiobacillus and Pseudomonas bacteria. PeerJ 2022, 10, e13801. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zeng, X.; Zheng, J.; Zou, Y.; Qiu, S.; Dai, Y. AHL-mediated quorum sensing to regulate bacterial substance and energy metabolism: A review. Microbiol. Res. 2022, 262, 127102. [Google Scholar] [CrossRef] [PubMed]
| (a) iron metabolism | ||||
| Microorganisms | Regulatory genes | Enzyme | Electron transfer chain | References |
| Acidithiobacillus ferrooxidans ATCC 23270 | rusA/B | Rusticyanin oxidase | 1. Fe2+ → Rusticyanin oxidase → Cyc1 → aa3 oxidase 2. Fe2+ → Rusticyanin oxidase → CycA1 → bc1 complex 3. Fe2+→Cyt579 → Cytochrome c | [6,19,20,21,22] |
| iro | Iron oxidase | |||
| A. ferrivorans s DSM 22755 | fox cluster | haem–copper terminal | ||
| Leptospirillum ferriphilum DSM 17947 | Cyc1 | |||
| Sulfobacillus acidophilus DSM 10332 | CycA1 | |||
| Sulfolobus tokodaii JCM10545 Acidiplasma aeolicum DSM 18409 Acidianus brierleyi DSM 1651 Metallosphaera sedula DSM 5348 | Cyt579 | |||
| (b) sulfur metabolism | ||||
| Microorganisms | Regulatory genes | Enzyme | Reaction | References |
| Acidithiobacillus caldus ATCC 51756 A. thiobacillus A01 A. ferrooxidans ATCC 23270 A. ferrivorans DSM 22755 Sulfobacillus acidophilus DSM 10332 Acidianus copahuensis ALE1 Metallosphaera sedula DSM 535 | tetH | Tetrathionate hydrolase | S4O62− → S2O32− + SO42− + S0 | |
| tsd | Thiosulfate dehydrogenase | S2O32− → S4O62− | ||
| sqr | Sulfide quinone reductase | H2S → S0 | [19,20,23,24,25] | |
| doxDA | Thiosulfate: quinone oxidoreductase | S2O32− → S4O62– | ||
| sor | Sulfur oxygenase reductase | S0 → H2S + SO32− + S2O32– | ||
| tst | Thiosulfate sulfurtransferase | S2O32− → SO32− + S0 | ||
| hdrABC | Heterodisulfide reductase complex | glutathione oxidized → glutathione + SO3 2− | ||
| sat/cysC | Sulfate adenylyltransferase/adenylylsulfate kinase | adenosine phosphosulfate → SO42− | ||
| sar | sulfite: acceptor oxidoreductase | SO3 2− → SO42− | ||
| SoxXYZAB | Sox system | S2−/S0/S2O32−/SO32− → SO4 2− | ||
| AIs | Microorganisms | Typical Structures | Regulatory Proteins | Functions | References |
|---|---|---|---|---|---|
| AHLs | A. ferrooxidans ATCC 23270 A. thiooxidans DSMZ 504 L. ferrooxidans DSM 2391 * A. ferrivorans SS3 * | 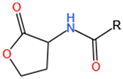 | LuxI LuxR Act | 1. Biofilm formation 2. Protein secretion 3. Flagellar movement | [17,38,39,43,44,45] |
| DSF | L. ferrooxidans DSM 2705 L. ferriphilum DSM 14647 |  | RpfG RpfF RpfC | 1. Biofilm formation 2. Resistance | [40] |
| c-di-GMP | A. ferrooxidans ATCC 23270 A. thiooxidans ATCC 51756 A. caldus ATCC 19377 | 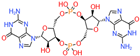 | DGCs PDEs | 1. Flagellar movement 2. Substrate adhesion 3. Biofilm formation | [46] |
| Exogenous Molecules | Acidophiles | Substrate | Results | References |
|---|---|---|---|---|
| OH-HSLs + oxo-HSLs | A. ferrooxidans ATCC 23270 | pyrite | reduce adhesion | [55] |
| -HSL + OH-HSL | A. ferrooxidans ATCC 23270 | sulfur | increase attachment | [98] |
| -HSL + OH-HSL + oxo-HSL/C14-HSL | A. ferrooxidans ATCC 23270 | sulfur/pyrite | promote biofilm formation | [98] |
| -HSL + oxo -HSLs + OH -HSL | A. ferrivorans SS3 | pyrite | 1. inhibit biofilm formation 2. reduce the leaching rate | [108] |
| -HSL + oxo-HSL + OH-HSL | L. ferrooxidans DSM 2391 | pyrite | 1. inhibit biofilm formation 2. reduce the leaching rate | [108] |
| C8-HSL/3-oxo-C8-HSL/C10-HSL | A. thiooxidans DSM 14887 | sulfur | promote biofilm formation | [17] |
| 3-sulfonylamide-C8-HSL | A. ferrooxidans ATCC 23270 | pyrite | increase attachment | [55] |
| 4-phenyl-3-oxo-HSL | A. ferrooxidans ATCC 23270 | pyrite | reduce adhesion | [55] |
| tetrazole | A. ferrooxidans ATCC 23270 | sulfur | 1. increase attachment 2. up-regulate afeI and zwf gene expression | [109] |
| tetrazole 9c | A. ferrooxidans ATCC 23270 | sulfur | increase attachment | [50] |
| DSF + BDSF | L. ferriphilum DSM 14647 | Fe2+/pyrite | 1. inhibit iron oxidation 2. reduce adhesion | [40,61] |
| DSF + BDSF | S. thermosulfidooxidans DSM 9293 | Fe2+/pyrite | 1. inhibit iron oxidation 2. reduce adhesion | [40,61] |
| DSF + BDSF | A. ferrooxidans ATCC 23270 | Fe2+ | inhibit iron oxidation | [40,61] |
| DSF + BDSF | A. caldus DSM 8584 | pyrite | reduce adhesion | [61] |
| DSF/BDSF + AHL | L. ferriphilum DSM 14647 | Fe2+ | reduce flrB transcription levels | [40] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, W.; Li, Y.; Chen, S.; Liang, Y.; Liu, X. Roles and Regulation of Quorum Sensing of Acidophiles in Bioleaching: A Review. Microorganisms 2024, 12, 422. https://doi.org/10.3390/microorganisms12030422
Luo W, Li Y, Chen S, Liang Y, Liu X. Roles and Regulation of Quorum Sensing of Acidophiles in Bioleaching: A Review. Microorganisms. 2024; 12(3):422. https://doi.org/10.3390/microorganisms12030422
Chicago/Turabian StyleLuo, Wang, Yiran Li, Shiqi Chen, Yili Liang, and Xueduan Liu. 2024. "Roles and Regulation of Quorum Sensing of Acidophiles in Bioleaching: A Review" Microorganisms 12, no. 3: 422. https://doi.org/10.3390/microorganisms12030422
APA StyleLuo, W., Li, Y., Chen, S., Liang, Y., & Liu, X. (2024). Roles and Regulation of Quorum Sensing of Acidophiles in Bioleaching: A Review. Microorganisms, 12(3), 422. https://doi.org/10.3390/microorganisms12030422





