Morpho-Anatomy, In Vitro Culture, and Phylogenetic Studies of Two Helicotylenchus Species from Southern Alberta, Canada
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nematode Isolation and Morphological Studies
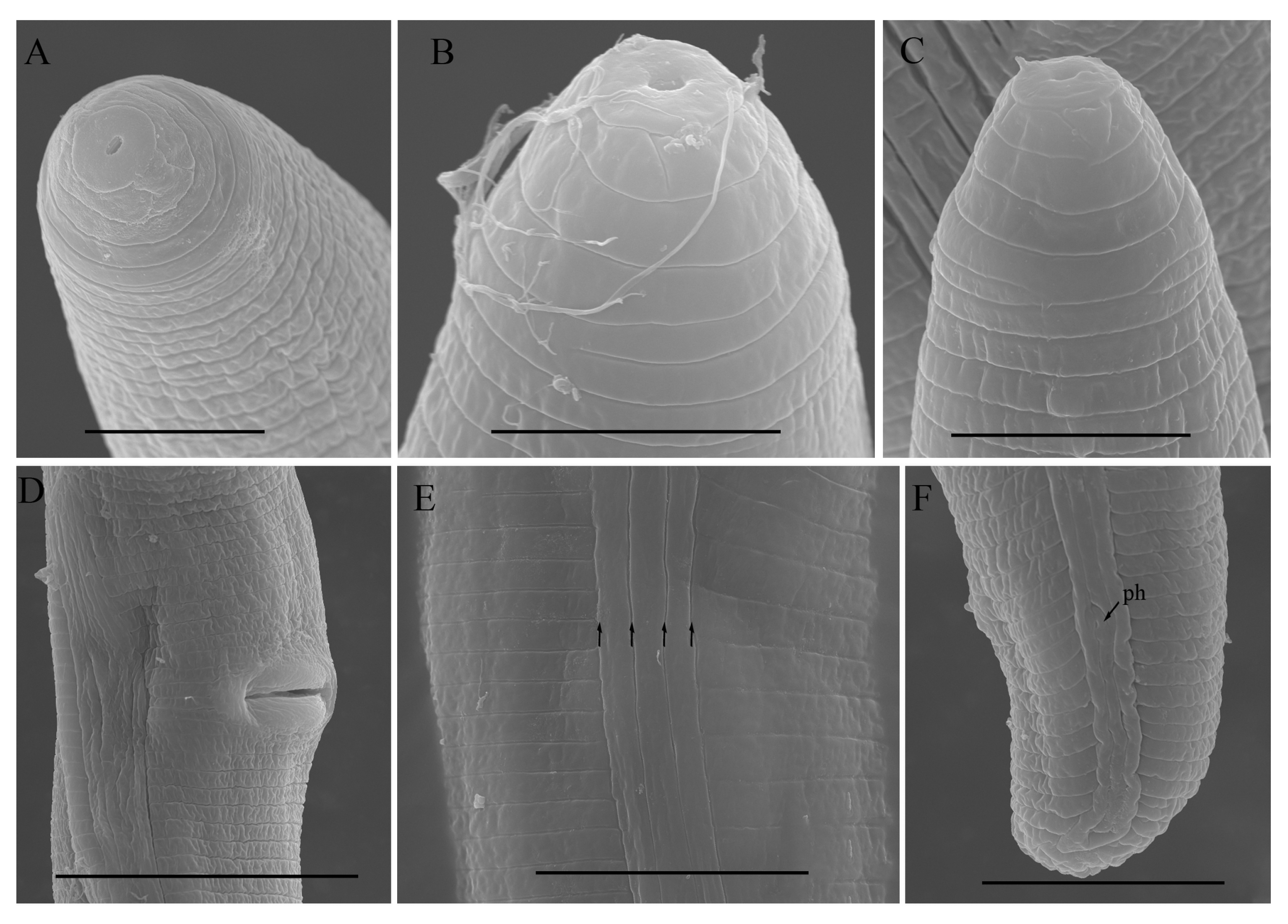
2.2. Scanning Electron Microscopy (SEM)
2.3. DNA Extraction, PCR and Sequencing
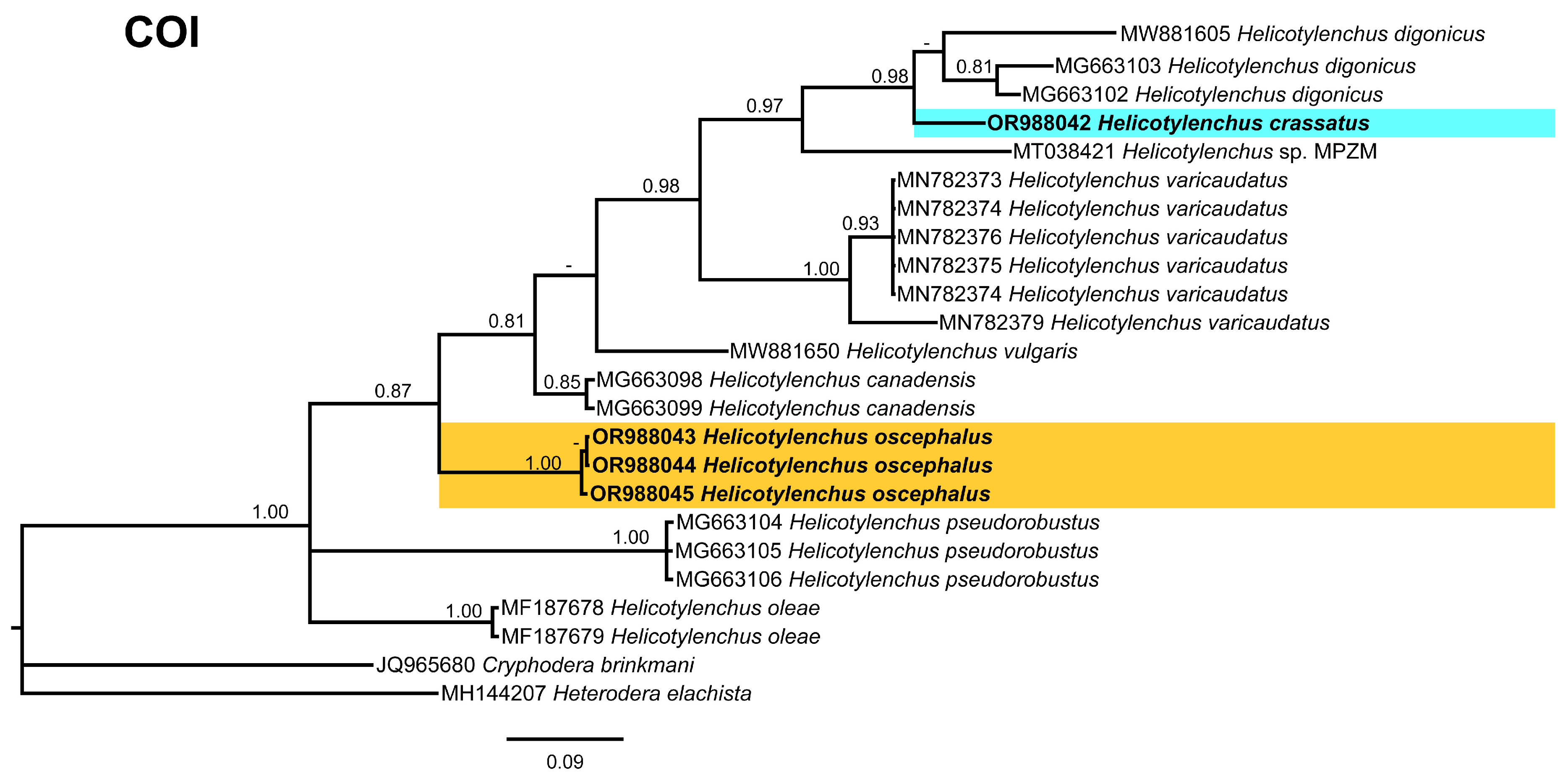
2.4. Phylogenetic Studies
3. Results
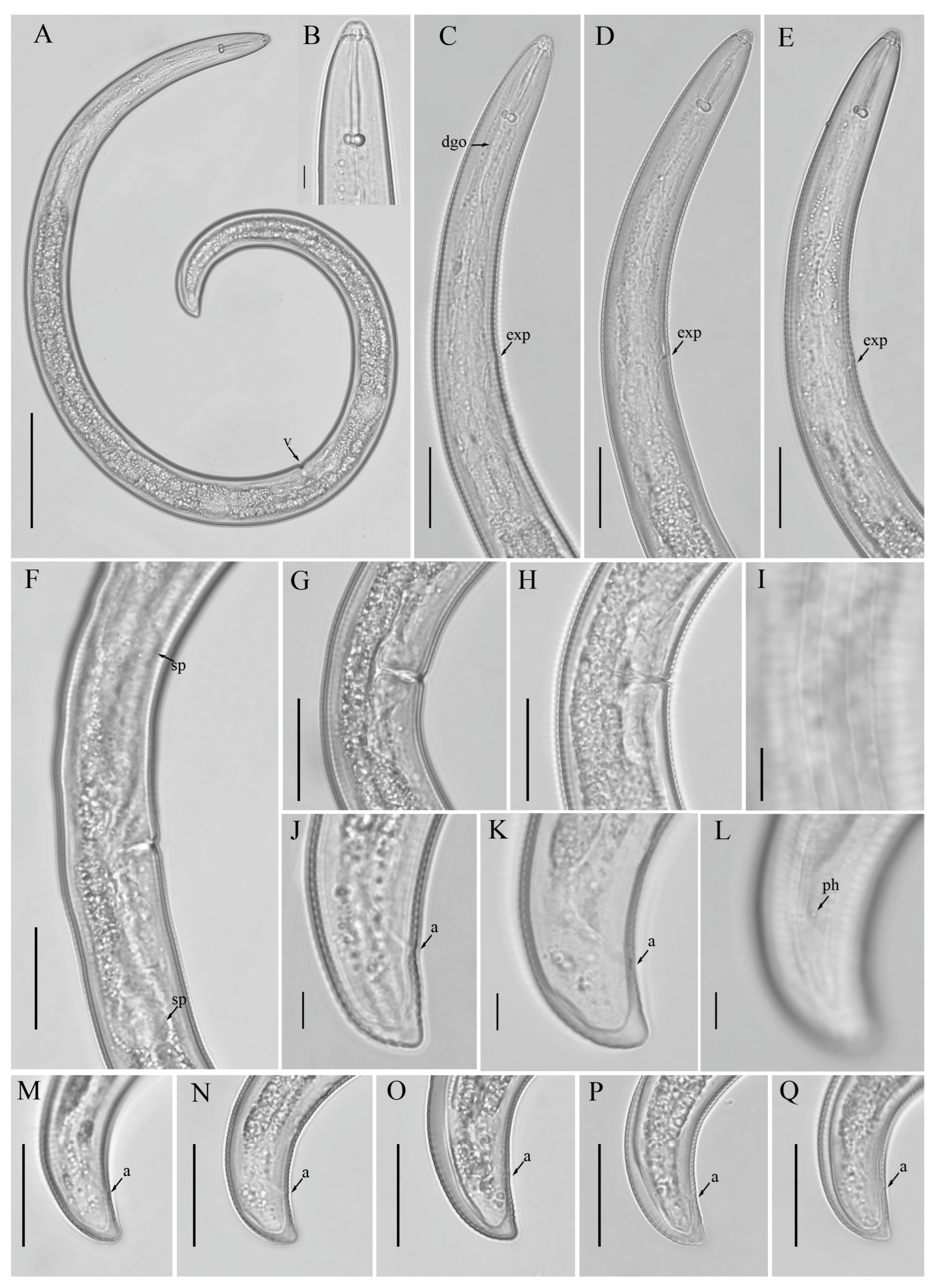
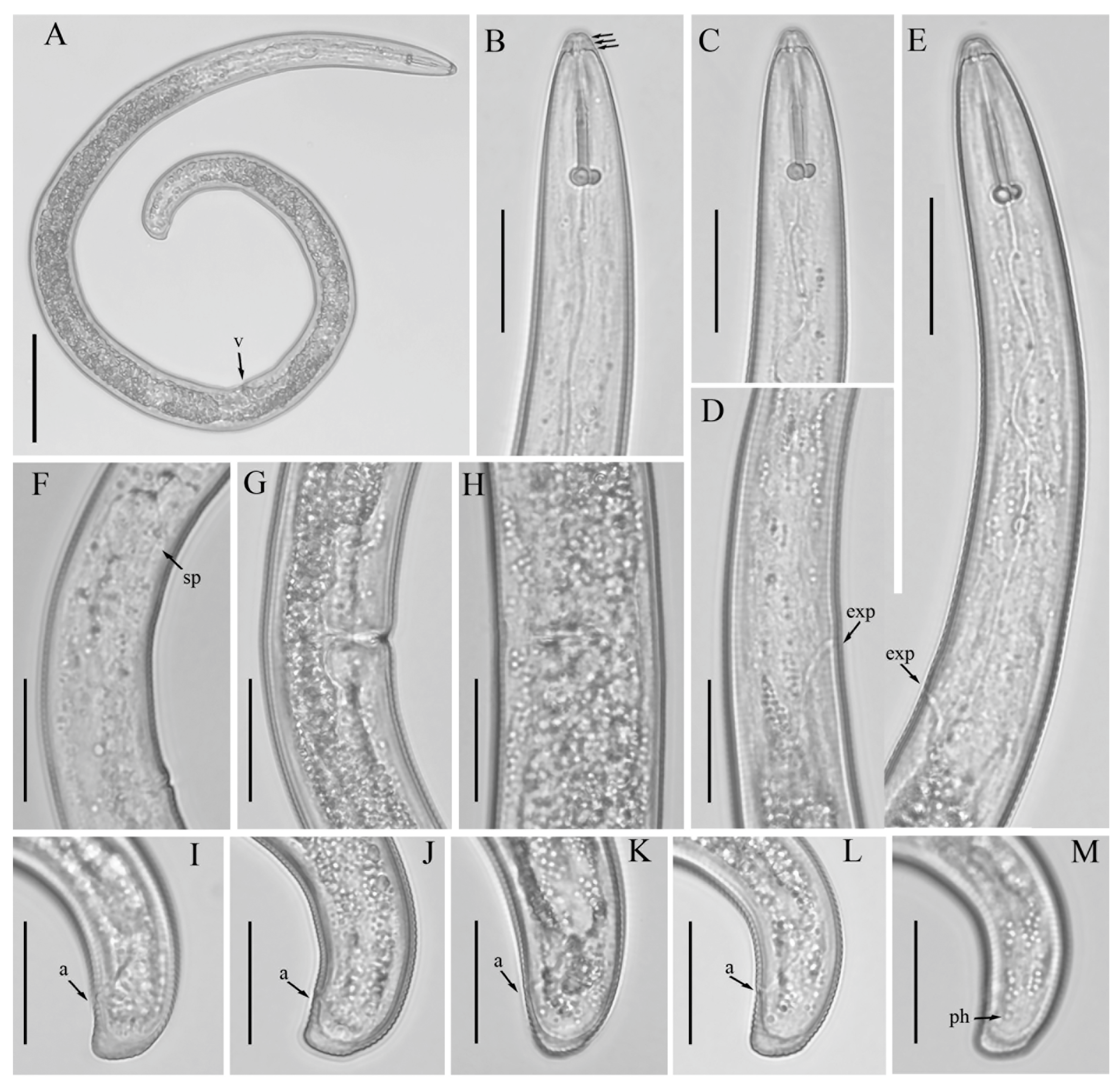
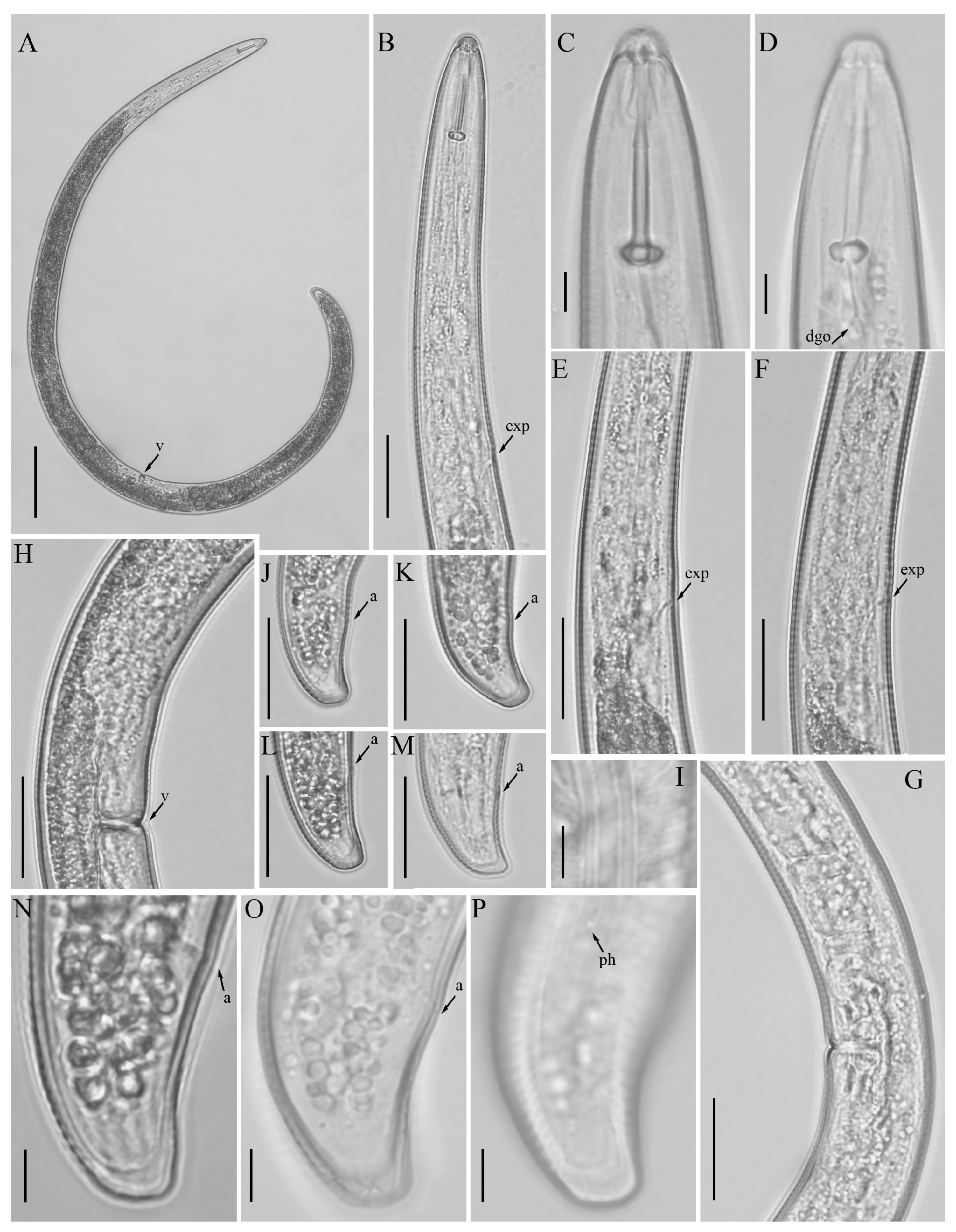
3.1. Description of Helicotylenchus crassatus Anderson [18] Morphotype 1
3.1.1. Description of Helicotylenchus crassatus Morphotype 2
3.1.2. Habitat, Locality and Host Associations
3.2. Designation of Helicotylenchus crassatus Morphotypes
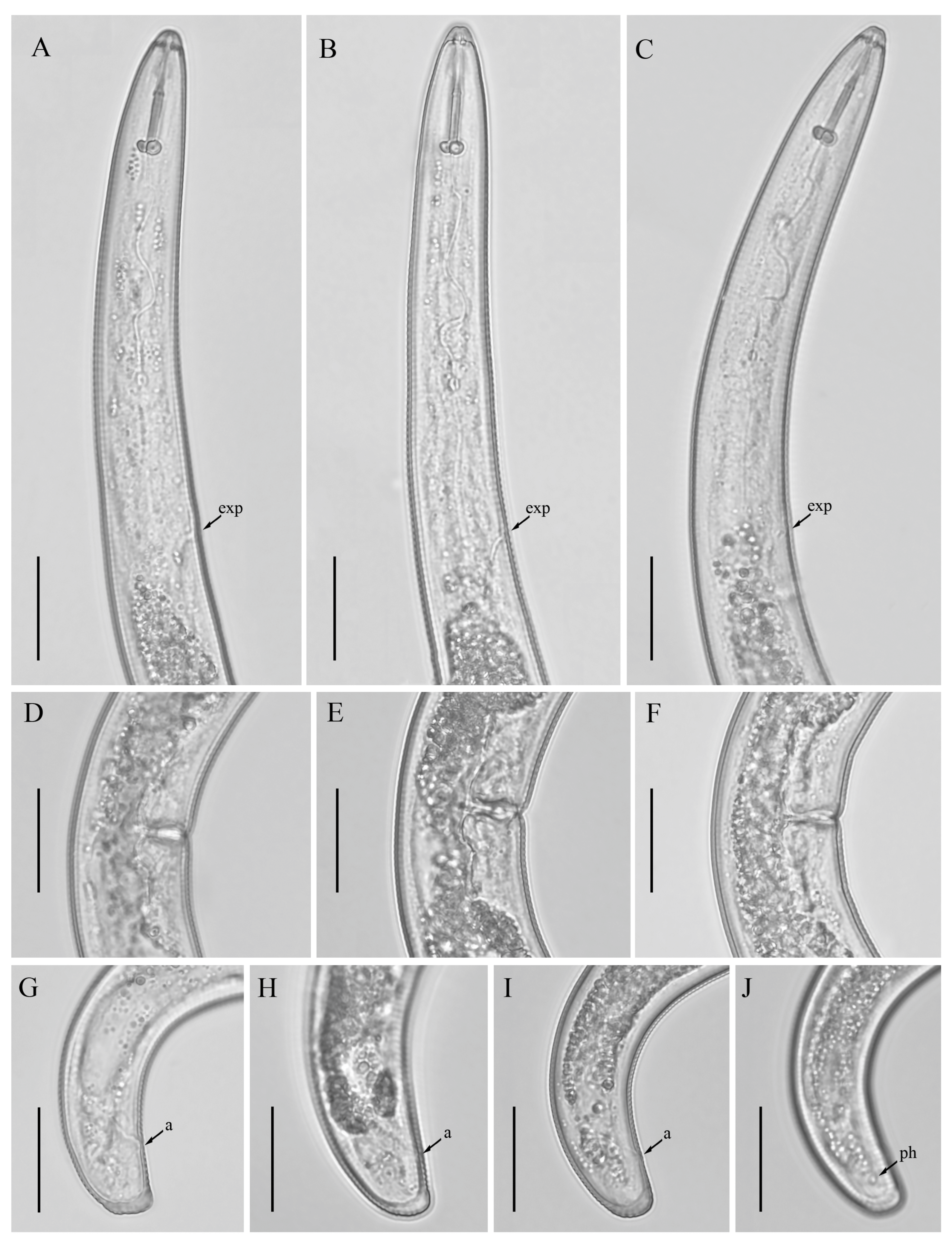
3.3. Description of Helicotylenchus oscephalus Anderson [16]
3.3.1. Habitat, Locality and Host Associations
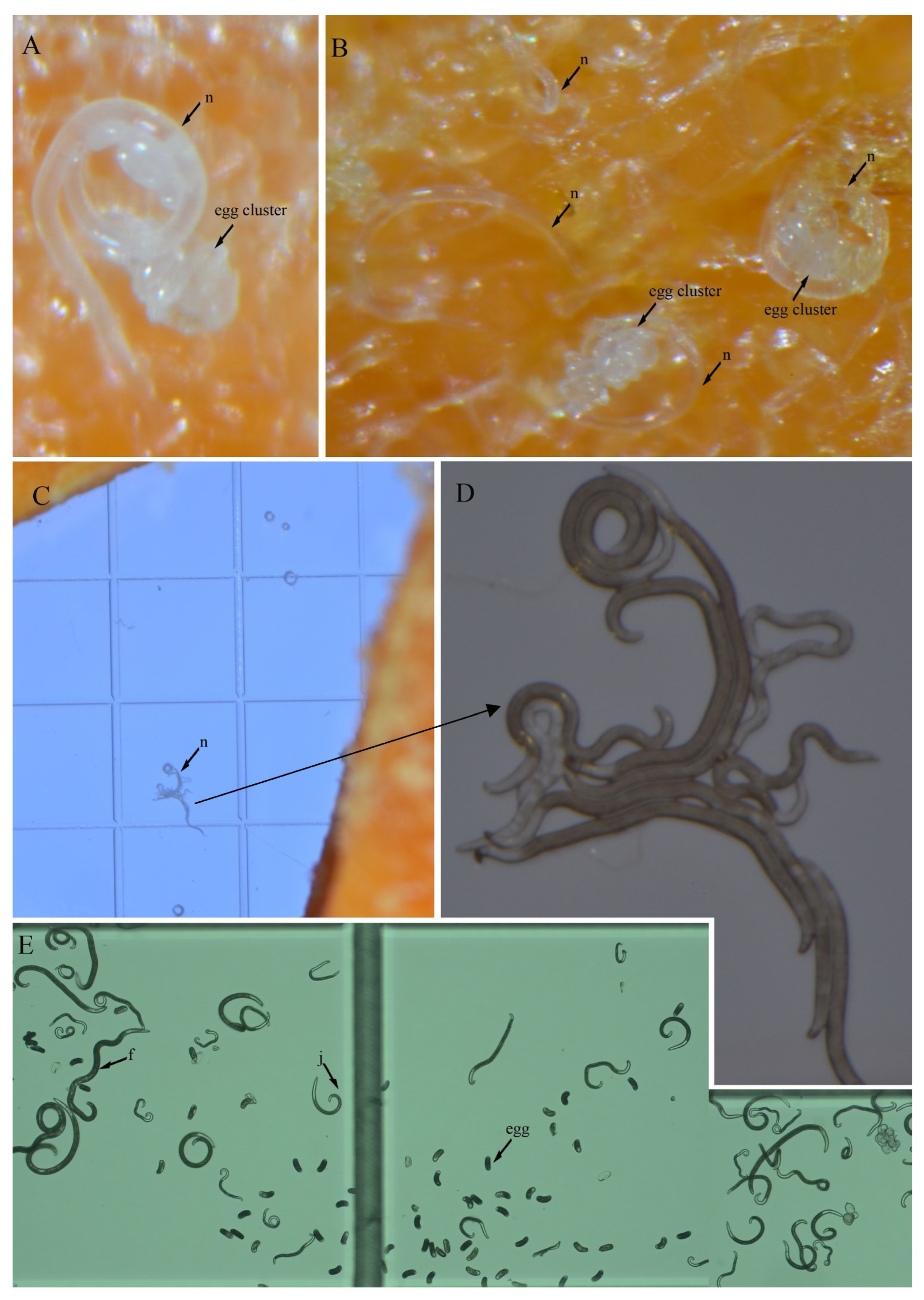
3.4. In Vitro Culture Suitability
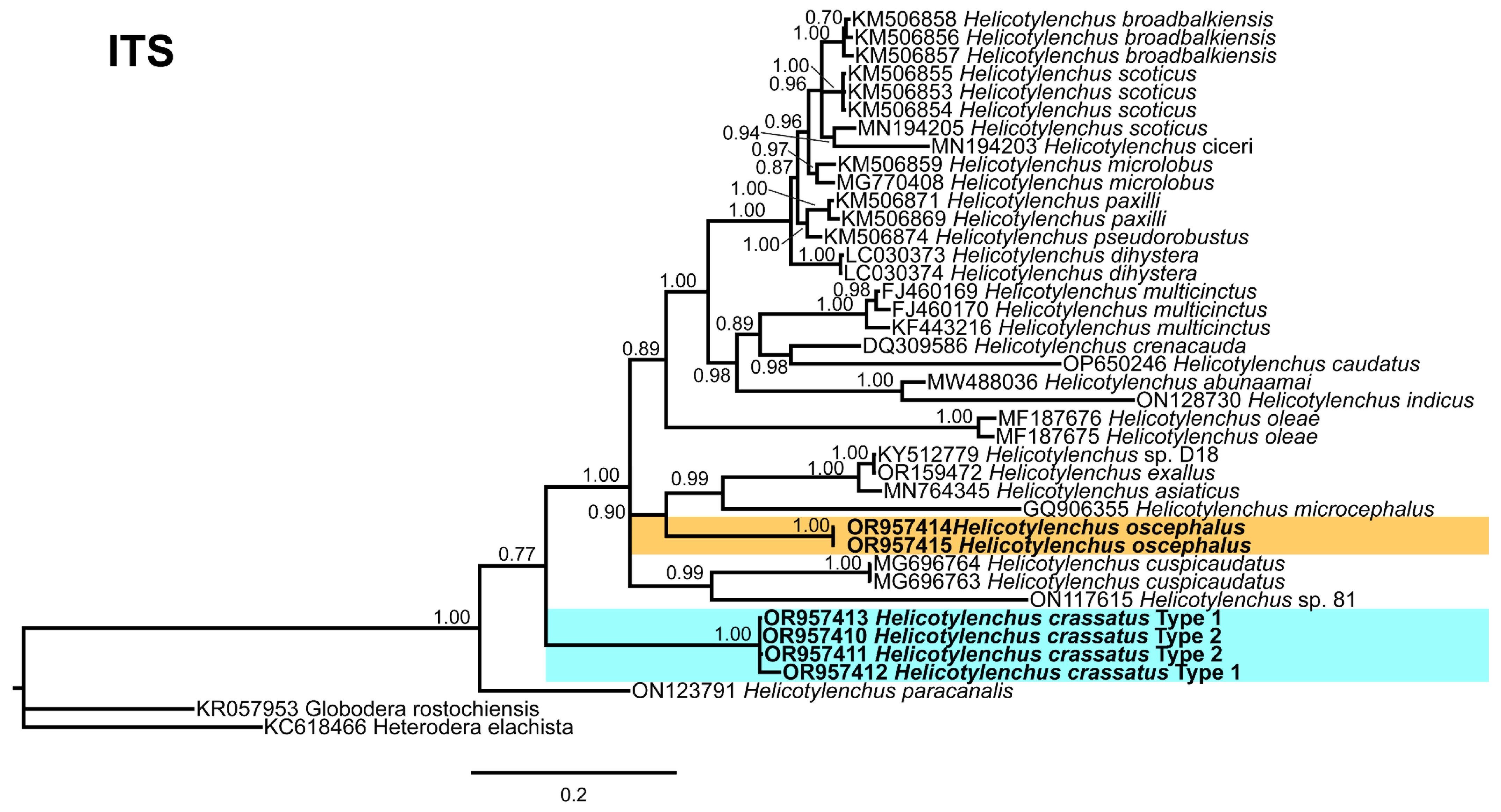
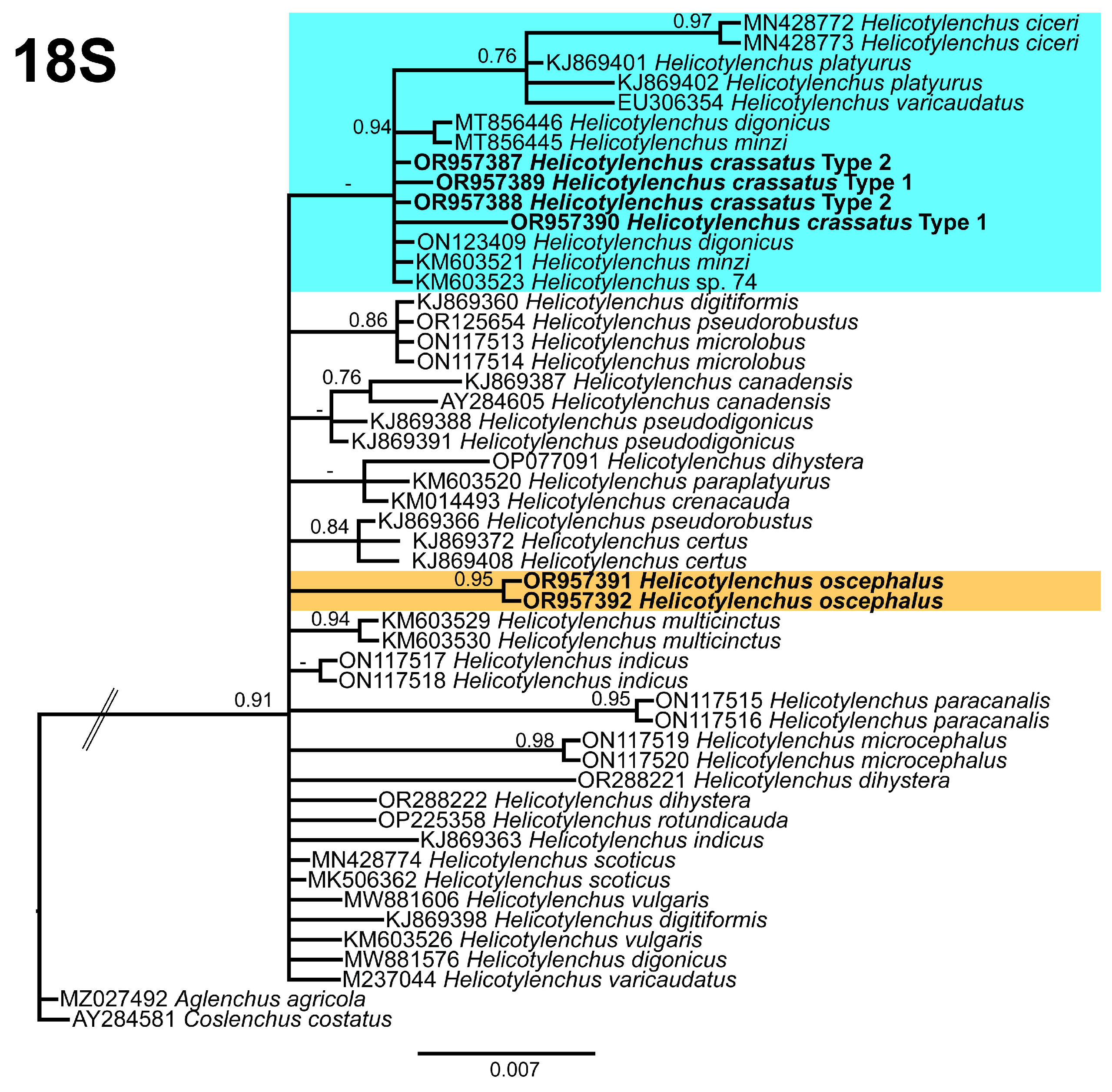
3.5. Phylogenetic Relationships of Detected Species with Other Helicotylenchus Species
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steiner, G. Helicotylenchus, a new genus of plant-parasitic nematodes and its relationship to Rotylenchus Filipjev. Proc. Helminthol. Soc. Wash. 1945, 12, 34–38. [Google Scholar]
- Siddiqi, M.R. Tylenchida: Parasites of Plants and Insects, 2nd ed.; CAB International: Wallingford, UK, 2000. [Google Scholar]
- Inserra, R.; Vovlas, N.; Golden, A.M. Helicotylenchus oleae n. sp. and H. neopaxilli n. sp.(Hoplolaimidae), two new spiral nematodes parasitic on olive trees in Italy. J. Nematol. 1979, 11, 56–62. [Google Scholar] [PubMed]
- Karakaş, M. Life cycle and mating behavior of Helicotylenchus multicinctus (Nematoda: Hoplolaimidae) on excised Musa cavendishii roots. Biologia 2007, 62, 320–322. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Inserra, R.N.; Marais, M.; Mullin, P.; Powers, T.O.; Roberts, P.A.; Van Den Berg, E.; Yeates, G.W.; Baldwin, J.G. Diversity and phylogenetic relationships within the spiral nematodes of Helicotylenchus Steiner, 1945 (Tylenchida: Hoplolaimidae) as inferred from analysis of the D2-D3 expansion segments of 28S rRNA gene sequences. Nematology 2011, 13, 333–345. [Google Scholar]
- Subbotin, S.A.; Vovlas, N.; Yeates, G.W.; Hallmann, J.; Kiewnick, S.; Chizhov, V.N.; Manzanilla-López, R.H.; Inserra, R.N.; Castillo, P. Morphological and molecular characterisation of Helicotylenchus pseudorobustus (Steiner, 1914) Golden, 1956 and related species (Tylenchida: Hoplolaimidae) with a phylogeny of the genus. Nematology 2015, 17, 27–52. [Google Scholar] [CrossRef]
- Cobb, N.A. Nematodes, Mostly Australian and Fijian; F. Cunninghame & Company, printers: Sydney, Australia, 1893. [Google Scholar]
- Golden, A.M. Taxonomy of the spiral nematodes (Rotylenchus and Helicotylenchus), and the developmental stages and host-parasite relationships of R. buxophilus, n. sp., attacking boxwood. Bull. Md. Agric. Exp. Stn. 1956, A-85, 28. [Google Scholar]
- Steiner, G. Freilebende nematoden aus der Schweiz. 2. Teil. einer vorlafifigen Mitteilung. Arch. Hydrobiol. Planktork. 1914, 9, 259–276. [Google Scholar]
- Yuen, P.H. Four new species of Helicotylenchus Steiner (Hoplolaiminae: Tylenchida) and a redescription of H. canadensis Waseem, 1961. Nematologica 1964, 10, 373–387. [Google Scholar] [CrossRef]
- McSorley, R.; Parrado, J. Nematological reviews: Helicotylenchus multicinctus on bananas: An international problem. Nematropica 1986, 16, 73–91. [Google Scholar]
- O’Bannon, J.; Inserra, R. Helicotylenchus Species as Crop Damaging Parasitic Nematodes. Nematology Circular 165; Florida Department of Agriculture and Consumer Services: Gainesville, FL, USA, 1989; pp. 1–3.
- Mwamula, A.O.; Na, H.; Kim, Y.H.; Kim, Y.H.; Han, G.; Lee, D.W. Characterization of a new spiral nematode, Helicotylenchus asiaticus n. sp. and three known species from Korea; with comments on the validity of Helicotylenchus microlobus Perry in Perry, Darling & Thorne, 1959. Eur. J. Plant Pathol. 2020, 157, 565–581. [Google Scholar]
- Anderson, R. Canadian species of the genus Helicotylenchus Steiner, 1945 (Nematoda: Hoplolaimidae), their identifying characteristics and descriptions of three new species. Can. J. Zool. 1974, 52, 1365–1381. [Google Scholar] [CrossRef]
- Anderson, R. Helicotylenchus urobelus n. sp.(Nematoda: Hoplolaimidae) from New Brunswick, Canada. Can. J. Zool. 1978, 56, 1232–1234. [Google Scholar] [CrossRef]
- Anderson, R. A supplemental key to species of Helicotylenchus Steiner, 1945 (Nematoda: Hoplolaimidae) described since 1972 and a description of H. oscephalus n. sp. Can. J. Zool. 1979, 57, 337–342. [Google Scholar] [CrossRef]
- Anderson, R.; Eveleigh, E. Description of Helicotylenchus amplius n. sp. and a key to the Canadian species of the genus (Nematoda: Hoplolaimidae). Can. J. Zool. 1982, 60, 318–321. [Google Scholar] [CrossRef]
- Anderson, R. Morphology and description of Helicotylenchus crassatus n. sp. (Nematoda: Hoplolaimidae) from eastern Canada. Can. J. Zool. 1973, 51, 1195–1200. [Google Scholar] [CrossRef]
- Forge, T.A.; Larney, F.J.; Kawchuk, L.M.; Pearson, D.C.; Koch, C.; Blackshaw, R.E. Crop rotation effects on Pratylenchus neglectus populations in the root zone of irrigated potatoes in southern Alberta. Can. J. Plant Pathol. 2015, 37, 363–368. [Google Scholar] [CrossRef]
- Jenkins, W. A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Dis. Rep. 1964, 48, 692. [Google Scholar]
- Seinhorst, J. A rapid method for the transfer of nematodes from fixative to anhydrous glycerin. Nematologica 1959, 4, 67–69. [Google Scholar] [CrossRef]
- Grisse, D. Redescription ou modification de quelques techniques utilisées dans l’étude des nématodes phytoparasitaires. Meded. Van De Rijks Fac. Landbouwwet. Gent 1969, 34, 351. [Google Scholar]
- Maria, M.; Powers, T.; Tian, Z.; Zheng, J. Distribution and description of criconematids from Hangzhou, Zhejiang Province. China J. Nematol. 2018, 50, 183–206. [Google Scholar]
- Holterman, M.; van der Wurff, A.; van den Elsen, S.; van Megen, H.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 2006, 23, 1792–1800. [Google Scholar] [CrossRef] [PubMed]
- De Ley, P.; Felix, M.-A.; Frisse, L.; Nadler, S.; Sternberg, P.; Thomas, W.K. Molecular and morphological characterisation of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae). Nematology 1999, 1, 591–612. [Google Scholar] [CrossRef]
- Vrain, T.; Lalik, B. Distribution and pathogenicity of the alfalfa stem nematode, Ditylenchus dipsaci, in British Columbia. Plant Dis. 1983, 67, 300–302. [Google Scholar] [CrossRef]
- Bowles, J.; Blair, D.; McManus, D.P. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 1992, 54, 165–173. [Google Scholar] [CrossRef]
- Derycke, S.; Remerie, T.; Vierstraete, A.; Backeljau, T.; Vanfleteren, J.; Vincx, M.; Moens, T. Mitochondrial DNA variation and cryptic speciation within the free-living marine nematode Pellioditis marina. Mar. Ecol. Prog. Ser. 2005, 300, 91–103. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tan, G.; Muffato, M.; Ledergerber, C.; Herrero, J.; Goldman, N.; Gil, M.; Dessimoz, C. Current methods for automated filtering of multiple sequence alignments frequently worsen single-gene phylogenetic inference. Syst. Biol. 2015, 64, 778–791. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Meth. 2012, 9, 772. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 21 December 2023).
- Uludamar, E.B.K.; Yildiz, Ş.; Imren, M.; Atilla, Ö.; ElekçİoĞlu, İ.H. Occurrence of plant parasitic nematode species in important crops in the Southeast Anatolia Region of Turkey. Turk. J. Entomol. 2018, 42, 63–74. [Google Scholar] [CrossRef]
- Mateille, T.; Tavoillot, J.; Martiny, B.; Dmowska, E.; Winiszewska, G.; Ferji, Z.; Msanda, F.; El Mousadik, A. Aridity or low temperatures: What affects the diversity of plant-parasitic nematode communities in the Moroccan argan relic forest. Appl. Soil Ecol. 2016, 101, 64–71. [Google Scholar] [CrossRef]
- Bahmani, J.; Khozeini, F.; Barooti, S.; Rezaee, S.; Ghaderi, R. Plant-parasitic nematodes associated with walnut in the Sanandej region of west Iran. J. Plant Prot. Res. 2013, 53, 404–408. [Google Scholar] [CrossRef]
- Ishaque, U.; Kazi, N. Some new records of Helicotylenchus (steiner, 1945) species from pakistan (nematoda: Hoplolaimidae). Plant Prot. 2022, 6, 113–120. [Google Scholar] [CrossRef]
- Castillo, P.; Trapero-Casas, J.; Jiménez-Díaz, R. Effect of time, temperature, and inoculum density on reproduction of Pratylenchus thornei in carrot disk cultures. J. Nematol. 1995, 27, 120. [Google Scholar] [PubMed]
- Luc, M. Nematodes and wilting in cotton in Southwestern Madagascar. Coton Et Fibres Trop. 1958, 13, 239–256. [Google Scholar]
- Cobb, N. New nematode genera found inhabiting fresh water and non-brackish soils. J. Wash. Acad. Sci. 1913, 3, 432–444. [Google Scholar] [CrossRef]
- Thorne, G. Notes on free-living and plant-parasitic nematodes, II. Proc. Helminthol. Soc. Wash. 1935, 2, 96–98. [Google Scholar]
- Whitehead, A. Rotylenchoides brevis ng, n. sp. (Rotylenchoidinae n. subfam.: Tylenchida). Nematologica 1958, 3, 327–331. [Google Scholar] [CrossRef]
- Fortuner, R. Morphometrical variability in Helicotylenchus Steiner, 1945. 6: Value of the characters used for specific identification. Rev. De Nématologie 1984, 7, 245–264. [Google Scholar]
- Sher, S. Revision of the hoplolaiminae (nematoda) VI. Helicotylenchus steiner, 1945 1. Nematologica 1966, 12, 1–56. [Google Scholar] [CrossRef]
- Perry, V.G.; Darling, H.M.; Thorne, G. Anatomy, taxonomy and control of certain spiral nematodes attacking blue grass in Wisconsin. Res. Bull. 1959, 207, 1–24. [Google Scholar]
- Wollenweber, H. Krankheiten und Beschädigungen der Kartoffel. Arbeiten des Forschungsinstitutes für Kartoffelbau, Heft 7; Paul Parey: Berlin, Germany, 1923. [Google Scholar]
- Skarbilovich, T. On the structure of systematics of nematodes order Tylenchida Thorne, 1949. Acta Parasitol. Pol. 1959, 7, 117–132. [Google Scholar]
- Ohshima, Y. Heterodera elachista n. sp., an upland rice cyst nematode from Japan. Jpn J. Nematol. 1974, 4, 51–56. [Google Scholar]
- de Man, J.G. Die, Frei in der Reinen Erde und im Süssen Wasser Lebenden Nematoden der Niederländischen Fauna: Eine Systematisch-faunistische Monographie; EJ Brill: Leiden, The Netherlands, 1884; Volume 1. [Google Scholar]
- Andrássy, I. Revision der Gattung Tylenchus Bastian, 1865 (Tylenchidae, Nematoda). Acta Zool. Hung 1954, 1, 5–42. [Google Scholar]
- de Man, J.G. Nouvelles recherches sur les nématodes libres terricoles de la Hollande. Capita Zool. 1921, I, 3–62. [Google Scholar]
- Siddiqi, M.R. The unusual position of the phasmids in Coslenchus costatus (de Man, 1921) gen. n., comb. n. and other Tylenchidae (Nematoda: Tylenchida). Nematologica 1978, 24, 449–455. [Google Scholar] [CrossRef]
- Karssen, G.; Van Aelst, A. Description of Cryphodera brinkmani n. sp.(Nematoda: Heteroderidae), a parasite of Pinus thunbergii Parlatore from Japan, including a key to the species of the genus Cryphodera Colbran, 1966. Nematology 1999, 1, 121–130. [Google Scholar] [CrossRef]
- Xia, Y.-H.; Li, J.; Xu, F.-F.; Lei, B.; Li, H.-L.; Wang, K.; Li, Y. Identification and a culture method for a Helicotylenchus microlobus from tomato in China. BMC Zool. 2022, 7, 1–11. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.M. Optimizing sampling and extraction methods for plant-parasitic and entomopathogenic nematodes. Plants 2021, 10, 629. [Google Scholar] [CrossRef]
- Munawar, M.; Yevtushenko, D.P.; Castillo, P. Overview of the genus Boleodorus and first reports of Boleodorus thylactus and B. volutus from southern Alberta, Canada. Animals 2021, 11, 1760. [Google Scholar] [CrossRef]
- Munawar, M.; Castillo, P.; Yevtushenko, D.P. Description of Filenchus species from agroecosystem of southern Alberta, Canada. Agronomy 2022, 12, 690. [Google Scholar] [CrossRef]





| This Study | Anderson [18] | |||||
|---|---|---|---|---|---|---|
| Location | Southern Alberta, Canada | Ontario, Canada | ||||
| Characteristics | Morphotype 1 | Morphotype 2 | ||||
| Population 93 | Population 50 | Population 46 | Population 60 | Population 61 | Original Population | |
| n | 17 | 17 | 10 | 10 | 10 | 20 |
| Body length (L) | 647.0 ± 42.2 (584.0–747.0) | 768.0 ± 66.0 (659.0–866.0) | 798 ± 51.2 (733.0–900.0) | 799.0 ± 45.8 (720.0–851.0) | 792.0 ± 75.7 (672.0–891.0) | 720 (646–779) |
| a | 26.0 ± 2.1 (21.0–30.0) | 31.0 ± 2.6 (28.0–37.0) | 29.0 ± 2.3 (25.0–32.0) | 29.0 ± 2.7 (26.0–34.0) | 27.5 ± 2.6 (22.0–31.0) | 31 (27–34) |
| b | 5.5 ± 0.5 (5.0–7.0) | 5.9 ± 0.5 (4.7–6.6) | 6.0 ± 0.4 (5.5–7.0) | 6.3 ± 0.3 (6.0–7.0) | 6.3 ± 0.6 (5.0–7.0) | 5.7 (5.2–6.2) |
| b’ | 4.7 ± 0.5 (4.0–6.0) | 5.0 ± 0.4 (4.0–5.5) | 5.4 ± 0.3 (5.0–6.0) | 5.0 ± 0.3 (5.0–6.0) | 5.0 ± 0.5 (4.2–6.0) | 4.7 (4.3–5.0) |
| c | 53.0 ± 7.0 (45.0–67.0) | 61.0 ± 4.5 (53.0–70.0) | 64.0 ± 4.5 (57.0–70.0) | 61.5 ± 6.6 (53.0–72.0) | 62.0 ± 9.8 (48.0–81.0) | 59 (45–71) |
| c’ | 1.1 ± 0.1 (0.8–1.4) | 0.9 ± 0.1 (0.7–1.1) | 0.8 ± 0.1 (0.7–1.0) | 0.8 ± 0.1 (0.8–1.0) | 0.8 ± 0.1 (0.6–1.0) | 0.79 (0.63–1.00) |
| O | 30.0 ± 3.6 (23.0–36.0) | 32.0 ± 2.9 (27.5–38.0) | 34.5 ± 3.5 (31.0–42.0) | 30.7 ± 2.0 (28.0–35.0) | 32.0 ± 3.4 (27.0–37.5) | 28 (20–36) |
| V | 62.0 ± 2.3 (57.0–67.0) | 61.0 ± 1.6 (57.0–63.0) | 63.0 ± 2.3 (59.0–66.0) | 62.0 ± 1.6 (58.5–64.0) | 62.0 ± 1.1 (60.0–63.0) | – |
| Lip region height | 3.5 ± 0.4 (3.0–4.0) | 3.5 ± 0.2 (3.0–3.8) | 3.4 ± 0.3 (3.0–3.8) | 3.7 ± 0.4 (3.0–4.2) | 3.7 ± 0.4 (3.2–4.3) | – |
| Lip region width | 7.0 ± 0.5 (6.0–8.0) | 6.8 ± 0.4 (6.0–7.5) | 6.8 ± 0.5 (6.0–7.4) | 6.9 ± 0.3 (6.3–7.4) | 6.9 ± 0.5 (6.0–7.3) | – |
| Stylet length | 26.0 ± 1.2 (24.0–28.0) | 27.0 ± 1.0 (25.0–28.0) | 26.0 ± 1.4 (24.0–28.0) | 27.0 ± 1.0 (25.0–28.0) | 27.0 ± 1.3 (24.5–28.0) | 28 (26–30) |
| Stylet knob height | 3.0 ± 0.4 (2.0–3.4) | 2.5 ± 0.2 (2.0–3.0) | 3.0 ± 0.3 (2.4–3.3) | 3.4 ± 0.4 (3.0–3.9) | 2.9 ± 0.3 (2.5–3.3) | – |
| Stylet knob width | 5.0 ± 0.3 (4.3–5.5) | 5.0 ± 0.4 (4.5–6.0) | 5.5 ± 0.2 (5.2–5.5) | 5.4 ± 0.3 (5.0–6.0) | 5.3 ± 0.3 (5.0–6.0) | – |
| DGO distance from stylet knob base | 8.0 ± 0.9 (6.5–9.0) | 8.8 ± 0.7 (7.5–10.0) | 8.9 ± 1.0 (8.0–10.5) | 8.0 ± 0.6 (7.5–9.5) | 8.8 ± 0.7 (7.5–10.0) | – |
| Median bulb length | 12.3 ± 1.0 (10.0–14.0) | 12.5 ± 1.0 (10.5–14.0) | 13.5 ± 0.9 (12.5–15.5) | 13.5 ± 1.1 (12.5–15.0) | 14.0 ± 0.8 (12.0–15.0) | – |
| Median bulb width | 10.0 ± 0.8 (8.0–11.5) | 10.0 ± 0.9 (8.0–11.0) | 11.0 ± 1.0 (9.0–12.5) | 11.0 ± 1.2 (9.0–13.0) | 11.0 ± 0.8 (10.0–12.5) | – |
| Anterior end to excretory pore | 101.0 ± 3.3 (91.0–106.0) | 118.5 ± 5.6 (112.0–129.0) | 118.0 ± 6.3 (110.0–128.0) | 119.0 ± 5.7 (111.0–129.0) | 120.0 ± 5.0 (111.0–131.0) | (106–125) |
| Pharynx length (till pharyngo-intestinal junction) | 119.0 ± 7.7 (95.0–128.0) | 130.0 ± 6.0 (121.0–147.0) | 126.5 ± 7.4 (118.5–140.0) | 126.0 ± 5.8 (117.0–136.0) | 127.0 ± 5.7 (115.5–133.0) | (114–134) |
| Pharynx length (till pharyngeal lobe) | 139.0 ± 9.0 (111.0–149.0) | 157.0 ± 10.4 (140.0–175.5) | 148.0 ± 7.2 (139.0–163.0) | 154.0 ± 6.7 (145.5–169.0) | 158.0 ± 8.9 (143.0–176.0) | (141–163) |
| Maximum body diam. | 25.0 ± 2.1 (20.0–29.0) | 24.5 ± 2.5 (21.0–30.7) | 27.5 ± 2.2 (24.5–32.0) | 27.0 ± 2.8 (22.0–31.0) | 29.0 ± 2.0 (25.0–31.0) | – |
| Vulva body diam. | 23.0 ± 2.1 (19.0–28.0) | 24.0 ± 2.6 (20.0–29.0) | 25.5 ± 2.3 (22.0–29.0) | 26.5 ± 1.7 (24.5–29.0) | 26.0 ± 1.7 (23.0–28.0) | – |
| Distance from vulva to tail terminus | 248.0 ± 28.2 (192.0–320.0) | 470.0 ± 39.5 (412.0–543.0) | 298.5 ± 32.0 (258.0–358.0) | 306.0 ± 24.3 (272.0–338.0) | 301.5 ± 28.8 (248.0–336.0) | – |
| Anal body diam. | 13.6 ± 1.0 (11.5–15.3) | 15.0 ± 1.6 (12.0–18.0) | 16.0 ± 1.8 (12.0–18.0) | 15.5 ± 1.5 (13.0–17.5) | 16.0 ± 1.9 (14.0–20.0) | – |
| Tail length | 12.3 ± 1.6 (10.0–15.0) | 12.6 ± 1.1 (11.0–15.0) | 12.5 ± 0.9 (11.0–14.0) | 13.0 ± 1.7 (10.0–15.0) | 12.5 ± 1.4 (11.0–15.0) | 12 (10–17) |
| Hyaline tail region | 3.9 ± 0.5 (3.0–4.6) | 2.7 ± 0.5 (2.3–4.0) | 3.6 ± 0.7 (2.6–4.5) | 3.8 ± 0.6 (3.0–4.9) | 4.0 ± 0.7 (3.0–4.6) | – |
| Characteristics | This Study | Anderson [16] |
|---|---|---|
| Population | Southern Alberta, Canada | British Columbia, Canada |
| n | 17 | 12 |
| Body length (L) | 876.6 ± 45.9 (802.0–963.0) | 857 (782–927) |
| a | 29.5 ± 1.5 (27.3–32.2) | 32 (28–35) |
| b | 6.5 ± 0.3 (6.0–7.0) | 6.2 (5.7–6.6) |
| b’ | 5.2 ± 0.2 (4.9–5.7) | 5.1 (4.6–5.5) |
| c | 37.4 ± 2.8 (31.2–41.5) | 33 (27–39) |
| c’ | 1.3 ± 0.1 (1.2–1.5) | 1.3 (1.1–1.7) |
| O | 27.8 ± 1.2 (26.5–30.0) | 28 (21–33) |
| V | 62.1 ± 1.4 (58.8–64.5) | 61 (58–63) |
| Lip region height | 35 ± 0.3 (3.0–4.0) | – |
| Lip region width | 7.4 ± 0.4 (7.0–8.3) | – |
| Stylet length | 29.5 ± 0.9 (28.0–31.0) | 27 (25–28) |
| Stylet knob height | 2.7 ± 0.5 (2.1–4.0) | – |
| Stylet knob width | 5.1 ± 0.3 (4.6–5.5) | – |
| DGO distance from stylet knob base | 8.3 ± 0.3 (8.0–9.0) | (6–9) |
| Median bulb length | 14.5 ± 0.7 (12.5–15.4) | – |
| Median bulb width | 11.3 ± 0.8 (10.0–12.4) | – |
| Anterior end to excretory pore | 137.5 ± 4.1 (130.0–144.0) | 135 (127–143) |
| Pharynx length (till pharyngo-intestinal junction) | 134.8 ± 4.4 (123.0–139.0) | 139 (128–153) |
| Pharynx length (till pharyngeal lobe) | 167.9 ± 8.6 (156.0–180.0) | 169 (156–185) |
| Maximum body diam. | 29.8 ± 2.1 (25.0–33.0) | (25–28) |
| Vulva body diam. | 27.4 ± 2.4 (24.0–31.0) | – |
| Distance from vulva to tail terminus | 331.0 ± 25.1 (284.0–371.0) | – |
| Anal body diam. | 17.8 ± 1.6 (15.0–20.0) | – |
| Tail length | 23.6 ± 2.3 (20.0–28.0) | 26 (21–29) |
| Phasmid position from tail terminus | 27.6 ± 1.3 (27.0–30.0) | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munawar, M.; Konschuh, M.; Castillo, P.; Yevtushenko, D.P. Morpho-Anatomy, In Vitro Culture, and Phylogenetic Studies of Two Helicotylenchus Species from Southern Alberta, Canada. Microorganisms 2024, 12, 497. https://doi.org/10.3390/microorganisms12030497
Munawar M, Konschuh M, Castillo P, Yevtushenko DP. Morpho-Anatomy, In Vitro Culture, and Phylogenetic Studies of Two Helicotylenchus Species from Southern Alberta, Canada. Microorganisms. 2024; 12(3):497. https://doi.org/10.3390/microorganisms12030497
Chicago/Turabian StyleMunawar, Maria, Michele Konschuh, Pablo Castillo, and Dmytro P. Yevtushenko. 2024. "Morpho-Anatomy, In Vitro Culture, and Phylogenetic Studies of Two Helicotylenchus Species from Southern Alberta, Canada" Microorganisms 12, no. 3: 497. https://doi.org/10.3390/microorganisms12030497
APA StyleMunawar, M., Konschuh, M., Castillo, P., & Yevtushenko, D. P. (2024). Morpho-Anatomy, In Vitro Culture, and Phylogenetic Studies of Two Helicotylenchus Species from Southern Alberta, Canada. Microorganisms, 12(3), 497. https://doi.org/10.3390/microorganisms12030497







