Assessing the Plastisphere from Floating Plastics in the Northwestern Mediterranean Sea, with Emphasis on Viruses
Abstract
:1. Introduction
2. Materials and Methods
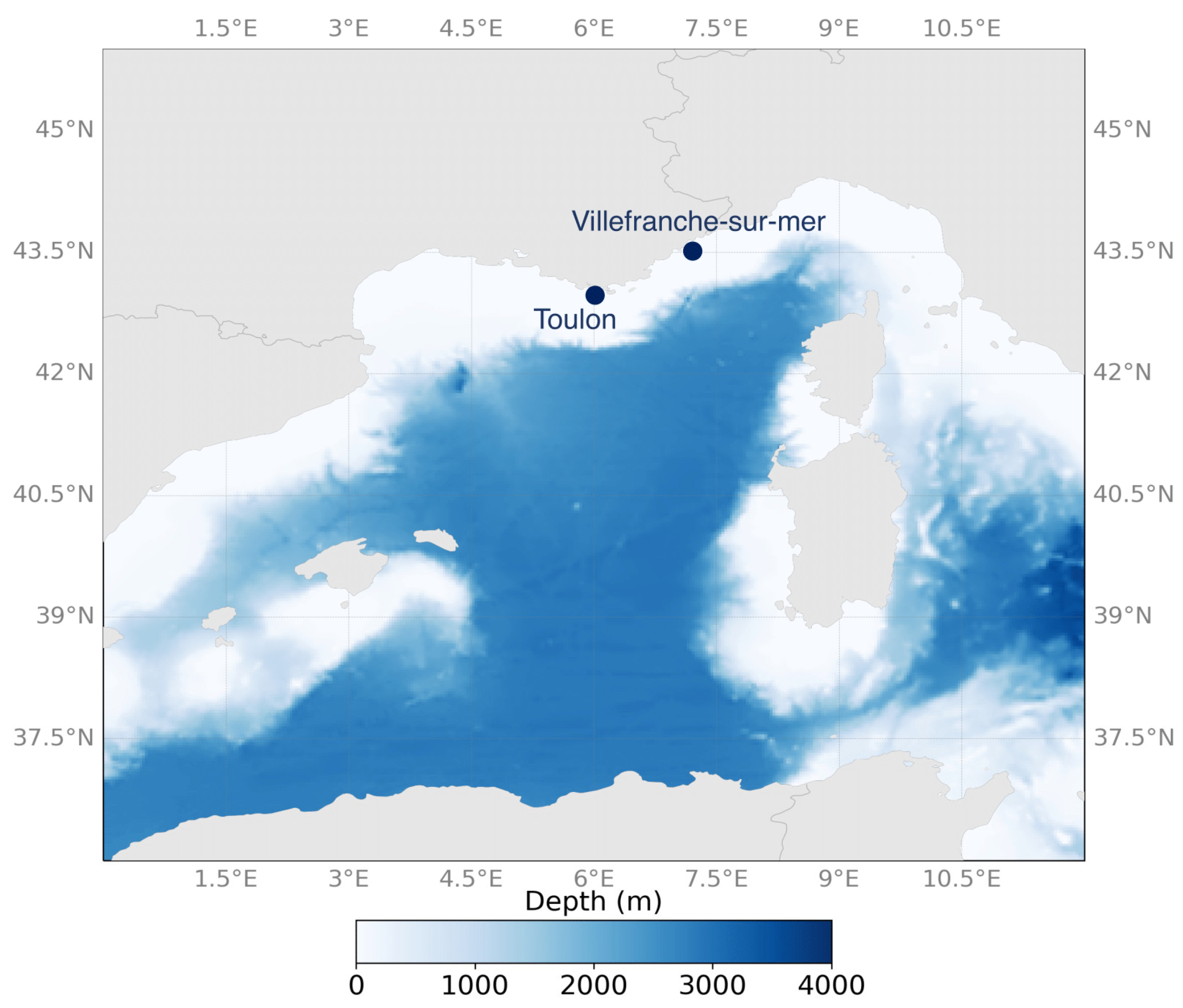
2.1. Sampling Sites
2.2. Sampling of Plastics at Sea
2.3. DNA Extraction
2.4. Identification of Polymer Composition
2.5. Assessment of the Plastisphere Composition
3. Results
3.1. Sequencing Metrics per Plastic Sample
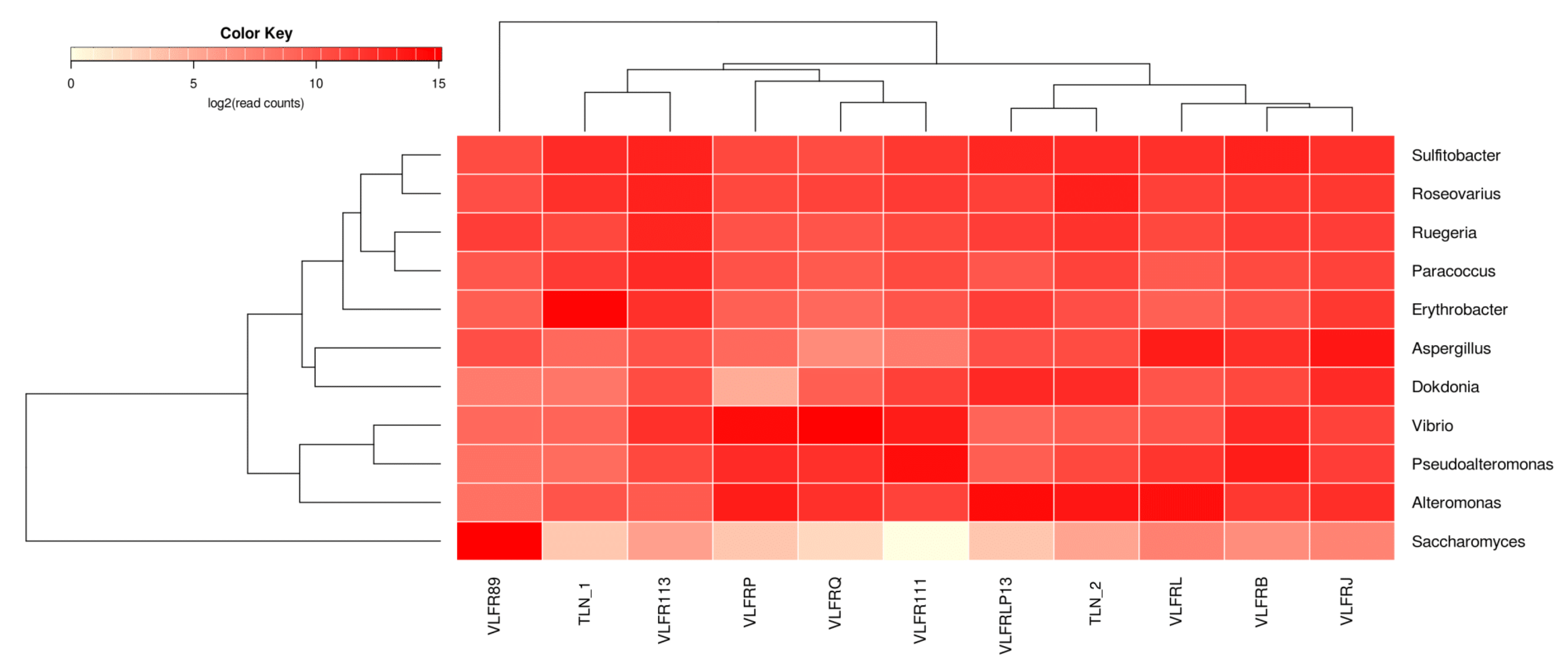
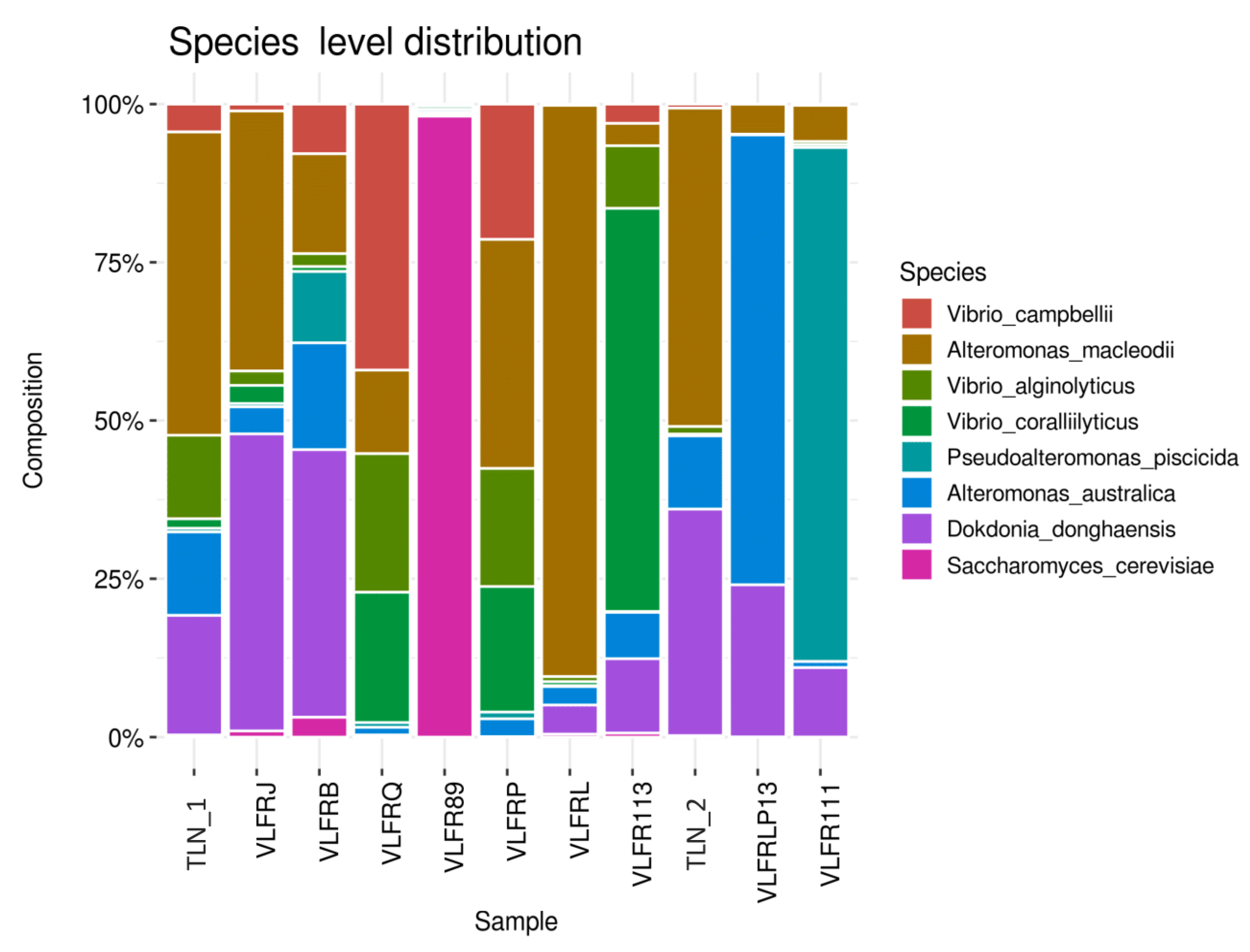
3.2. Taxonomic Abundance and Diversity of the Plastisphere in the Northwestern Mediterranean Sea
3.3. Plastisphere Composition According to Polymer Type and Shape
3.4. Viruses in the Mediterranean Plastisphere
3.5. Other Plastisphere Organisms with Important Ecological Functions
4. Discussion
4.1. Full Diversity and Potential Ecological Role of the Mediterranean Plastisphere
4.2. Viruses in the Marine Plastisphere
4.3. Other Potential Pathogens within the Mediterranean Plastisphere
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UNEP. United Nations Environment Programme. From Pollution to Solution. A Global Assesment of Marine Litter and Plastic Pollution Nairobi. 2021. Available online: https://www.unep.org/ (accessed on 20 August 2023).
- CIEL–Center for International Environmental Law. Fossils, Plastics and Petrochemical Feedstocks. Fueling Plastics CIEL Series. 2017, p. 5. Available online: https://www.ciel.org/reports/fuelingplastics/ (accessed on 10 August 2023).
- Ford, H.V.; Jones, N.H.; Davies, A.J.; Godley, B.J.; Jambeck, J.R.; Napper, I.E.; Suckling, C.C.; Williams, G.J.; Woodall, L.C.; Koldewey, H.J. The fundamental links between climate change and marine plastic pollution. Sci. Total Environ. 2022, 806, 150392. [Google Scholar] [CrossRef]
- Lopez, B.E.; Allen, J.M.; Dukes, J.S.; Lenoir, J.; Vil, M.; Blumenthal, D.M.; Beaury, E.M.; Fuscoa, E.J.; Laginhasi, B.B.; Morellia, T.L.; et al. Global environmental changes more frequently offset than intensify detrimental effects of biological invasions. Proc. Natl. Acad. Sci. USA 2023, 119, 2117389119. [Google Scholar] [CrossRef]
- Du, Y.; Liu, X.; Dong, X.; Yin, Z. A review on marine plastisphere: Biodiversity, formation, and role in degradation. Comput. Struct. Biotechnol. J. 2022, 20, 975–988. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Zhang, X.H.; Yu, M. Microbial colonization and degradation of marine microplastics in the plastisphere: A review. Front. Microbiol. 2023, 14, 1127308. [Google Scholar] [CrossRef] [PubMed]
- Messer, L.F.; Lee, C.E.; Wattiez, R.; Matallana-Surget, S. Novel functional insights into the microbiome inhabiting marine plastic debris: Critical considerations to counteract the challenges of thin biofilms using multi-omics and comparative metaproteomics. Microbiome 2023. [Google Scholar] [CrossRef]
- Bowley, J.; Baker-Austin, C.; Porter, A.; Hartnell, R.; Lewis, C. Oceanic Hitchhikers–Assessing Pathogen Risks from Marine Microplastic. Trends Microbiol. 2021, 29, 107–116. [Google Scholar] [CrossRef]
- Metcalf, R.; Oliver, D.M.; Moresco, V.; Quilliam, R.S. Quantifying the importance of plastic pollution for the dissemination of human pathogens: The challenges of choosing an appropriate ‘control’ material. Sci. Total Environ. 2022, 810, 152292. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.G.; Machovsky-capuska, G.E.; Andrades, R. Plastic ingestion as an evolutionary trap: Toward a holistic understanding. Science 2021, 60, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Wright, R.; Erni-Cassola, G.; Zadjelovic, V.; Latva, M.; Christie-Oleza, J.A. Marine Plastic Debris: A New Surface for Microbial Colonization. Environ. Sci. Technol. 2020, 54, 11657–11672. [Google Scholar] [CrossRef] [PubMed]
- Delacuvellerie, A.; Ballerini, T.; Frère, L.; Matallana-Surget, S.; Dumontet, B.; Wattiez, R. From rivers to marine environments: A constantly evolving microbial community within the plastisphere. Mar. Pollut. Bull. 2022, 179, 113660. [Google Scholar] [CrossRef]
- Zettler, E.R.; Mincer, T.J.; Amaral-Zettler, L.A. Life in the “Plastisphere”: Microbial communities on plastic marine debris. Environ. Sci. Technol. 2013, 47, 7137. [Google Scholar] [CrossRef]
- Barros, J.; Seena, S. Plastisphere in freshwaters: An emerging concern. Environ. Pollutio. 2021, 290, 118123. [Google Scholar] [CrossRef]
- MacLean, J.; Mayanna, S.; Benning, L.G.; Horn, F.; Bartholomäus, A.; Wiesner, Y.; Wagner, D.; Liebner, S. The Terrestrial Plastisphere: Diversity and Polymer-Colonizing Potential of Plastic-Associated Microbial Communities in Soil. Microorganisms 2021, 9, 1876. [Google Scholar] [CrossRef]
- Scales, B.S.; Cable, R.N.; Duhaime, M.B.; Gerdts, G.; Fischer, F.; Fischer, D.; Oberbeckmann, S. Cross-hemisphere study reveals geographically ubiquitous, plastic-specific bacteria emerging from the rare and unexplored biosphere. MSphere 2021, 6, 10–1128. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Mincer, T.J. Ecology of the plastisphere. Nat. Rev. Microbiol. 2020, 18, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, A.L.d.F.; Taylor, J.D.; Rodrigues, L.d.S.; Kessler, F.; Secchi, E.; Proietti, M.C. Floating plastics and their associated biota in the Western South Atlantic. Sci. Total Environ. 2022, 805, 150186. [Google Scholar] [CrossRef] [PubMed]
- Moresco, V.; Oliver, D.M.; Weidmann, M.; Matallana-Surget, S.; Quilliam, R.S. Survival of human enteric and respiratory viruses on plastics in soil, freshwater, and marine environments. Environ. Res. 2021, 199, 111367. [Google Scholar] [CrossRef] [PubMed]
- Moresco, V.; Charatzidou, A.; Oliver, D.M.; Weidmann, M.; Matallana-Surget, S.; Quilliam, R.S. Binding, recovery, and infectiousness of enveloped and non-enveloped viruses associated with plastic pollution in surface water. Environ. Pollut. 2022, 308, 119594. [Google Scholar] [CrossRef]
- UNEP-MAP. United Nations Environment Programme/Mediterranean Action Plan and Plan Bleu; State of the Environment and Development in the Mediterranean: Nairobi, Kenya, 2020. [Google Scholar]
- Dussud, C.; Meistertzheim, A.L.; Conan, P.; Pujo-Pay, M.; George, M.; Fabre, P.; Coudane, J.; Higgs, P.; Elineau, A.; Pedrotti, M.L.; et al. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding sea waters. Environ. Pollut. 2018, 236, 807–816. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Kreikemeyer, B.; and Labrenz, M. Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 2018, 8, 12. [Google Scholar] [CrossRef]
- Davidov, K.; Iankelevich-Kounio, E.; Yakovenko, I.; Koucherov, Y.; Rubin-Blum, M.; Oren, M. Identification of plastic-associated species in the Mediterranean Sea using DNA metabarcoding with Nanopore MinION. Sci. Rep. 2020, 10, 17533. [Google Scholar] [CrossRef]
- Basili, M.; Quero, G.M.; Giovannelli, D.; Manini, E.; Vignaroli, C.; Avio, C.G.; De Marco, R.; Luna, G.M. Major Role of Surrounding Environment in Shaping Biofilm Community Composition on Marine Plastic Debris. Front. Mar. Sci. 2020, 7, 262. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.; Ballerini, T.; Zettler, E.R.; Asbun, A.A.; Adame, A.; Casotti, R.; Dumontet, B.; Donnarumma, V.; Engelmann, J.C.; Frère, L.; et al. Diversity and predicted inter- and intra-domain interactions in the Mediterranean Plastisphere. Environ. Pollut. 2021, 286, 117439. [Google Scholar] [CrossRef] [PubMed]
- Pedrotti, M.L.; Lacerda, A.L.d.F.; Petit, S.; Ghiglione, J.F.; Gorsky, G. Vibrio spp and other potential pathogenic bacteria associated to microfibers in the North-Western Mediterranean Sea. PLoS ONE 2022, 17, e0275284. [Google Scholar] [CrossRef] [PubMed]
- Karkanorachaki, K.; Syranidou, E.; Kalogerakis, N. Extreme weather events as an important factor for the evolution of plastisphere but not for the degradation process. Water Res. 2023, 246, 120687. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses–Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- de Walle, F.B.; Nikolopoulou-Tamvakli, M.; Heinen, W.J. Environmental Condition of the Mediterranean Sea: European Community Countries; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2013; Volume 5, pp. 1–32. [Google Scholar]
- Briand, M.J.; Herlory, O.; Briant, N.; BrachPapa, C.; Boissery, P.; Bouchoucha, M. The French Mussel Watch: More than two decades of chemical contamination survey in Mediterranean coastal waters. Mar. Pollut. Bull. 2023, 191, 114901. [Google Scholar] [CrossRef]
- Rossi, N.; Jamet, J.-L. In situ heavy metals (copper, lead and cadmium) in different plankton compartments and suspended particulate matter in two coupled Mediterranean coastal ecosystems (Toulon Bay, France). Mar. Pollut. Bull. 2008, 56, 1862–1870. [Google Scholar] [CrossRef]
- Pougnet, F.; Schäfer, J.; Dutruch, L.; Garnier, C.; Tessier, E.; Dang, D.H.; Lanceleur, L.; Mullot, J.U.; Lenoble, V.; Blanc, G. Sources and historical record of tin and butyl-tin species in a Mediterranean bay (Toulon Bay, France). Environ. Sci. Pollut. Res. 2014, 21, 6640–6651. [Google Scholar] [CrossRef]
- Araújo, D.F.; Ponzevera, E.; Briant, N.; Knoery, J.; Bruzac, S.; Sireau, T.; Brach-Papa, C. Copper, zinc and lead isotope signatures of sediments from a Mediterranean coastal bay impacted by naval activities and urban sources. Appl. Geochem. 2019, 111, 104440. [Google Scholar] [CrossRef]
- Jung, M.R.; Horgen, F.D.; Orski, S.V.; Rodriguez, C.V.; Beers, K.L.; Balazs, G.H.; Jones, T.T.; Work, T.M.; Brignac, K.C. Validation of ATR FT-IR to identify polymers of plastic marine debris, including those ingested by marine organisms. Mar. Pollut. Bull. 2018, 127, 704–716. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn for enhanced metagenomic taxonomic profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Baker, D.N.; Salzberg, S.L. KrakenUniq: Confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. 2014. Available online: http://ccb.jhu.edu/software/kraken/ (accessed on 30 October 2022).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package vegan. Community Ecol. Package 2013, 2, 1–295. [Google Scholar]
- Orata, F.D.; Hedreyda, C.T.; Cabrera, E.C. Isolation and sequence analysis of the full-length toxR gene of type strain Vibrio campbellii and use of the toxR gene sequence to evaluate variation and relatedness with other Vibrio species. Philipp. Sci. Lett. 2011, 4, 70–80. [Google Scholar]
- Oramadike, C.; Ogunbanwo, S.T. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from seafoods in Lagos Lagoon Nigeria. Cogent Food Agric. 2015, 1, 1041349. [Google Scholar] [CrossRef]
- Arfatahery, N.; Mirshafiey, A.; Abedimohtasab, T.P.; Zeinolabedinizamani, M. Study of the Prevalence of Staphylococcus aureus in Marine and Farmed Shrimps in Iran Aiming the Future Development of a Prophylactic Vaccine. Procedia Vaccinol. 2015, 9, 44–49. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R. Marine biofilms: A successful microbial strategy with economic implications. Front. Mar. Sci. 2018, 5, 126. [Google Scholar] [CrossRef]
- De Tender, C.; Devriese, L.I.; Haegeman, A.; Maes, S.; Vangeyte, J.; Cattrijsse, A.; Dawyndt, P.; Ruttink, T. Temporal Dynamics of Bacterial and Fungal Colonization on Plastic Debris in the North Sea. Environ. Sci. Technol. 2017, 51, 7350–7360. [Google Scholar] [CrossRef]
- Lacerda, A.L.d.F.; Proietti, M.C.; Secchi, E.R.; Taylor, J.D. Diverse groups of fungi are associated with plastics in the surface waters of the Western South Atlantic and the Antarctic Peninsula. Mol. Ecol. 2020, 29, 1903–1918. [Google Scholar] [CrossRef]
- Philippe, A.; Noël, C.; Eyheraguibel, B.; Briand, J.F.; Paul-Pont, I.; Ghiglione, J.F.; Coton, E.; Burgaud, G. Fungal Diversity and Dynamics during Long-Term Immersion of Conventional and Biodegradable Plastics in the Marine Environment. Diversity 2023, 15, 579. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Levold, F.; Aallgaier, M.; Simon, M.; Brinkhoff, T. Marine diatom species harbor distinct bacterial communities. Environ. Microbiol. 2005, 7, 860–873. [Google Scholar] [CrossRef]
- Sapp, M.; Schwaderer, A.; Wiltshire, K.H.; Hoppe, H.G.; Gerdts, G.; Wichels, A. Species-specific bacterial communities in the phycosphere of microalgae? Microb. Ecol. 2007, 53, 683–699. [Google Scholar] [CrossRef]
- Sapp, M.; Wichels, A.; Gerdts, G. Impacts of cultivation of marine diatoms on the associated bacterial community. Appl. Environ. Microbiol. 2007, 73, 3117–3120. [Google Scholar] [CrossRef]
- Koch, H.; Germscheid, N.; Freese, H.M.; Noriega-Ortega, B.; Lücking, D.; Berger, M.; Qiu, G.; Marzinelli, E.M.; Campbell, A.H.; Steinberg, P.D.; et al. Genomic, metabolic and phenotypic variability shapes ecological differentiation and intraspecies interactions of Alteromonas macleodii. Sci. Rep. 2020, 10, 809. [Google Scholar] [CrossRef]
- Cusick, K.D.; Dale, J.R.; Little, B.J.; Biffinger, J.C. Draft genome sequences of four Alteromonas macleodii strains isolated from copper coupons and grown long-term at elevated copper levels. Genome Announc. 2016, 4, e01311-16. [Google Scholar] [CrossRef]
- Briand, J.-F.; Pollet, T.; Misson, B.; Garnier, C.; Lejars, M.; Maintenay, M.; Barry-Martinet, R.; Portas, A.; Ghiglione, J.-F.; Bressy, C. Surface Characteristics Together with Environmental Conditions Shape Marine Biofilm Dynamics in Coastal NW Mediterranean Locations. Front. Mar. Sci. 2022, 8, 746383. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, K.; Xu, S.; Yan, M.; Tao, D.; Chen, L.; Wei, Y.; Wu, C.; Liu, G.; Lam, P.K.S. Heavy metals in the “plastisphere” of marine microplastics: Adsorption mechanisms and composite risk. Gondwana Res. 2021, 108, 171–180. [Google Scholar] [CrossRef]
- Cornejo-D’Ottone, M.; Molina, V.; Pavez, J.; Silva, N. Greenhouse gas cycling by the plastisphere: The sleeper issue of plastic pollution. Chemosphere 2020, 246, 125709. [Google Scholar] [CrossRef]
- Seeley, M.E.; Song, B.; Passie, R.; Hale, R.C. Microplastics Affect Sedimentary Microbial Communities and Nitrogen Cycling. Nat. Commun. 2020, 11, 2372. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, R.; Shan, D.; Shao, Z. Indigenous oil-degrading bacteria in crude oil-contaminated seawater of the Yellow Sea, China. Appl. Microbiol. Biotechnol. 2014, 98, 7281. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, Q.; Shao, Z. Thioclava nitratireducens sp nov.; isolated from surface seawater. Int. J. Syst. Evol. Microbiol. 2017, 67, 2109–2113. [Google Scholar] [CrossRef]
- Suarez, C.; Hackl, T.; Wilen, B.M.; Persson, F.; Hagelia, P.; Jetten, M.S.M.; Martins, P.D. Novel and unusual genes for nitrogen and metal cycling in Planctomycetota and KSB1-affiliated metagenome-assembled genomes reconstructed from a marine subsea tunnel. FEMS Microbiol. Lett. 2023, 370, 1–10. [Google Scholar] [CrossRef]
- Audrézet, F.; Pochon, X.; Floerl, O.; Le Guen, M.J.; Trochel, B.; Gambarini, V.; Lear, G.; Zaiko, A. Eco-Plastics in the Sea: Succession of Micro- and Macro-Fouling on a Biodegradable Polymer Augmented with Oyster Shell. Front. Mar. Sci. 2022, 9, 891183. [Google Scholar] [CrossRef]
- Denger, K.; Mayer, J.; Buhmann, M.; Weinitschke, S.; Smits, T.H.M.; Cook, A.M. Bifurcated degradative pathway of 3-sulfolactate in Roseovarius nubinhibens ISM via sulfoacetaldehyde acetyltransferase and (S)-cysteate sulfolyase. J. Bacteriol. 2009, 191, 5648–5656. [Google Scholar] [CrossRef]
- Sosa, O.A.; Repeta, D.J.; Ferrón, S.; Bryant, J.A.; Mende, D.R.; Karl, D.M.; DeLong, E.F. Isolation and characterization of bacteria that degrade phosphonates in marine dissolved organic matter. Front. Microbiol. 2017, 8, 1786. [Google Scholar] [CrossRef]
- Eriksen, M.; Lebreton, L.C.M.; Carson, H.S.; Thiel, M.; Moore, C.J.; Borerro, J.C.; Galgani, F.; Ryan, P.G.; Reisser, J. Plastic Pollution in the World’s Oceans: More than 5 Trillion Plastic Pieces Weighing over 250,000 Tons Afloat at Sea. PLoS ONE 2014, 9, e111913. [Google Scholar] [CrossRef]
- Catao, E.; Gallois, N.; Fay, F.; Misson, B.; Briand, J.-F. Metal resistance genes enrichment in marine biofilm communities selected by biocide-containing surfaces in temperate and tropical coastal environments. Env. Poll. 2021, 268, 115835. [Google Scholar] [CrossRef]
- Tian, B.C.; Liu, G.L.; Chi, Z.; Hu, Z.; Chi, Z.M. Occurrence and Distribution of Strains of Saccharomyces cerevisiae in China Seas. J. Mar. Sci. Eng. 2021, 9, 590. [Google Scholar] [CrossRef]
- Delacuvellerie, A.; Cyriaque, V.; Gobert, S.; Benali, S.; Wattiez, R. The plastisphere in marine ecosystems hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J. Hazard Mat. 2019, 380, 120899. [Google Scholar] [CrossRef]
- Ogunbayo, A.O.; Olanipekun, O.O.; Adamu, I.A. Preliminary Studies on the Microbial Degradation of Plastic Waste Using Aspergillus niger and Pseudomonas sp. J. Environ. Prot. 2019, 10, 625–631. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Labrenz, M. Marine Microbial Assemblages on Microplastics: Diversity, Adaptation, and Role in Degradation. Annu. Rev. Mar. Sci. 2020, 12, 209–232. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Bargiela, R.; Golyshin, P.N. Calm and Frenzy: Marine obligate hydrocarbonoclastic bacteria sustain ocean wellness. Curr. Opin. Biotechnol. 2022, 73, 337–345. [Google Scholar] [CrossRef]
- Khan, Z.A.; Kumar, R.; Dasgupta, I. CRISPR/Cas-Mediated Resistance against Viruses in Plants. Int. J. Mol. Sci. 2022, 23(4), 2303. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses Executive Committee. The new scope of virus taxonomy: Partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020, 5, 668–674. [Google Scholar] [CrossRef]
- Weinbauer, M.G.; Rowe, J.M.; Wilhelm, S.W. Determining rates of virus production in aquatic systems by the virus reduction approach. In Manual of Aquatic Virus Ecology; Wilhelm, S.W., Weinbauer, M.G., Suttle, C.A., Eds.; ASLO. Association for the Sciences of Limnology and Oceanography: Washington, DC, USA, 2010; pp. 1–8. [Google Scholar]
- Lu, J.; Yu, Z.; Ngiam, L.; Guo, J. Microplastics as potential carriers of viruses could prolong virus survival and infectivity. Water Res. 2022, 225, 119115. [Google Scholar] [CrossRef]
- Li, R.; Zhu, L.; Cui, L.; Zhu, Y.-G. Viral diversity and potential environmental risk in microplastic at watershed scale: Evidence from metagenomic analysis of the plastisphere. Environ. Int. 2022, 161, 107146. [Google Scholar] [CrossRef]
- Stentiford, G.D.; Lightner, D.V. Cases of White Spot Disease (WSD) in European shrimp farms. Aquaculture 2011, 319, 302–306. [Google Scholar] [CrossRef]
- Feijó, R.G.; Kamimura, M.T.; Oliveira-Neto, J.M.; Vila-Nova, C.M.V.M.; Gomes, A.C.S.; Maria das Graças, L.C.; Vasconcelos, R.F.; Gesteira, T.C.V.; Marins, L.F.; Maggioni, R. Infectious myonecrosis virus and white spot syndrome virus co-infection in Pacific white shrimp (Litopenaeus vannamei) farmed in Brazil. Aquaculture 2013, 380–383, 1–5. [Google Scholar] [CrossRef]
- Yaemkasem, S.; Boonyawiwat, V.; Sukmak, M.; Thongratsakul, S.; Poolkhet, C. Spatial and temporal patterns of white spot disease in Rayong Province, Thailand, from October 2015 to September 2018. Prev. Vet. Med. 2022, 199, 105560. [Google Scholar] [CrossRef]
- Patil, P.K.; Geetha, R.; Ravisankar, T.; Avunje, S.; Solanki, H.G.; Abraham, T.J.; Vinoth, S.P.; Jithendran, K.P.; Alavandi, S.V.; Vijayan, K.K. Economic loss due to diseases in Indian shrimp farming with special reference to Enterocytozoon hepatopenaei (EHP) and white spot syndrome virus (WSSV). Aquaculture 2021, 533, 736231. [Google Scholar] [CrossRef]
- Lightner, D.V.; Redman, R.M.; Pantoja, C.R.; Tang, K.F.J.; Noble, B.L.; Schofield, P.; Mohney, L.L.; Nunan, L.M.; Navarro, S.A. Historic emergence, impact and current status of shrimp pathogens in the Americas. J. Invertebr. Pathol. 2012, 110, 174–183. [Google Scholar] [CrossRef]
- Srisala, J.; Thaiue, D.; Saguanrut, P.; Taengchaiyaphum, S.; Flegel, T.W.; Sritunyalucksana, K. Wenzhou shrimp virus 8(WzSV8) detection by unique inclusions in shrimp hepatopancreatic E-cells and by RT-PCR. Aquaculture 2023, 572, 739483. [Google Scholar] [CrossRef]
- Johnson, P.T. Viral diseases of marine invertebrates. Helgol. Mar. Res. 1984, 37, 65–98. [Google Scholar] [CrossRef]
- Field, C. Viral Diseases of Marine Mammals. Marine Mammal Center–MSD VetManual. 2022. Available online: https://www.msdvetmanual.com/exotic-and-laboratory-animals/marine-mammals/viral-diseases-of-marine-mammals (accessed on 14 June 2022).
- Van Bressem, M.F.; Waerebeek, K.V.; Aznar, F.J.; Raga, J.A.; Jepson, P.D.; Duignan, P.; Deaville, R.; Flach, L.; Viddi, F.; Baker, J.R.; et al. Epidemiological pattern of tattoo skin disease: A potential general health indicator for cetaceans. Dis. Aquat. Org. 2009, 85, 225–237. [Google Scholar] [CrossRef]
- Van Etten, J.; Graves, M.; Müller, D.; Bolland, W.; Delaroque, N. Phycodnaviridae—Large DNA algal viruses. Arch. Virol. 2002, 147, 1479–1516. [Google Scholar] [CrossRef] [PubMed]
- Dunigan, D.D.; Fitzgerald, L.A.; Van Etten, J.L. Phycodnaviruses: A peek at genetic diversity. Virus Res. 2006, 117, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Ma, X.; Guo, R.; Jiang, T.; Wang, T.; Shao, H.; He, H.; Wang, H.; Liang, Y.; McMinn, A.; et al. Characterization and genomic analysis of a novel Synechococcus phage S-H9–2 belonging to Bristolvirus genus isolated from the Yellow Sea. Virus Res. 2023, 328, 199072. [Google Scholar] [CrossRef] [PubMed]
- Ormsby, M.J.; Woodford, L.; Quilliam, R.S. Can plastic pollution drive the emergence and dissemination of novel zoonotic diseases? Environ. Res. 2024, 246, 118172. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Wu, M.; Sonne, C.; Lam, S.S.; Kwong, R.W.; Jiang, Y.; Ren, H. The hidden risk of microplastic-associated pathogens in aquatic environments. Eco-Environ. Health 2023, 2, 142–151. [Google Scholar] [CrossRef]
- Keswani, A.; Oliver, D.M.; Gutierrez, T.; Quilliam, R.S. Microbial hitchhikers on marine plastic debris: Human exposure risks at bathing waters and beach environments. Mar. Environ. Res. 2016, 118, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Mincer, T.J.; Bos, R.P.; Zettler, E.R.; Zhao, S.; Asbun, A.A.; Orsi, W.D.; Guzzetta, V.S.; Amaral-Zettler, L.A. Sargasso Sea Vibrio bacteria: Underexplored potential pathovars in a perturbed habitat. Water Res. 2023, 242, 120033. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.M.; Rabadão, E.M.; Ventura, C.; Da Cunha, S.; Côrte-Real, R.; Meliço-Silvestre, A.A. A Case of Vibrio alginolyticus Bacteremia and Probable Sphenoiditis Following a Dive in the Sea. Clin. Infect. Dis. 1993, 17, 299–300. [Google Scholar] [CrossRef] [PubMed]
- Reilly, G.D.; Reilly, C.A.; Smith, E.G.; Baker-Austin, C. Vibrio alginolyticus-associated wound infection acquired in British waters, Guernsey. Euro Surveill. 2011, 16, 19994. [Google Scholar] [CrossRef] [PubMed]
- Citil, B.E.; Derin, S.; Sankur, F.; Sahan, M.; Citil, M.U. Vibrio alginolyticus Associated Chronic Myringitis Acquired in Mediterranean Waters of Turkey. Case Rep. Infect. Dis. 2015, 187212. [Google Scholar] [CrossRef]
- Noguchi, T.; Arakawa, O. Tetrodotoxin–Distribution and accumulation in aquatic organisms, and cases of human intoxication. Mar. Drugs 2008, 6, 220–242. [Google Scholar] [CrossRef] [PubMed]
- Meron, D.; Efrony, R.; Johnson, W.R.; Schaefer, A.L.; Morris, P.J.; Rosenberg, E.; Greenberg, E.P.; Banin, E. Role of flagella in virulence of the coral pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 2009, 75, 5704–5707. [Google Scholar] [CrossRef]
- Ushijima, B.; Richards, G.P.; Watson, M.A.; Schubiger, C.B.; Häse, C.C. Factors affecting infection of corals and larval oysters by Vibrio coralliilyticus. PLoS ONE 2018, 13, e0199475. [Google Scholar] [CrossRef]
- Austin, B.; Austin, D.; Sutherland, R.; Thompson, F.; Swings, J. Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii. Environ. Microbiol. 2005, 7, 1488–1495. [Google Scholar] [CrossRef]
- Naudet, J.; d’Orbcastel, E.R.; Bouvier, T.; Godreuil, S.; Dyall, S.; Bouvy, S.; Rieuvilleneuve, F.; Restrepo-Ortiz, C.X.; Bettarel, Y.; Auguet, J.C. Identifying macroplastic pathobiomes and antibiotic resistance in a subtropical fish farm. Mar. Pollut. Bull. 2023, 194, 115267. [Google Scholar] [CrossRef] [PubMed]
- Kirstein, I.V.; Kirmizi, S.; Wichels, A.; Garin-Fernandez, A.; Erler, R.; Löder, M.; Gerdts, G. Dangerous hitchhikers? Evidence for potentially pathogenic Vibrio spp. on microplastic particles. Mar. Environ Res. 2016, 120, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Billaud, M.; Seneca, F.; Tambutté, E.; Czerucka, D. An Increase of Seawater Temperature Upregulates the Expression of Vibrio parahaemolyticus Virulence Factors Implicated in Adhesion and Biofilm Formation. Front. Microbiol. 2022, 13, 840628. [Google Scholar] [CrossRef]
- Copernicus. European State of the Climate–Summary. 2022. Available online: https://climate.copernicus.eu/esotc/2022/european-state-climate-2022-summary (accessed on 22 July 2023).
- Kimes, N.E.; Grim, C.J.; Johnson, W.R.; Hasan, N.A.; Tall, B.D.; Kothary, M.H.; Kiss, H.; Munk, A.C.; Tapia, R.; Green, L.; et al. Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 2012, 6, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, X.; Ma, M.; Zhuang, Y.; Chang, L.; Xiao, L.; Wang, G. Extracellular Compounds from Pathogenic Bacterium Pseudoalteromonas piscicida X-8 Cause Bleaching Disease, Triggering Active Defense Responses in Commercially Farmed Saccharina japonica. Biology 2022, 12, 47. [Google Scholar] [CrossRef]
- Kaboré, O.D.; Godreuil, S.; Drancourt, M. Planctomycetes as Host-Associated Bacteria: A Perspective That Holds Promise for Their Future Isolations, by Mimicking Their Native Environmental Niches in Clinical Microbiology Laboratories. Front. Cell. Infect. Microbiol. 2020, 10, 519301. [Google Scholar] [CrossRef]
- Kim, K.; Weiss, L.M. Toxoplasma gondii: The model apicomplexan. Int. J. Parasitol. 2004, 34, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.M.; Sander, V.A. Editorial: Apicomplexa Epidemiology, Control, Vaccines and Their Role in Host-Pathogen Interaction. Front. Vet. Sci. 2022, 9, 885181. [Google Scholar] [CrossRef]
- Halle, A.T.; Ghiglione, J.-F. Nanoplastics: A Complex, Polluting Terra Incognita. Environ. Sci. Technol. 2021, 55, 14466–14469. [Google Scholar] [CrossRef]
- Shi, X.; Chen, Z.; Wei, W.; Chen, J.; Ni, B.-J. Toxicity of micro/nanoplastics in the environment: Roles of plastisphere and eco-corona. Soil Environ. Health 2023, 1, 100002. [Google Scholar] [CrossRef]
- Liu, S.; Junaid, M.; Liao, H.; Liu, X.; Wu, Y.; Wang, J. Eco-corona formation and associated ecotoxicological impacts of nanoplastics in the environment. Sci. Total Environ. 2022, 836, 155703. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lacerda, A.L.; Briand, J.-F.; Lenoble, V.; Oreste, E.Q.; Kessler, F.; Pedrotti, M.L. Assessing the Plastisphere from Floating Plastics in the Northwestern Mediterranean Sea, with Emphasis on Viruses. Microorganisms 2024, 12, 444. https://doi.org/10.3390/microorganisms12030444
Lacerda AL, Briand J-F, Lenoble V, Oreste EQ, Kessler F, Pedrotti ML. Assessing the Plastisphere from Floating Plastics in the Northwestern Mediterranean Sea, with Emphasis on Viruses. Microorganisms. 2024; 12(3):444. https://doi.org/10.3390/microorganisms12030444
Chicago/Turabian StyleLacerda, Ana Luzia, Jean-François Briand, Véronique Lenoble, Eliézer Quadro Oreste, Felipe Kessler, and Maria Luiza Pedrotti. 2024. "Assessing the Plastisphere from Floating Plastics in the Northwestern Mediterranean Sea, with Emphasis on Viruses" Microorganisms 12, no. 3: 444. https://doi.org/10.3390/microorganisms12030444
APA StyleLacerda, A. L., Briand, J.-F., Lenoble, V., Oreste, E. Q., Kessler, F., & Pedrotti, M. L. (2024). Assessing the Plastisphere from Floating Plastics in the Northwestern Mediterranean Sea, with Emphasis on Viruses. Microorganisms, 12(3), 444. https://doi.org/10.3390/microorganisms12030444







