Isolation of Thermophilic Bacteria from Extreme Environments in Northern Chile
Abstract
:1. Introduction
2. Materials and Methods
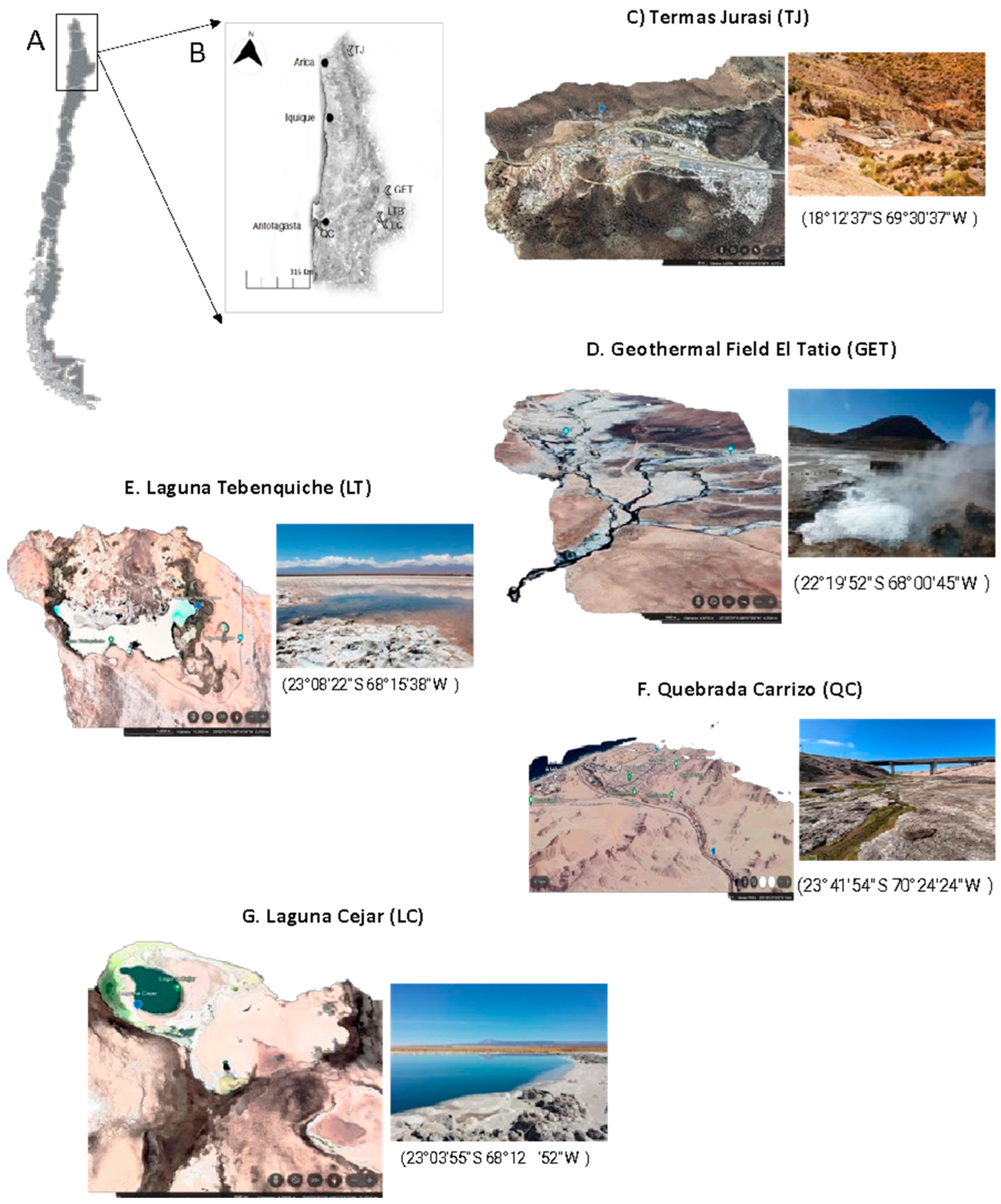
2.1. Sample Collection
2.2. Culture Media and Isolation of Thermophilic Bacteria
2.3. Determination of Cultivation Parameters
2.4. Determination of Hydrolytic Enzymes Activities
2.4.1. Hydrolysis of Sugars
2.4.2. Starch Hydrolysis
2.4.3. Lipid Hydrolysis
2.4.4. Protein Hydrolysis
2.4.5. Cellulolytic Hydrolysis
2.5. Sporulation Tests
2.6. DNA Extraction
2.7. Taxonomic Identification of the Isolates
| Culture Medium | Organic Source | Mineral Salts | Reference |
|---|---|---|---|
| TT medium | 0.4% Yeast extract 0.8% Peptone | 0.2% (w/v) NaCl | [38] |
| YPS | 0.1% Yeast extract 0.4% Peptone | 0.5% Artificial seawater | [39] |
| Miller | 2 g/L Yeast extract | 1.5 g/L KCl 0.5 g/L K2HPO4 0.5 g/L KH2PO4 | [40] |
| Kunaemneni | Solution A: 0.75% Glucose 0.5% Peptone | Solution B: 0.5% (w/v) MgSO4 0.5% (w/v) KH2PO4 0.01% (w/v) FeSO4. 7 H2O Mix 0.5% v/v of solution A in solution B. | [41] |
| Huang | 0.5% Yeast extract 1% Peptone 0.5 g/L Glucose | 0.4 g/L Na2HPO4 0.085% (w/v) Na2CO3 0.02 g/L ZnSO4 0.02 g/L MgSO4 0.02 g/L CaCl2 | [42] |
| ATM 1 | 0.3% Yeast extract 0.25% Peptone | AMT salt solution: 832.42 mg/L CaCl2 8.22 mg/L MgCl2 334.85 mg/L NaCl 4697.4 mg/L KCl 16.763 mg/L SiO2 77.832 mg/L NaHSO4 14.377 mg/L CsCl 0.030 mg/L FeSO4 0.025 mg/L CuSO4 | Designed based on the results of Fernandez-Turiel et al. (2005) [28,43]. |
| ATM 2 | 0.1% Yeast extract 0.4% peptone | ATM salt solution: Same as ATM1. | Designed based on the results of Fernandez-Turiel et al. (2005) [28,43]. |
2.8. Depositions of Strains
3. Results
3.1. Sample Characteristics
3.2. Cultivation and Isolation of Thermophilic Microorganisms
3.3. Taxonomic Identification Based on 16S rRNA Sequencing and Phylogenetic Analysis
3.4. Determination of Hydrolytic Enzymes Activities in Isolated Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Madigan, M.T.; Martinko, J.M.; Brock, T. Brock Biology of Microorganisms, 11th ed.; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2006. [Google Scholar]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the Extremes: Extremophiles and the Limits of Life in a Planetary Context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef]
- Satyanarayana, T.; Raghukumar, C.; Shivaji, S. Extremophilic microbes: Diversity and perspectives. Curr. Sci. 2005, 89, 78–90. Available online: http://www.jstor.org/stable/24110434 (accessed on 15 February 2023).
- Rothschild, L.J.; Mancinelli, R.L. Life in extreme environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002, 13, 253–261. [Google Scholar] [CrossRef]
- Van den Burg, B. Extremophiles as a source for novel enzymes. Curr. Opin. Microbiol. 2003, 6, 213–218. [Google Scholar] [CrossRef] [PubMed]
- MacElroy, R. Some comments on the evolution od extremophiles. Biosystems 1974, 6, 74–75. [Google Scholar] [CrossRef]
- Kumar, L.; Awasthi, G.; Singh, B. Extremophiles: A Novel Source of Industrially Important Enzymes. Biotechnology 2011, 10, 121–135. [Google Scholar] [CrossRef]
- Sysoev, M.; Grötzinger, S.W.; Renn, D.; Eppinger, J.; Rueping, M.; Karan, R. Bioprospecting of Novel Extremozymes from Prokaryotes—The Advent of Culture-Independent Methods. Front. Microbiol. 2021, 12, 630013. [Google Scholar] [CrossRef]
- Gupta, G.N.; Srivastava, S.; Khare, S.K.; Prakash, V. Extremophiles: An overview of microorganisms from extreme environment. Int. J. Agric. Environ. Biotechnol. 2014, 7, 371–380. [Google Scholar] [CrossRef]
- Mesbah, N.M. Industrial Biotechnology Based on Enzymes from Extreme Environments. Front. Bioeng. Biotechnol. 2022, 10, 870083. [Google Scholar] [CrossRef]
- Niehaus, F.; Bertoldo, C.; Kähler, M.; Antranikian, G. Extremophiles as a source of novel enzymes for industrial application. Appl. Microbiol. Biotechnol. 1999, 51, 711–729. [Google Scholar] [CrossRef]
- Antranikian, G.; Vorgias, C.E.; Bertoldo, C. Extreme environments as a resource for microorganisms and novel biocatalysts. In Marine Biotechnology I; Springer: Berlin/Heidelberg, Germany, 2005; pp. 219–262. [Google Scholar]
- Demirjian, D.C.; Morís-Varas, F.; Cassidy, C.S. Enzymes from extremophiles. Curr. Opin. Chem. Biol. 2001, 5, 144–151. [Google Scholar] [CrossRef]
- Mangrola, A.V.; Patel, R.K.; Dudhagara, P.; Gandhi, H.; Ghelani, A.; Jain, K.R.; Shah, H.; Mevada, V.A. Thermophiles: Physiology, Metabolism, Enzymology, and Adaptation Mechanisms. In Physiology, Genomics, and Biotechnological Applications of Extremophiles; Gunjal, A., Thombre, R., Parray, J., Eds.; IGI Global: Hershey, PA, USA, 2022; pp. 65–93. [Google Scholar] [CrossRef]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef]
- Stetter, K.O. Hyperthermophilic prokaryotes. FEMS Microbiol. Rev. 1996, 18, 149–158. [Google Scholar] [CrossRef]
- Fiala, G.; Stetter, K.O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 °C. Arch. Microbiol. 1986, 145, 56–60. [Google Scholar] [CrossRef]
- Takai, K.; Nakamura, K.; Toki, T.; Tsunogai, U.; Miyazaki, M.; Miyazaki, J.; Hirayama, H.; Nakagawa, S.; Nunoura, T.; Horikoshi, K. Cell proliferation at 122 C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. USA 2008, 105, 10949–10954. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Xiao, X.; Xu, J. An integrative genomic island affects the adaptations of the piezophilic hyperthermophilic archaeon Pyrococcus yayanosii to high temperature and high hydrostatic pressure. Front. Microbiol. 2016, 7, 1927. [Google Scholar] [CrossRef] [PubMed]
- Straub, C.T.; Counts, J.A.; Nguyen, D.M.N.; Wu, C.H.; Zeldes, B.M.; Crosby, J.R.; Conway, J.M.; Otten, J.K.; Lipscomb, G.L.; Schut, G.J.; et al. Biotechnology of extremely thermophilic archaea. FEMS Microbiol. Rev. 2018, 42, 543–578. [Google Scholar] [CrossRef]
- Haki, G.D.; Rakshit, S.K. Developments in industrially important thermostable enzymes: A review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Arbab, S.; Ullah, H.; Khan, M.I.U.; Khattak, M.N.K.; Zhang, J.; Li, K. Diversity and distribution of thermophilic microorganisms and their applications in biotechnology. J. Basic Microbiol. 2022, 62, 95–108. [Google Scholar] [CrossRef]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef]
- Glennon, J.A.; Pfaff, R.M. The extraordinary thermal activity of El Tatio Geyser Field, Antofagasta Region, Chile. GOSA Trans. 2003, 8, 31–78. [Google Scholar]
- Lizama, C.; Monteoliva-Sánchez, M.; Prado, B.; Ramos-Cormenzana, A.; Weckesser, J.; Campos, V. Taxonomic study of extreme halophilic archaea isolated from the “Salar de Atacama”, Chile. Syst. Appl. Microbiol. 2001, 24, 464–474. [Google Scholar] [CrossRef]
- Castro, T.B.; Gajardo, G.; Castro, J.M.; Castro, G.M. A biometric and ecologic comparison between Artemia from Mexico and Chile. Saline Syst. 2006, 2, 13. [Google Scholar] [CrossRef]
- Valenzuela, B.; Solís-Cornejo, F.; Araya, R.; Zamorano, P. Isolation and Characterization of Thermus thermophilus Strain ET-1: An Extremely Thermophilic Bacterium with Extracellular Thermostable Proteolytic Activity Isolated from El Tatio Geothermal Field, Antofagasta, Chile. Int. J. Mol. Sci. 2023, 24, 14512. [Google Scholar] [CrossRef] [PubMed]
- Porter, K.G.; Feig, Y.S. The use of DAPI for identifying and counting aquatic microflora 1. Limnol. Oceanogr. 1980, 25, 943–948. [Google Scholar] [CrossRef]
- Connon, S.A.; Giovannoni, S.J. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 2002, 68, 3878–3885. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, P.; Murray, R.G.E.; Wood, W.A.; Krieg, N.R. Methods for General and Molecular Bacteriology; American Society for Microbiology: Washington, DC, USA, 1994. [Google Scholar]
- Scheldeman, P.; Pil, A.; Herman, L.; De Vos, P.; Heyndrickx, M. Incidence and diversity of potentially highly heat-resistant spores isolated at dairy farms. Appl. Environ. Microbiol. 2005, 71, 1480–1494. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M.D., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Demirtas, M.U.; Kolhatkar, A.; Kilbane, J.J., II. Effect of aeration and agitation on growth rate of Thermus thermophilus in batch mode. J. Biosci. Bioeng. 2003, 95, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Charbonnier, F.; Erauso, G.; Barbeyron, T.; Prieur, D.; Forterre, P. Evidence that a plasmid from a hyperthermophilic archaebacterium is relaxed at physiological temperatures. J. Bacteriol. 1992, 174, 6103–6108. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; San, K.Y. Continuous production of cell-free recombinant proteins using Escherichia coli. Biotechnol. Prog. 1993, 9, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Kunamneni, A.; Permaul, K.; Singh, S. Amylase production in solid state fermentation by the thermophilic fungus Thermomyces lanuginosus. J. Biosci. Bioeng. 2005, 100, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Guangrong, H.; Tiejing, Y.; Po, H.; Jiaxing, J. Purification and characterization of a protease from Thermophilic bacillus strain HS08. Afr. J. Biotechnol. 2006, 5, 2433–2438. [Google Scholar]
- Fernandez-Turiel, J.L.; Garcia-Valles, M.; Gimeno-Torrente, D.; Saavedra-Alonso, J.; Martinez-Manent, S. The hot spring and geyser sinters of El Tatio, Northern Chile. Sediment. Geol. 2005, 180, 125–147. [Google Scholar] [CrossRef]
- Madigan, M.T.; Orent, A. Thermophilic and halophilic extremophiles. Curr. Opin. Microbiol. 1999, 2, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Kita, K.; Ishida, A.; Tanaka, K.; Ishikawa, S.; Yoshida, K.-I. Complete Genome Sequence of Thermophilic Bacterium Aeribacillus pallidus PI8. Microbiol. Resour. Announc. 2020, 9, e00224-20. [Google Scholar] [CrossRef]
- Sathiyanarayanan, G.; Filippidou, S.; Junier, T.; Rufatt, P.M.; Jeanneret, N.; Wunderlin, T.; Sieber, N.; Dorador, C.; Junier, P. Manganese-II oxidation and Copper-II resistance in endospore forming Firmicutes isolated from uncontaminated environmental sites. AIMS Environ. Sci. 2016, 3, 220–238. [Google Scholar] [CrossRef]
- Orellana, R.; Macaya, C.; Bravo, G.; Dorochesi, F.; Cumsille, A.; Valencia, R.; Rojas, C.; Seeger, M. Living at the frontiers of life: Extremophiles in Chile and their potential for bioremediation. Front. Microbiol. 2018, 9, 2309. [Google Scholar] [CrossRef]
- Brock, T.D. Thermophilic Microorganisms and Life at High Temperatures; Springer: New York, NY, USA, 1978. [Google Scholar]
- Khalil, A. Screening and characterization of thermophilic bacteria (lipase, cellulase and amylase producers) from hot springs in Saudi Arabia. J. Food Agric. Environ. 2011, 9, 672–675. [Google Scholar]
- Sung, M.H.; Kim, H.; Bae, J.W.; Rhee, S.K.; Jeon, C.O.; Kim, K.; Kim, J.J.; Hong, S.P.; Lee, S.G.; Yoon, J.H.; et al. Geobacillus toebii sp. nov., a novel thermophilic bacterium isolated from hay compost. Int. J. Syst. Evol. Microbiol. 2002, 52, 2251–2255. [Google Scholar]
- Feng, L.; Wang, W.; Cheng, J.; Ren, Y.; Zhao, G.; Gao, C.; Tang, Y.; Liu, X.; Han, W.; Peng, X.; et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. USA 2007, 104, 5602–5607. [Google Scholar] [CrossRef]
- Tai, S.K.; Lin, H.P.P.; Kuo, J.; Liu, J.K. Isolation and characterization of a cellulolytic Geobacillus thermoleovorans T4 strain from sugar refinery wastewater. Extremophiles 2004, 8, 345–349. [Google Scholar] [CrossRef]
- Donachie, S.P.; Bowman, J.P.; Alam, M. Nesiotobacter exalbescens gen. nov., sp. nov., a moderately thermophilic alphaproteobacterium from an Hawaiian hypersaline lake. Int. J. Syst. Evol. Microbiol. 2006, 56, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Marchant, R.; Banat, I.M.; Rahman, T.J.; Berzano, M. The frequency and characteristics of highly thermophilic bacteria in cool soil environments. Environ. Microbiol. 2002, 4, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M.; Marchant, R.; Rahman, T.J. Geobacillus debilis sp. nov., a novel obligately thermophilic bacterium isolated from a cool soil environment, and reassignment of Bacillus pallidus to Geobacillus pallidus comb. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 2197–2201. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Jean, J.-S.; Kar, S.; Chou, M.-L.; Chen, C.-Y. Screening of plant growth-promoting traits in arsenic-resistant bacteria isolated from agricultural soil and their potential implication for arsenic bioremediation. J. Hazard. Mater. 2014, 272, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Harwood, C.R.; Mouillon, J.-M.; Pohl, S.; Arnau, J. Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 2018, 42, 721–738. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, T.; Sharma, A.; Mehta, D.; Puri, A.K.; Kumar, V.; Nisha, M.; Joshi, S. Biotechnological Applications of Biocatalysts from the Firmicutes Bacillus and Geobacillus Species. In Microorganisms in Sustainable Agriculture and Biotechnology; Springer: Dordrecht, The Netherlands, 2012; pp. 343–379. [Google Scholar]
- Pikuta, E.; Lysenko, A.; Chuvilskaya, N.; Mendrock, U.; Hippe, H.; Suzina, N.; Nikitin, D.; Osipov, G.; Laurinavichius, K. Anoxybacillus pushchinensis gen. nov., sp. nov., a novel anaerobic, alkaliphilic, moderately thermophilic bacterium from manure, and description of Anoxybacillus flavitherms comb. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 2109–2117. [Google Scholar] [CrossRef]
- Da Rosa, D.F.; Macedo, A.J. The genus Anoxybacillus: An emerging and versatile source of valuable biotechnological products. Extremophiles 2023, 27, 22. [Google Scholar] [CrossRef]
- Nakamichi, T.; Nakashima, T.; Fujisaki, H.; Takamatsu, N.; Muramatsu, T.; Takahashi, Y.; Ishibashi, Y. Characteristics of Anoxybacillus sp. MU3 isolated from a hot spring and its application to the hyper thermal solubilization of sewage sludge. Environ. Eng. Sci. 2010, 27, 993–999. [Google Scholar] [CrossRef]
- Acer, Ö.; Bekler, F.M.; Pirinççioğlu, H.; Güven, R.G.; Güven, K. Purification and characterization of thermostable and detergent-stable α-amylase from Anoxybacillus sp. AH1. Food Technol. Biotechnol. 2016, 54, 70. [Google Scholar] [CrossRef]
- Rodas, A.M.; Ferrer, S.; Pardo, I. 16S-ARDRA, a Tool for Identification of Lactic Acid Bacteria Isolated from Grape Must and Wine. Syst. Appl. Microbiol. 2003, 26, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Cihan, A.C. Taxonomic classification of Anoxybacillus isolates from geothermal regions in Turkey by 16S rRNA gene sequences and ARDRA, ITS-PCR, Rep-PCR analyses. Pol. J. Microbiol. 2013, 62, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Karaoglu, H.; Yanmis, D.; Sal, F.A.; Celik, A.; Canakci, S.; Belduz, A.O. Biochemical characterization of a novel glucose isomerase from Anoxybacillus gonensis G2 T that displays a high level of activity and thermal stability. J. Mol. Catal. B Enzym. 2013, 97, 215–224. [Google Scholar] [CrossRef]
- De Almeida, P.Z.; de Oliveira, T.B.; de Lucas, R.C.; Salgado, J.C.S.; Perez, M.M.; Galan, B.; Garcia, J.L.; Polizeli, M.D.L.D. Heterologous production and biochemical characterization of a new highly glucose tolerant GH1 β-glucosidase from Anoxybacillus thermarum. Process. Biochem. 2020, 99, 1–8. [Google Scholar] [CrossRef]
- Wang, J.; Chang, F.; Tang, X.; Li, W.; Yin, Q.; Yang, Y.; Hu, Y. Bacterial laccase of Anoxybacillus ayderensis SK3-4 from hot springs showing potential for industrial dye decolorization. Ann. Microbiol. 2020, 70, 51. [Google Scholar] [CrossRef]
- Poli, A.; Esposito, E.; Lama, L.; Orlando, P.; Nicolaus, G.; De Appolonia, F.; Gambacorta, A.; Nicolaus, B. Anoxybacillus amylolyticus sp. nov., a thermophilic amylase producing bacterium isolated from Mount Rittmann (Antarctica). Syst. Appl. Microbiol. 2006, 29, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Belduz, A.O.; Dulger, S.; Demirbag, Z. Anoxybacillus gonensis sp. nov., a moderately thermophilic, xylose-utilizing, endospore-forming bacterium. Int. J. Syst. Evol. Microbiol. 2003, 53, 1315–1320. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; D’Souza, D.R.; Bhandari, D.; Parashar, V.; Capalash, N. Demonstration of the Principles of Restriction Endonuclease Cleavage Reactions Using Thermostable Bfl I from Anoxybacillus flavithermus. Biochem. Mol. Biol. Educ. 2003, 31, 392–396. [Google Scholar] [CrossRef]
- Yildirim, M.; Col, M.; Colak, A.; Güner, S.; Dülger, S.; Beldüz, A.O. Diphenolases from Anoxybacillus kestanbolensis strains K1 and K4 T. World J. Microbiol. Biotechnol. 2005, 21, 501–507. [Google Scholar] [CrossRef]
- Ertunga, N.S.; Colak, A.; Belduz, A.O.; Canakci, S.; Karaoglu, H.; Sandalli, C. Cloning, expression, purification and characterization of fructose-1, 6-bisphosphate aldolase from Anoxybacillus gonensis G2. J. Biochem. 2007, 141, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Omeroglu, M.A.; Baltaci, M.O.; Adiguzel, A. Anoxybacillus: An overview of a versatile genus with recent biotechnological applications. World J. Microbiol. Biotechnol. 2023, 39, 139. [Google Scholar] [CrossRef]
- Nazina, T.N.; Tourova, T.P.; Poltaraus, A.B.; Novikova, E.V.; A Grigoryan, A.; Ivanova, A.; Lysenko, A.M.; Petrunyaka, V.V.; Osipov, A.G.; Belyaev, S.S.; et al. Taxonomic study of aerobic thermophilic bacilli: Descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int. J. Syst. Evol. Microbiol. 2001, 51, 433–446. [Google Scholar] [CrossRef]
- Khaswal, A.; Chaturvedi, N.; Mishra, S.K.; Kumar, P.R.; Paul, P.K. Current status and applications of genus Geobacillus in the production of industrially important products—A review. Folia Microbiol. 2022, 67, 389–404. [Google Scholar] [CrossRef]
- Hussein, A.H.; Lisowska, B.K.; Leak, D.J. The genus Geobacillus and their biotechnological potential. Adv. Appl. Microbiol. 2015, 92, 1–48. [Google Scholar]
- Aliyu, H.; Lebre, P.; Blom, J.; Cowan, D.; De-Maayer, P. Phylogenomic re-assessment of the thermophilic genus Geobacillus. Syst. Appl. Microbiol. 2016, 39, 527–533. [Google Scholar] [CrossRef]
- Minana-Galbis, D.; Pinzon, D.L.; Loren, J.G.; Manresa, A.; Oliart-Ros, R.M. Reclassification of Geobacillus pallidus (Scholz et al. 1988) Banat et al. 2004 as Aeribacillus pallidus gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 1600–1604. [Google Scholar] [CrossRef]

| Site | Temperature (°C) | pH | Classification | Location |
|---|---|---|---|---|
| Termas Jurasi | 48–52 | 7.5–8.2 | Thermal affluent | 18°12′37″ S 69°30′37″ W 4053 m |
| El Tatio Geothermal Field | 55–82 | 7.5–8.2 | Geothermal field | 22°19′52″ S 68°00′45″ W 4278 m |
| Laguna Tebenquiche | 12–22 | 7.8–8.2 | Hypersaline lagoon, Atacama Desert | 23°08′22″ S 68°15′38″ W 2316 m |
| Laguna Cejar | 22–32 | 7.4–8.6 | Hypersaline lagoon, Atacama Desert | 23°03′55″ S 68°12′52″ W 2334 m |
| Quebrada Carrizo | 18–22 | 6.8–7 | Fresh water spring, Coastal Desert | 23°41′54″ S 70°24′24″ W 95 m |
| Source | Iso-late | Media | Temperature Range/Optimal (°C) | pH Range/Optimal | Morphology | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | YPS | Miller | Kunaemnemi | Huang | ATM1 | ATM2 | Gram | Colony | ||||
| Termas Jurasi (2 isolated) | CJ1 | - | - | ++ | - | ++ | +++ | + | 55–70/65 | 6–8/7 | Positives Bacillaries | Whitish, edges uniform, convex surface |
| CJ5 | - | - | + | - | - | +++ | - | 45–60/60 | 6–9/8 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| -El Tatio Geothermal Field (9 isolated) | CT1 | - | - | - | - | - | +++ | - | 40–65/55 | 6–9/8 | Positives Bacillaries | Whitish, edges uniform, convex surface |
| 702B | - | - | + | - | + | +++ | - | 50–70/60 | 6–9/8 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| 2A55 | - | - | - | - | + | +++ | - | 50–65/55 | 6–9/7 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| 2B55 | - | - | + | - | + | +++ | - | 50–65/60 | 6–9/8 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| SA | - | - | + | - | ++ | +++ | + | 45–70/55 | 6–8/6 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| 2*55 | - | - | + | - | + | +++ | - | 45–65/60 | 6–8/8 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| M8 | - | - | + | - | + | +++ | - | 50–70/50 | 6–8/8 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| 3A55 | - | - | + | - | + | +++ | + | 40–65/55 | 6–8/7 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| Laguna Tebenquiche (2 isolated) | TB4 | - | - | + | - | + | +++ | + | 50–65/55 | 6–8/8 | Positives Bacillaries | Whitish, edges uniform, convex surface |
| TB5 | - | - | + | - | + | +++ | + | 45–60/55 | 6–8/7 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| Laguna Cejar (1 isolated) | C3 | - | - | + | - | + | +++ | + | 45–65/60 | 6–8/8 | Positives Bacillaries | Whitish, edges uniform, convex surface |
| Quebrada Carrizo (2 isolated) | M3 | - | - | ++ | - | + | +++ | + | 45–60/50 | 6–9/7 | Positives Bacillaries | whitish, edges sawn, surface convex |
| Q1 | - | - | + | - | + | +++ | - | 45–70/60 | 6–9/8 | Positives Bacillaries | Whitish, edges uniform, convex surface | |
| Source | Isolate | Closest Strain | Similarity (%) | Deposited Number |
|---|---|---|---|---|
| Termas Jurasi (2 isolated) | CJ1 | Aeribacillus pallidus PI8 | 99.9 | AP022323.1 |
| CJ5 | Anoxybacillus kamchatkensis strain G10 | 99.6 | CP025535.1 | |
| El Tatio Geothermal Field (8 isolated) | CT1 | Anoxybacillus kualawohkensis strain ET103 | 99.5 | KJ722465.1 |
| 702B | Anoxybacillus kualawohkensis strain ET103 | 99.2 | KJ722465.1 | |
| 2A55 | Anoxybacillus tepidamans strain R-35643 | 98.4 | NR_116985.1 | |
| 2B55 | Anoxybacillus kualawohkensis strain ET103 | 98.4 | KJ722465.1 | |
| SA | Anoxybacillus flavithermus clone LK4 | 99.8 | EU816689.1 | |
| 2*55 | Anoxybacillus kualawohkensis strain ET103 | 99.4 | KJ722465.1 | |
| M8 | Anoxybacillus kualawohkensis strain ET103 | 99.2 | KJ722465.1 | |
| 3A55 | Geobacillus thermoleovorans strain SURF-48B | 99.9 | CP061472.1 | |
| Laguna Tebenquiche (2 isolated) | TB4 | Aeribacillus pallidus PI8 | 99.8 | AP022323.1 |
| TB5 | Geobacillus thermoleovorans strain SURF-48B | 99.8 | CP061472.1 | |
| Laguna Cejar (1 isolated) | C3 | Parageobacillus thermoglucosidasius strain TM242 | 99.8 | CP016916.1 |
| Quebrada Carrizo (2 isolated) | M3 | Aeribacillus pallidus PI8 | 100 | AP022323.1 |
| Q1 | Aeribacillus pallidus strain 21KAM21 | 100 | KJ722451.1 |
| Source | Isolated Strain | Closest Strain | Protease Activity | Amylase Activity | Lipase Activity | Cellulase Activity |
|---|---|---|---|---|---|---|
| Termas Jurasi (2 isolated) | CJ1 | Aeribacillus pallidus PI8 | ++ | - | - | - |
| CJ5 | Anoxybacillus kamchatkensis strain G10 | + | - | - | - | |
| El Tatio Geothermal Field (8 isolated) | CT1 | Anoxybacillus kualawohkensis strain ET103 | - | - | - | - |
| 702B | Anoxybacillus kualawohkensis strain ET103 | ++ | + | + | - | |
| 2A55 | Anoxybacillus tepidamans strain R-35643 | - | ++ | - | - | |
| 2B55 | Anoxybacillus kualawohkensis strain ET103 | - | - | - | - | |
| SA | Anoxybacillus flavithermus clone LK4 | - | +++ | - | - | |
| 2*55 | Anoxybacillus kualawohkensis strain ET103 | - | - | - | - | |
| M8 | Anoxybacillus kualawohkensis strain ET103 | + | + | - | - | |
| 3A55 | Geobacillus thermoleovorans strain SURF-48B | + | ++ | +++ | - | |
| Laguna Tebenquiche (2 isolated) | TB4 | Aeribacillus pallidus PI8 | + | - | - | - |
| TB5 | Geobacillus thermoleovorans strain SURF-48B | ++ | - | - | - | |
| Laguna Cejar (1 isolated) | C3 | Parageobacillus thermoglucosidasius strain TM242 | ++ | +++ | - | - |
| Quebrada Carrizo (2 isolated) | M3 | Aeribacillus pallidus PI8 | - | + | - | - |
| Q1 | Aeribacillus pallidus strain 21KAM21 | - | + | + | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valenzuela, B.; Solís-Cornejo, F.; Araya, R.; Zamorano, P. Isolation of Thermophilic Bacteria from Extreme Environments in Northern Chile. Microorganisms 2024, 12, 473. https://doi.org/10.3390/microorganisms12030473
Valenzuela B, Solís-Cornejo F, Araya R, Zamorano P. Isolation of Thermophilic Bacteria from Extreme Environments in Northern Chile. Microorganisms. 2024; 12(3):473. https://doi.org/10.3390/microorganisms12030473
Chicago/Turabian StyleValenzuela, Bernardita, Francisco Solís-Cornejo, Rubén Araya, and Pedro Zamorano. 2024. "Isolation of Thermophilic Bacteria from Extreme Environments in Northern Chile" Microorganisms 12, no. 3: 473. https://doi.org/10.3390/microorganisms12030473
APA StyleValenzuela, B., Solís-Cornejo, F., Araya, R., & Zamorano, P. (2024). Isolation of Thermophilic Bacteria from Extreme Environments in Northern Chile. Microorganisms, 12(3), 473. https://doi.org/10.3390/microorganisms12030473






