Presence of Zonula Occludens Toxin-Coding Genes among Vibrio parahaemolyticus Isolates of Clinical and Environmental Origin
Abstract
1. Introduction
2. Materials and Methods
2.1. Recollection of Seafood from Market Points and Harvest Areas in Chile
2.2. Environmental Strains Isolation
2.3. V. parahaemolyticus Identification
2.4. Phylogeny Analyses of Zot Sequence in Database
2.5. Primers Design, Amplification and Sequencing
2.6. Presence of Zot in Isolates from the Adriatic Sea (Italy)
2.7. Genome Analysis of Clinical Isolates
3. Results
3.1. Availability of Zot Sequences Described in V. parahaemolyticus
3.2. Identification of Conserved Regions in V. parahaemolyticus Zot-Sequences and Primers Design
3.3. Primers Specificity and Amplicon Analysis
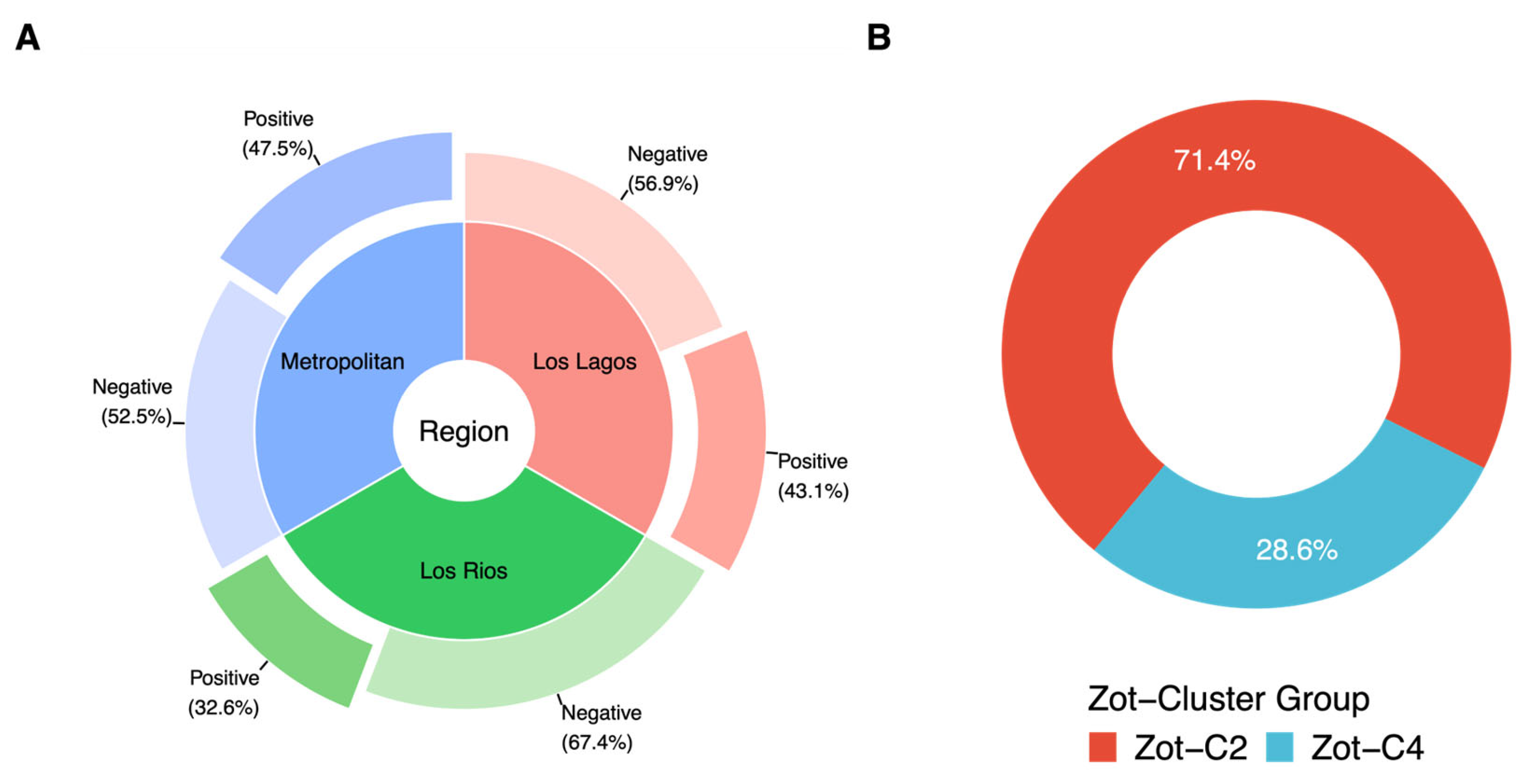
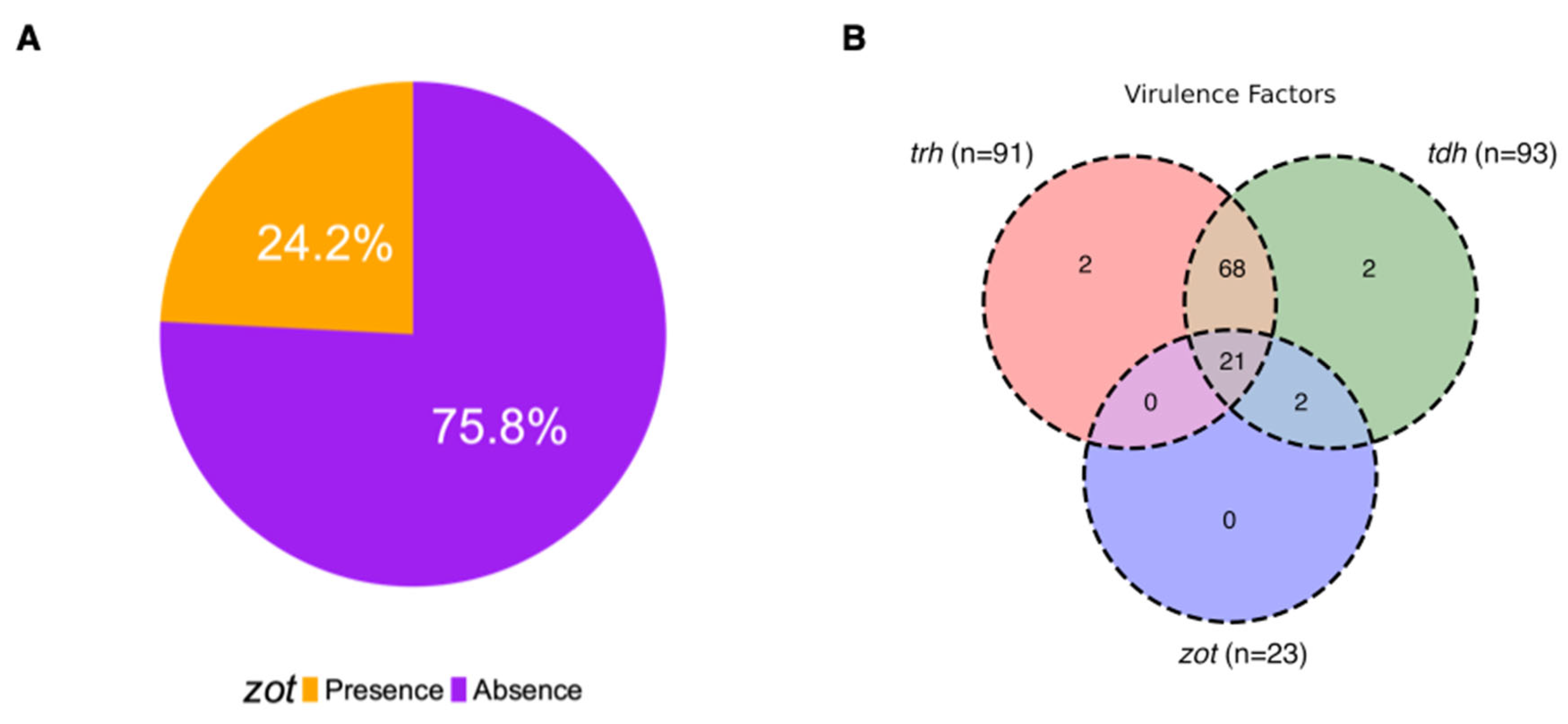
3.4. Presence of Zot Gene in Environmental V. parahaemolyticus Chilean Isolates
3.5. Presence of Zot Gene in Environmental V. parahaemolyticus Italian Isolates
3.6. Presence of Zot Gene in Clinical V. parahaemolyticus Genomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Letchumanan, V.; Chan, K.-G.; Lee, L.-H. Vibrio parahaemolyticus: A Review on the Pathogenesis, Prevalence, and Advance Molecular Identification Techniques. Front. Microbiol. 2014, 5, 705. [Google Scholar] [CrossRef]
- Raghunath, P. Roles of Thermostable Direct Hemolysin (TDH) and TDH-Related Hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 2014, 5, 805. [Google Scholar] [CrossRef]
- Ding, G.; Zhao, L.I.; Xu, J.; Cheng, J.; Cai, Y.; Du, H.; Xiao, G.; Zhao, F. Quantitative Risk Assessment of Vibrio parahaemolyticus in Shellfish from Retail to Consumption in Coastal Cities of Eastern China. J. Food Prot. 2022, 85, 1320–1328. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, H.; Osei-Adjei, G.; Zhang, Y.; Yang, W.; Yang, H.; Yin, Z.; Huang, X.; Zhou, D. Transcriptional Regulation of the Type VI Secretion System 1 Genes by Quorum Sensing and ToxR in Vibrio parahaemolyticus. Front. Microbiol. 2017, 8, 2005. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, L.; Osei-Adjei, G.; Zhang, Y.; Yang, W.; Yin, Z.; Lu, R.; Sheng, X.; Yang, R.; Huang, X.; et al. Autoregulation of ToxR and Its Regulatory Actions on Major Virulence Gene Loci in Vibrio parahaemolyticus. Front. Cell. Infect. Microbiol. 2018, 8, 291. [Google Scholar] [CrossRef]
- Ritchie, J.M.; Rui, H.; Zhou, X.; Iida, T.; Kodoma, T.; Ito, S.; Davis, B.M.; Bronson, R.T.; Waldor, M.K. Inflammation and Disintegration of Intestinal Villi in an Experimental Model for Vibrio parahaemolyticus-Induced Diarrhea. PLoS Pathog. 2012, 8, e1002593. [Google Scholar] [CrossRef]
- Hubbard, T.P.; Chao, M.C.; Abel, S.; Blondel, C.J.; Abel Zur Wiesch, P.; Zhou, X.; Davis, B.M.; Waldor, M.K. Genetic Analysis of Vibrio parahaemolyticus Intestinal Colonization. Proc. Natl. Acad. Sci. USA 2016, 113, 6283–6288. [Google Scholar] [CrossRef]
- Castillo, D.; Pérez-Reytor, D.; Plaza, N.; Ramírez-Araya, S.; Blondel, C.J.; Corsini, G.; Bastías, R.; Loyola, D.E.; Jaña, V.; Pavez, L.; et al. Exploring the Genomic Traits of Non-Toxigenic Vibrio parahaemolyticus Strains Isolated in Southern Chile. Front. Microbiol. 2018, 9, 161. [Google Scholar] [CrossRef]
- Mahoney, J.C.; Gerding, M.J.; Jones, S.H.; Whistler, C.A. Comparison of the Pathogenic Potentials of Environmental and Clinical Vibrio parahaemolyticus Strains Indicates a Role for Temperature Regulation in Virulence. Appl. Environ. Microbiol. 2010, 76, 7459–7465. [Google Scholar] [CrossRef]
- Wagley, S.; Borne, R.; Harrison, J.; Baker-Austin, C.; Ottaviani, D.; Leoni, F.; Vuddhakul, V.; Titball, R.W. Galleria Mellonella as an Infection Model to Investigate Virulence of Vibrio parahaemolyticus. Virulence 2017, 9, 197–207. [Google Scholar] [CrossRef]
- Lynch, T.; Livingstone, S.; Buenaventura, E.; Lutter, E.; Fedwick, J.; Buret, A.G.; Graham, D.; DeVinney, R. Vibrio parahaemolyticus Disruption of Epithelial Cell Tight Junctions Occurs Independently of Toxin Production. Infect. Immun. 2005, 73, 1275–1283. [Google Scholar] [CrossRef]
- Meparambu Prabhakaran, D.; Ramamurthy, T.; Thomas, S. Genetic and Virulence Characterisation of Vibrio parahaemolyticus Isolated from Indian Coast. BMC Microbiol. 2020, 20, 62. [Google Scholar] [CrossRef]
- Pérez-Reytor, D.; Pavón, A.; Lopez-Joven, C.; Ramírez-Araya, S.; Peña-Varas, C.; Plaza, N.; Alegría-Arcos, M.; Corsini, G.; Jaña, V.; Pavez, L.; et al. Analysis of the Zonula Occludens Toxin Found in the Genome of the Chilean Non-Toxigenic Vibrio parahaemolyticus Strain PMC53.7. Front. Cell. Infect. Microbiol. 2020, 10, 482. [Google Scholar] [CrossRef]
- Fasano, A. Toxins and the Gut: Role in Human Disease. Gut 2002, 50, iii9–iii14. [Google Scholar] [CrossRef]
- Mauritzen, J.J.; Castillo, D.; Tan, D.; Svenningsen, S.L.; Middelboe, M. Beyond Cholera: Characterization of Zot-Encoding Filamentous Phages in the Marine Fish Pathogen Vibrio anguillarum. Viruses 2020, 12, 730. [Google Scholar] [CrossRef]
- Levine, M.M.; Kaper, J.B.; Herrington, D.; Losonsky, G.; Morris, J.G.; Clements, M.L.; Black, R.E.; Tall, B.; Hall, R. Volunteer Studies of Deletion Mutants of Vibrio cholerae O1 Prepared by Recombinant Techniques. Infect. Immun. 1988, 56, 161–167. [Google Scholar] [CrossRef]
- Fasano, A.; Fiorentini, C.; Donelli, G.; Uzzau, S.; Kaper, J.B.; Margaretten, K.; Ding, X.; Guandalini, S.; Comstock, L.; Goldblum, S.E. Zonula Occludens Toxin Modulates Tight Junctions through Protein Kinase C-Dependent Actin Reorganization, in Vitro. J. Clin. Investig. 1995, 96, 710–720. [Google Scholar] [CrossRef]
- Fu, S.; Wang, Q.; Wang, R.; Zhang, Y.; Lan, R.; He, F.; Yang, Q. Horizontal Transfer of Antibiotic Resistance Genes within the Bacterial Communities in Aquacultural Environment. Sci. Total Environ. 2022, 820, 153286. [Google Scholar] [CrossRef]
- Danso, E.K.; Asare, P.; Otchere, I.D.; Akyeh, L.M.; Asante-Poku, A.; Aboagye, S.Y.; Osei-Wusu, S.; Opare, D.; Ntoumi, F.; Zumla, A.; et al. A Molecular and Epidemiological Study of Vibrio cholerae Isolates from Cholera Outbreaks in Southern Ghana. PLoS ONE 2020, 15, e0236016. [Google Scholar] [CrossRef]
- Bacian, C.; Verdugo, C.; García, K.; Perez-Larruscain, J.; de Blas, I.; Cachicas, V.; Lopez-Joven, C. Longitudinal Study of Total and Pathogenic Vibrio parahaemolyticus (Tdh+ and/or Trh+) in Two Natural Extraction Areas of Mytilus chilensis in Southern Chile. Front. Microbiol. 2021, 12, 621737. [Google Scholar] [CrossRef]
- Bej, A.K.; Patterson, D.P.; Brasher, C.W.; Vickery, M.C.; Jones, D.D.; Kaysner, C.A. Detection of Total and Hemolysin-Producing Vibrio parahaemolyticus in Shellfish Using Multiplex PCR Amplification of Tlh, Tdh and Trh. J. Microbiol. Methods 1999, 36, 215–225. [Google Scholar] [CrossRef]
- Kim, Y.B.; Okuda, J.; Matsumoto, C.; Takahashi, N.; Hashimoto, S.; Nishibuchi, M. Identification of Vibrio parahaemolyticus Strains at the Species Level by PCR Targeted to the toxR Gene. J. Clin. Microbiol. 1999, 37, 1173–1177. [Google Scholar] [CrossRef]
- Taniguchi, H.; Hirano, H.; Kubomura, S.; Higashi, K.; Mizuguchi, Y. Comparison of the Nucleotide Sequences of the Genes for the Thermostable Direct Hemolysin and the Thermolabile Hemolysin from Vibrio parahaemolyticus. Microb. Pathog. 1986, 1, 425–432. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Owczarzy, R.; Moreira, B.G.; You, Y.; Behlke, M.A.; Walder, J.A. Predicting Stability of DNA Duplexes in Solutions Containing Magnesium and Monovalent Cations. Biochemistry 2008, 47, 5336–5353. [Google Scholar] [CrossRef]
- ISO 21872-1:2017; Microbiology of the Food Chain Horizontal Method for the Determination of Vibrio spp.—Part 1: Detection of Potentially Enteropathogenic Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. International Organization for Standardization: London, UK, 2017.
- Chen, Y.; Stine, O.C.; Badger, J.H.; Gil, A.I.; Nair, G.B.; Nishibuchi, M.; Fouts, D.E. Comparative Genomic Analysis of Vibrio parahaemolyticus: Serotype Conversion and Virulence. BMC Genom. 2011, 12, 294. [Google Scholar] [CrossRef]
- Gonzalez-Escalona, N.; Strain, E.A.; De Jesús, A.J.; Jones, J.L.; Depaola, A. Genome Sequence of the Clinical O4:K12 Serotype Vibrio parahaemolyticus Strain 10329. J. Bacteriol. 2011, 193, 3405–3406. [Google Scholar] [CrossRef]
- Kalburge, S.S.; Polson, S.W.; Boyd Crotty, K.; Katz, L.; Turnsek, M.; Tarr, C.L.; Martinez-Urtaza, J.; Boyd, E.F. Complete Genome Sequence of Vibrio parahaemolyticus Environmental Strain UCM-V493. Genome Announc. 2014, 2, e00159-14. [Google Scholar] [CrossRef]
- Lüdeke, C.H.M.; Kong, N.; Weimer, B.C.; Fischer, M.; Jones, J.L. Complete Genome Sequences of a Clinical Isolate and an Environmental Isolate of Vibrio parahaemolyticus. Genome Announc. 2015, 3, e00216-15. [Google Scholar] [CrossRef]
- Kim, S.; Chung, H.Y.; Lee, D.-H.; Lim, J.G.; Kim, S.K.; Ku, H.-J.; Kim, Y.-T.; Kim, H.; Ryu, S.; Lee, J.-H.; et al. Complete Genome Sequence of Vibrio parahaemolyticus Strain FORC_008, a Foodborne Pathogen from a Flounder Fish in South Korea. Pathog. Dis. 2016, 74, ftw044. [Google Scholar] [CrossRef][Green Version]
- Nasu, H.; Iida, T.; Sugahara, T.; Yamaichi, Y.; Park, K.S.; Yokoyama, K.; Makino, K.; Shinagawa, H.; Honda, T. A Filamentous Phage Associated with Recent Pandemic Vibrio parahaemolyticus O3:K6 Strains. J. Clin. Microbiol. 2000, 38, 2156–2161. [Google Scholar] [CrossRef]
- Chung, H.Y.; Lee, B.; Na, E.J.; Lee, K.-H.; Ryu, S.; Yoon, H.; Lee, J.-H.; Kim, H.B.; Kim, H.; Jeong, H.G.; et al. Potential Survival and Pathogenesis of a Novel Strain, Vibrio parahaemolyticus FORC_022, Isolated from a Soy Sauce Marinated Crab by Genome and Transcriptome Analyses. Front. Microbiol. 2018, 9, 1504. [Google Scholar] [CrossRef]
- Ahn, S.; Chung, H.Y.; Lim, S.; Kim, K.; Kim, S.; Na, E.J.; Caetano-Anolles, K.; Lee, J.-H.; Ryu, S.; Choi, S.H.; et al. Complete Genome of Vibrio parahaemolyticus FORC014 Isolated from the Toothfish. Gut Pathog. 2016, 8, 59. [Google Scholar] [CrossRef]
- Xu, F.; Ilyas, S.; Hall, J.A.; Jones, S.H.; Cooper, V.S.; Whistler, C.A. Genetic Characterization of Clinical and Environmental Vibrio parahaemolyticus from the Northeast USA Reveals Emerging Resident and Non-Indigenous Pathogen Lineages. Front. Microbiol. 2015, 6, 272. [Google Scholar] [CrossRef]
- Castillo, D.; Kauffman, K.; Hussain, F.; Kalatzis, P.; Rørbo, N.; Polz, M.F.; Middelboe, M. Widespread distribution of prophage-encoded virulence factors in marine Vibrio communities. Sci. Rep. 2018, 8, 9973. [Google Scholar] [CrossRef]
- Feng, J.; Russel, M.; Model, P. A Permeabilized Cell System That Assembles Filamentous Bacteriophage. Proc. Natl. Acad. Sci. USA 1997, 94, 4068–4073. [Google Scholar] [CrossRef]
- Makino, K.; Oshima, K.; Kurokawa, K.; Yokoyama, K.; Uda, T.; Tagomori, K.; Iijima, Y.; Najima, M.; Nakano, M.; Yamashita, A.; et al. Genome Sequence of Vibrio parahaemolyticus: A Pathogenic Mechanism Distinct from That of V. cholerae. Lancet Lond. Engl. 2003, 361, 743–749. [Google Scholar] [CrossRef]
- Okada, N.; Iida, T.; Park, K.-S.; Goto, N.; Yasunaga, T.; Hiyoshi, H.; Matsuda, S.; Kodama, T.; Honda, T. Identification and Characterization of a Novel Type III Secretion System in Trh-Positive Vibrio parahaemolyticus Strain TH3996 Reveal Genetic Lineage and Diversity of Pathogenic Machinery beyond the Species Level. Infect. Immun. 2009, 77, 904–913. [Google Scholar] [CrossRef]
- Xu, F.; Gonzalez-Escalona, N.; Haendiges, J.; Myers, R.A.; Ferguson, J.; Stiles, T.; Hickey, E.; Moore, M.; Hickey, J.M.; Schillaci, C.; et al. Sequence Type 631 Vibrio parahaemolyticus, an Emerging Foodborne Pathogen in North America. J. Clin. Microbiol. 2017, 55, 645–648. [Google Scholar] [CrossRef]
- Turner, J.W.; Malayil, L.; Guadagnoli, D.; Cole, D.; Lipp, E.K. Detection of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae with Respect to Seasonal Fluctuations in Temperature and Plankton Abundance. Environ. Microbiol. 2014, 16, 1019–1028. [Google Scholar] [CrossRef]
- Jesser, K.J.; Valdivia-Granda, W.; Jones, J.L.; Noble, R.T. Clustering of Vibrio parahaemolyticus Isolates Using MLST and Whole-Genome Phylogenetics and Protein Motif Fingerprinting. Front. Public Health 2019, 7, 66. [Google Scholar] [CrossRef]
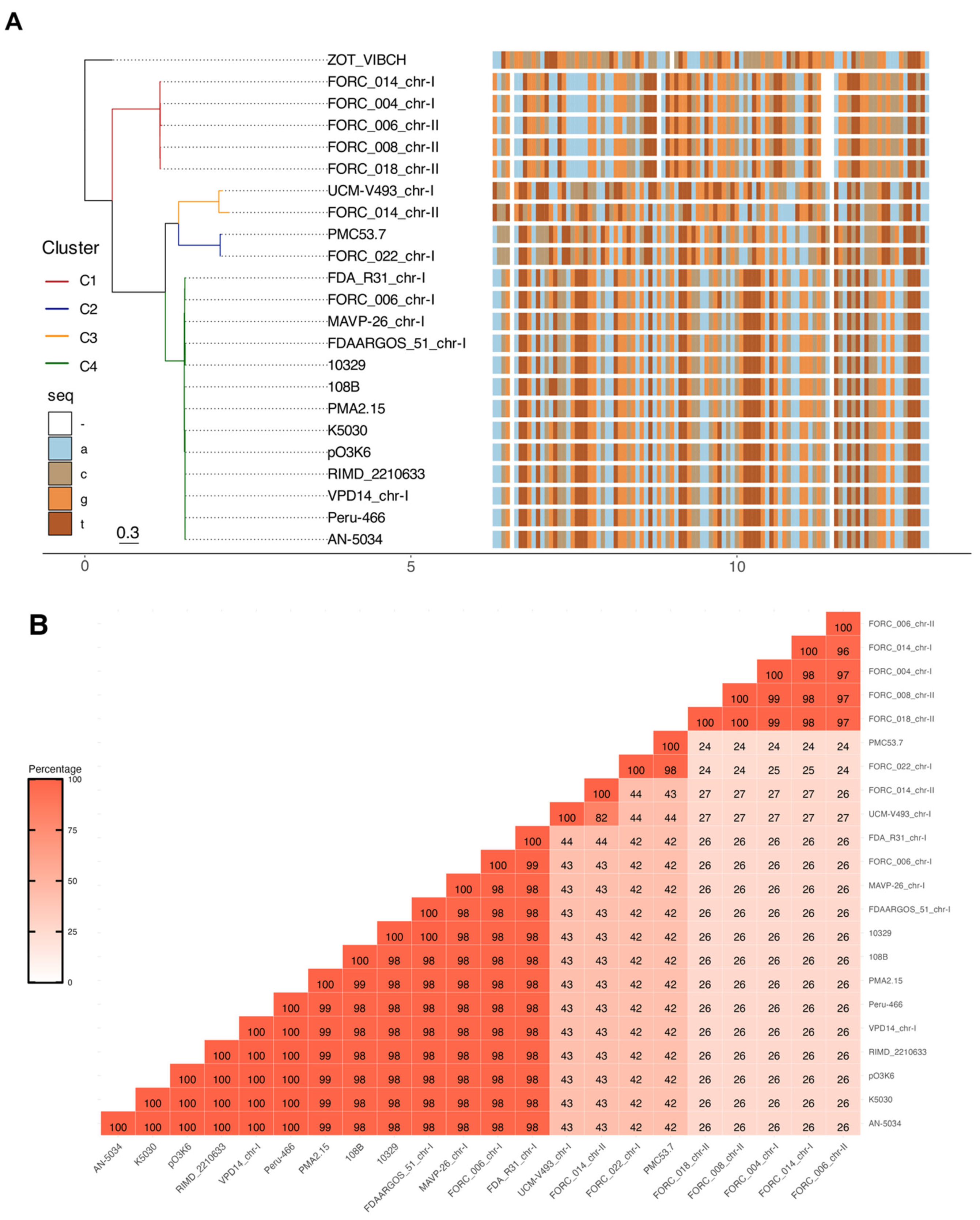

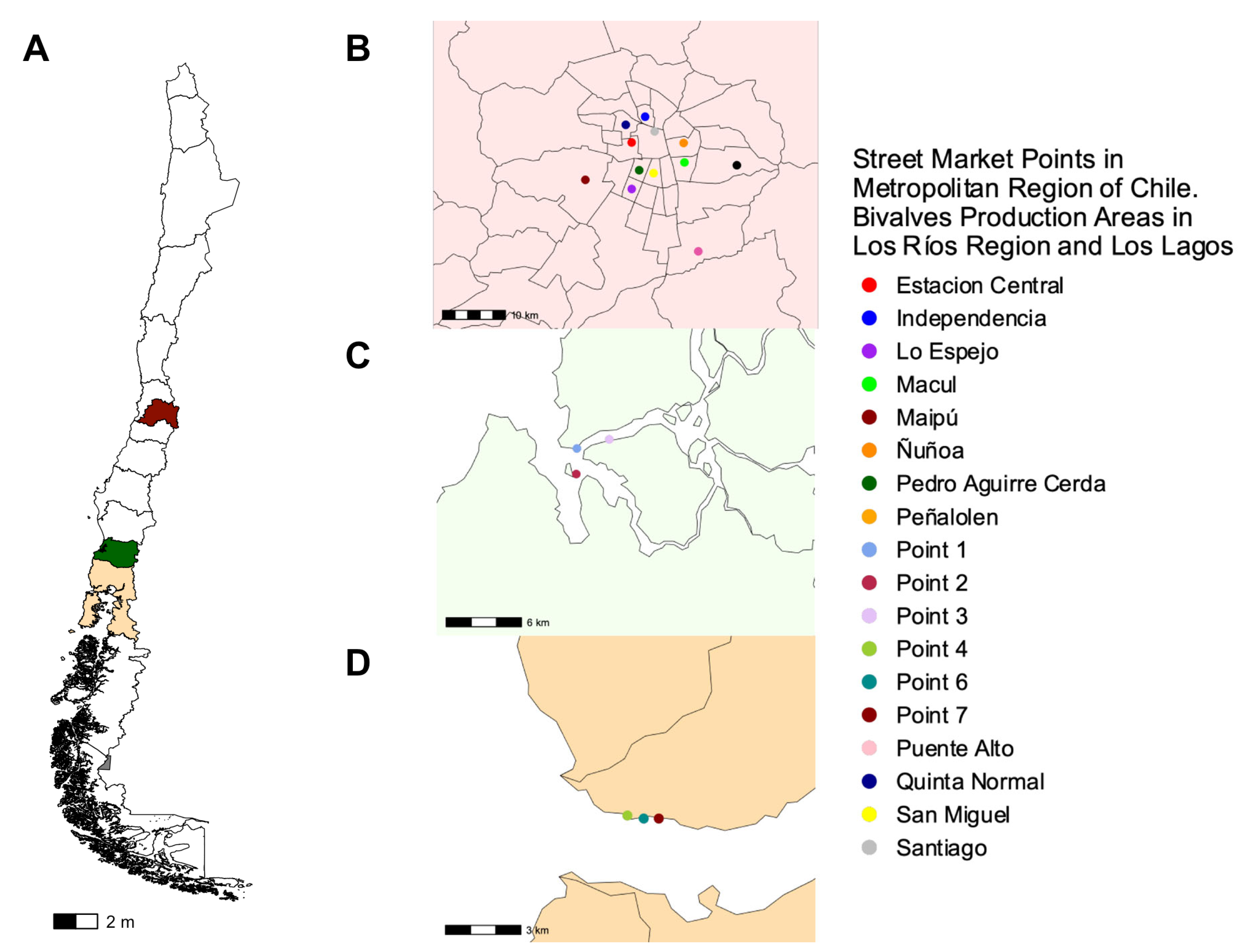

| GeneBank ID | Strain | Year 1 | Country | Isolation Source | Locus Tag | Description | Ref. |
|---|---|---|---|---|---|---|---|
| NZ_MKQF01000006.1 | PMC53.7 | 2007 | Chile | Clinical | BJL74_RS17965 | Zot | [8] |
| ACFO01000029.1 | AN5034 | 1998 | Bangladesh | Clinical | VIPARAN5034_0529 | Zot family | [34] |
| ACKB01000029.1 | K5030 | 2005 | India | Clinical | VIPARK5030_1517 | Zot family | [34] |
| ACFM01000008.1 | Peru-466 | 1996 | Peru | Clinical | VIPARP466_1523 | Zot family | [34] |
| AFBW01000009.1 | 10329 | 1998 | USA | Clinical | VP10329_13410 | Zot | [35] |
| CP007004.1 | UCM-V493 | 2002 | Spain | Environmental | VPUCM_1658 | Zot | [36] |
| CP009765.1 | FORC_006 | 2014 | KOR | Environmental | FORC6_1425 | Zot | Unpublished |
| CP006004.1 | 01:Kukstr.FDA_R31 | 2007 | USA | Environmental | M634_09525 | Toxin | [37] |
| NZ_CP009847.1 | FORC_004 | 2014 | KOR | Environmental | FORC4_1454 | Zot | Unpublished |
| CP009766.1 | FORC_006 | 2014 | KOR | Environmental | FORC6_3837 | Zot | Unpublished |
| NZ_CPOO9983.1 | FORC_008 | 2004 | KOR | Environmental | FORC8_3762 | Zot | [38] |
| MKQT01000002.1 | PMA2.15 | 2015 | Chile | Environmental | BJL84_RS00850 | Toxin | [8] |
| AP000581.2 | pO3K6 | 2016 | Japan | Clinical | orf7 | Zot like protein | [39] |
| BA000031.2 | RIMD2210633 | 1996 | Japan | Clinical | VP1558 | bacteriophage f237 ORF7 | [39] |
| CP013248.1 | FORC_022 | 2015 | KOR | Environmental | FORC22_1571 | Zot | [40] |
| CP013827.1 | FORC_018 | 2016 | KOR | Environmental | FORC18_3769 | Zot | Unpublished |
| CP011406.1 | FORC_014 | 2015 | KOR | Environmental | FORC14_1633 | Zot | [41] |
| CP011407.1 | FORC_014 | 2015 | KOR | Environmental | FORC14_3924 | Zot | [41] |
| CP023248.1 | MAVP-26 | 2013 | USA | Clinical | YA91_16975 | Toxin | [42] |
| QPIY01000007.1 | 108B | 2018 | USA | Environmental | DET53_107186 | Zot | Unpublished |
| CP031781.1 | VPD14 | 2012 | China | Environmental | D0853_08290 | Hyp protein | Unpublished |
| CP026041.1 | FDAARGOS_51 | 1998 | USA | Clinical | RK51_011060 | Toxin | Unpublished |
| N° of Isolates Containing Toxins | |||||||
|---|---|---|---|---|---|---|---|
| Region/Year | N° Total Isolates | tlh | tdh | trh | zot | tdh-zot * | trh-zot * |
| Metropolitan | 59 | 59 | 0 | 0 | 28 | 0 | 0 |
| Los Rios | 212 | 212 | 23 | 4 | 69 | 3 | 1 |
| Los Lagos | 65 | 65 | 0 | 1 | 28 | 0 | 0 |
| Total | 336 | 336 | 23 | 5 | 125 | 3 | 1 |
| 2017 | 173 | 173 | 17 | 5 | 57 | 2 | 1 |
| 2018 | 128 | 128 | 6 | 0 | 49 | 1 | 0 |
| 2020 | 35 | 35 | 0 | 0 | 19 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iribarren, C.; Plaza, N.; Ramírez-Araya, S.; Pérez-Reytor, D.; Urrutia, Í.M.; Suffredini, E.; Vicenza, T.; Ulloa, S.; Fernández, J.; Navarrete, P.; et al. Presence of Zonula Occludens Toxin-Coding Genes among Vibrio parahaemolyticus Isolates of Clinical and Environmental Origin. Microorganisms 2024, 12, 504. https://doi.org/10.3390/microorganisms12030504
Iribarren C, Plaza N, Ramírez-Araya S, Pérez-Reytor D, Urrutia ÍM, Suffredini E, Vicenza T, Ulloa S, Fernández J, Navarrete P, et al. Presence of Zonula Occludens Toxin-Coding Genes among Vibrio parahaemolyticus Isolates of Clinical and Environmental Origin. Microorganisms. 2024; 12(3):504. https://doi.org/10.3390/microorganisms12030504
Chicago/Turabian StyleIribarren, Cristian, Nicolás Plaza, Sebastián Ramírez-Araya, Diliana Pérez-Reytor, Ítalo M. Urrutia, Elisabetta Suffredini, Teresa Vicenza, Soledad Ulloa, Jorge Fernández, Paola Navarrete, and et al. 2024. "Presence of Zonula Occludens Toxin-Coding Genes among Vibrio parahaemolyticus Isolates of Clinical and Environmental Origin" Microorganisms 12, no. 3: 504. https://doi.org/10.3390/microorganisms12030504
APA StyleIribarren, C., Plaza, N., Ramírez-Araya, S., Pérez-Reytor, D., Urrutia, Í. M., Suffredini, E., Vicenza, T., Ulloa, S., Fernández, J., Navarrete, P., Jaña, V., Pavez, L., del Pozo, T., Corsini, G., Lopez-Joven, C., & García, K. (2024). Presence of Zonula Occludens Toxin-Coding Genes among Vibrio parahaemolyticus Isolates of Clinical and Environmental Origin. Microorganisms, 12(3), 504. https://doi.org/10.3390/microorganisms12030504










