Isolation and Identification of Chicken-Derived Lactic Acid Bacteria: In Vitro Probiotic Properties and Antagonistic Effects against Salmonella pullorum, Staphylococcus aureus, and Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Pathogenic Bacteria, Medium, Simulated Gastric, and Small Intestinal Fluids
2.2. Isolation of Lactic Acid Bacteria (LAB)
2.3. Antimicrobial Activity
2.4. Molecular Identification
2.5. Growth Curve and Lactic Acid Production
2.6. Stress Tolerance
2.6.1. Acid Tolerance
2.6.2. Bile Salt Tolerance
2.6.3. Tolerance to Simulated Gastric and Intestinal Fluid
2.7. Antagonistic Effect
2.8. Statistical Analysis
3. Results
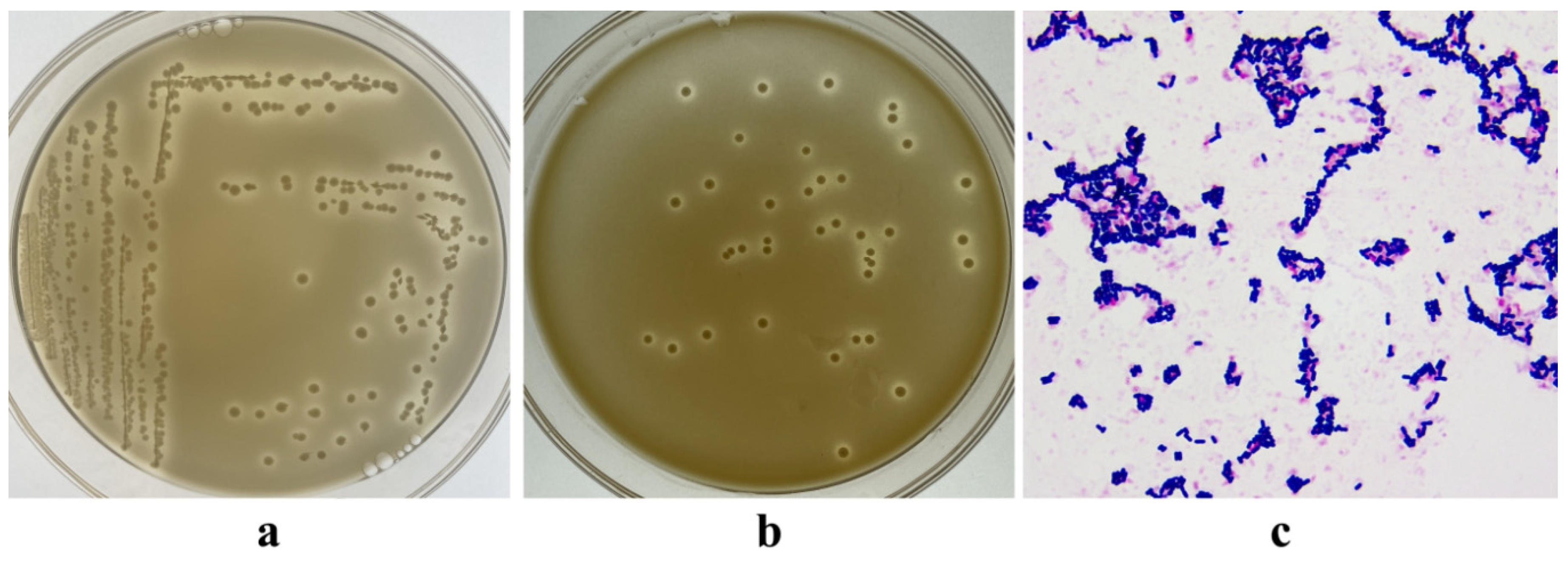
3.1. Isolation of LAB
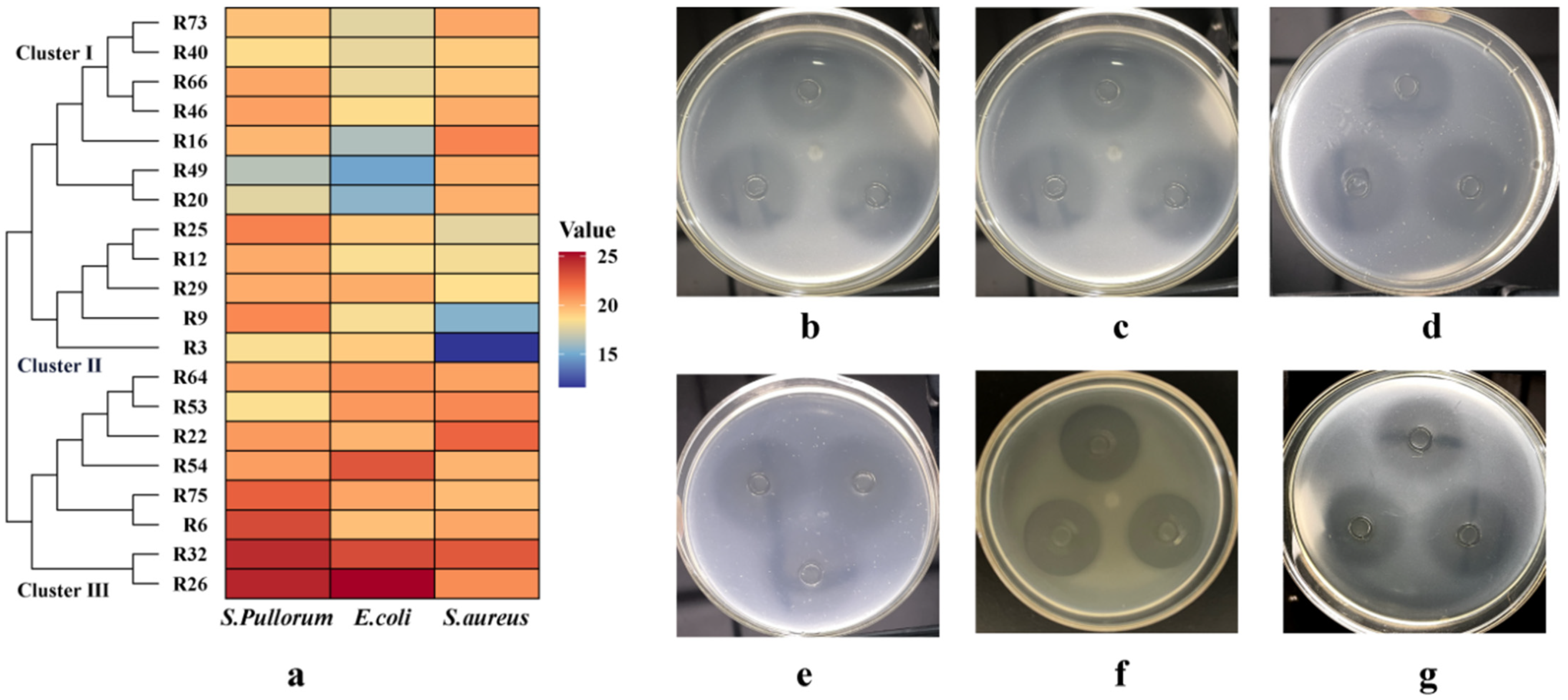
3.2. Antibacterial Activity
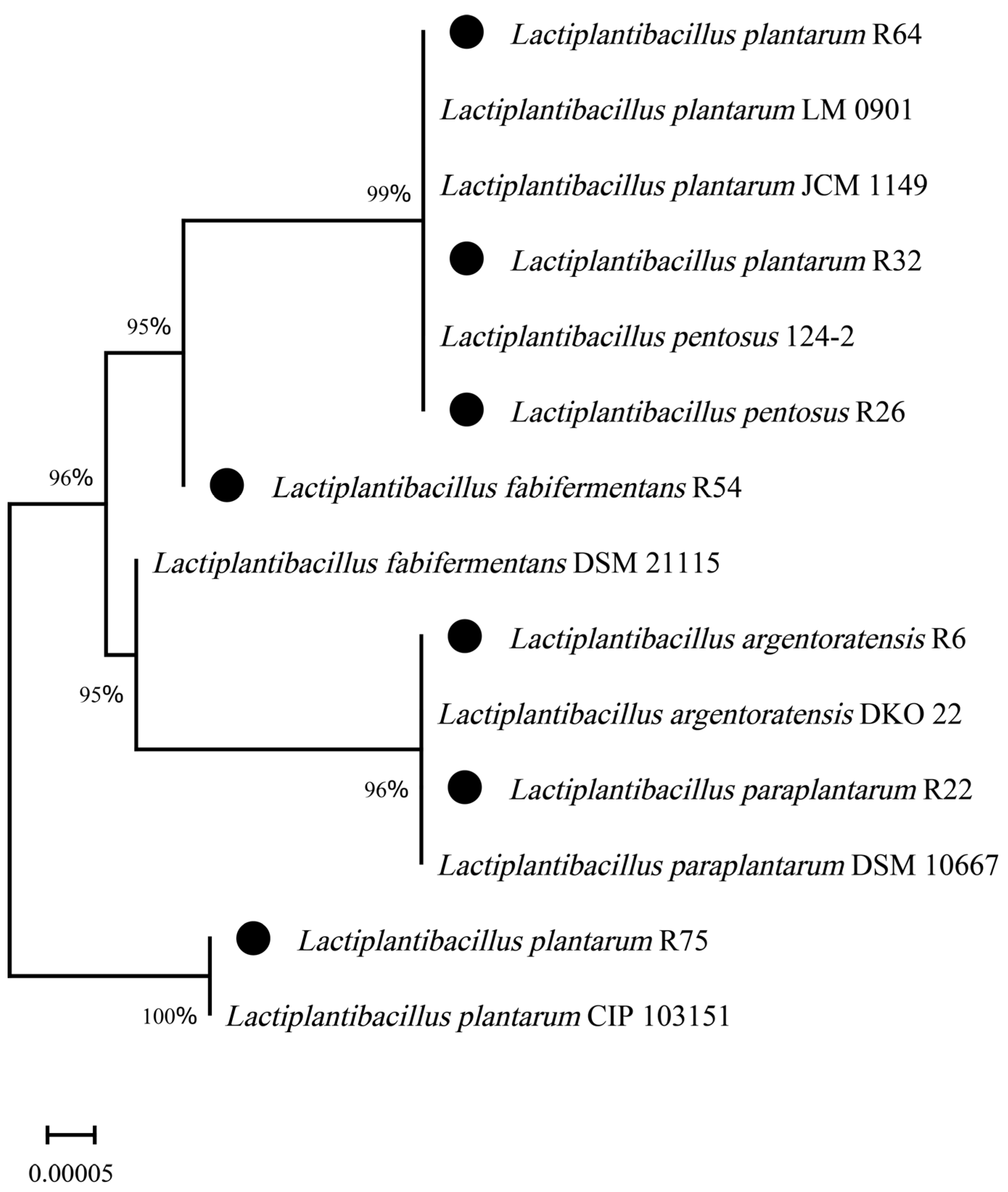
3.3. Molecular Identification
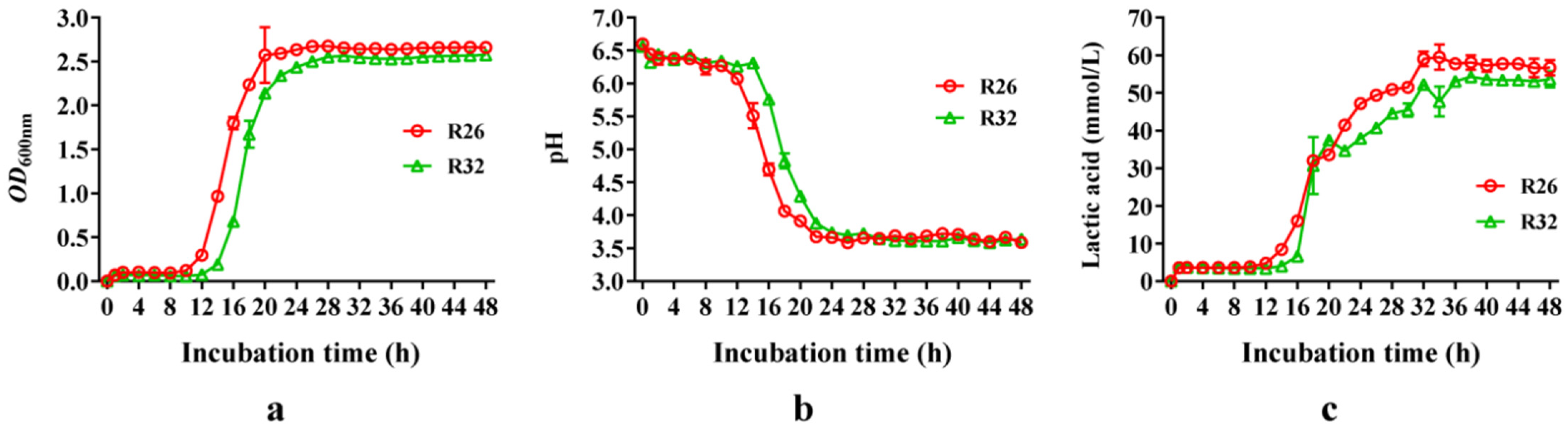
3.4. Growth Curve and Lactic Acid Production of L. pentosus R26 and L. plantarum R32
3.5. Acid Tolerance
3.6. Bile Salt Tolerance
3.7. Tolerance to Simulated Gastric and Intestinal Fluid
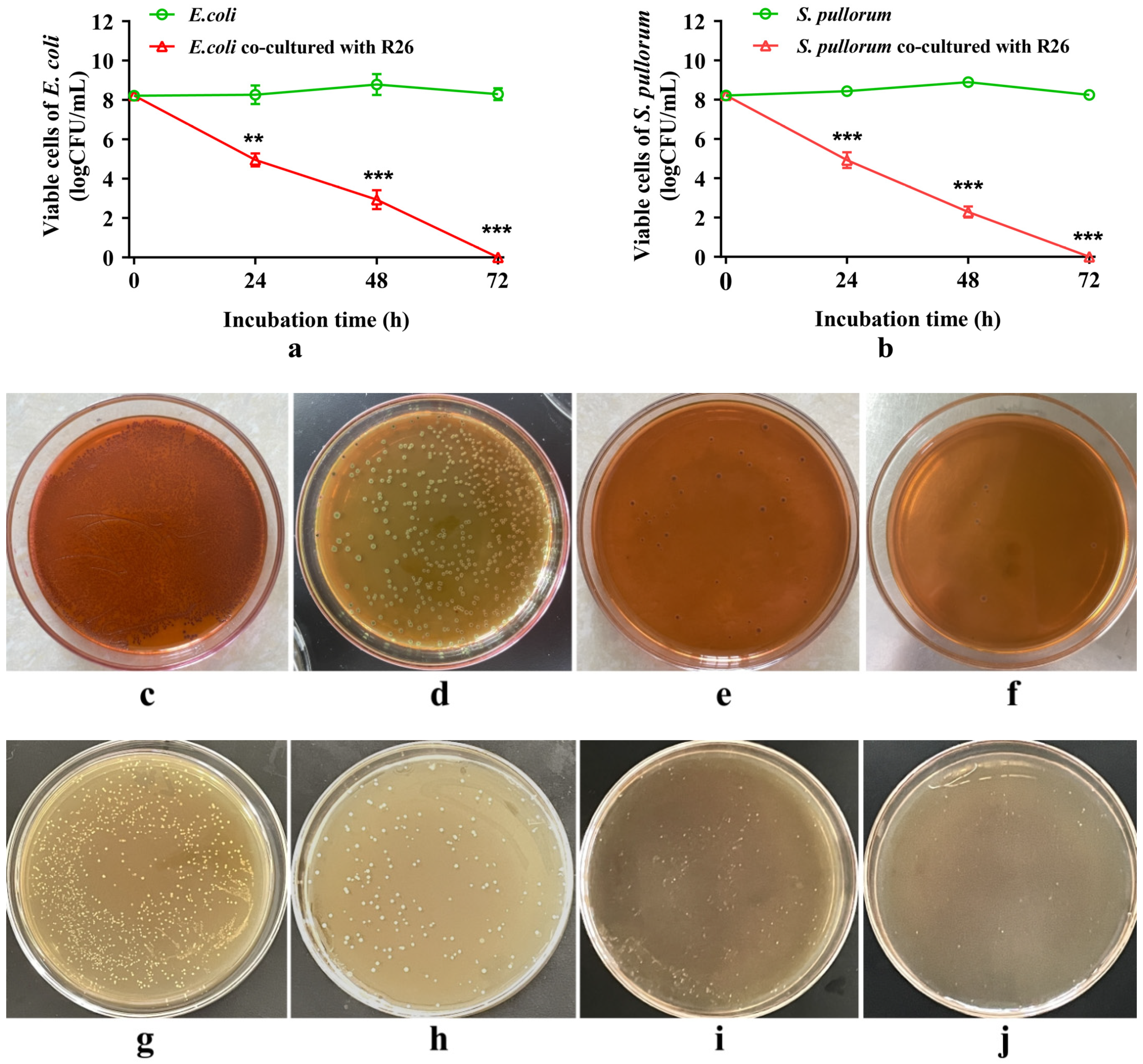
3.8. Antagonistic Effect
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Minj, J.; Chandra, P.; Paul, C.; Sharma, R.K. Bio-functional properties of probiotic Lactobacillus: Current applications and research perspectives. Crit. Rev. Food Sci. Nutr. 2021, 61, 2207–2224. [Google Scholar] [CrossRef] [PubMed]
- Wieërs, G.; Belkhir, L.; Enaud, R.; Leclercq, S.; Philippart de Foy, J.M.; Dequenne, I.; de Timary, P.; Cani, P.D. How probiotics affect the microbiota. Front. Cell Infect. Microbiol. 2020, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics regulate gut microbiota: An effective method to improve immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef] [PubMed]
- Gieryńska, M.; Szulc-Dąbrowska, L.; Struzik, J.; Mielcarska, M.B.; Gregorczyk-Zboroch, K.P. Integrity of the intestinal barrier: The involvement of epithelial cells and microbiota-a mutual relationship. Animals 2022, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Sun, L.; Gonzalez, F.J. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe 2022, 30, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast Milk, a source of beneficial microbes and associated benefits for infant health. Nutrients 2020, 12, 1039. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Wongkuna, S.; Ghimire, S.; Kumar, R.; Antony, L.; Doerner, K.C.; Singery, A.; Nelson, E.; Woyengo, T.; Chankhamhaengdecha, S.; et al. Gut microbial dynamics during conventionalization of germfree chicken. mSphere 2019, 4, e00035-19. [Google Scholar] [CrossRef]
- Rychlik, I. Composition and function of chicken gut microbiota. Animals 2020, 10, 103. [Google Scholar] [CrossRef]
- Darbandi, A.; Asadi, A.; Mahdizade Ari, M.; Ohadi, E.; Talebi, M.; Halaj Zadeh, M.; Darb Emamie, A.; Ghanavati, R.; Kakanj, M. Bacteriocins: Properties and potential use as antimicrobials. J. Clin. Lab. Anal. 2022, 36, e24093. [Google Scholar] [CrossRef]
- Zhang, H.; Pertiwi, H.; Hou, Y.; Majdeddin, M.; Michiels, J. Protective effects of Lactobacillus on heat stress-induced intestinal injury in finisher broilers by regulating gut microbiota and stimulating epithelial development. Sci. Total Environ. 2024, 918, 170410. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, K.; Zhang, A.; Chang, W.; Zheng, A.; Chen, Z.; Cai, H.; Liu, G. Effects of Lactobacillus acidophilus on the growth performance, immune response, and intestinal barrier function of broiler chickens challenged with Escherichia coli O157. Poult. Sci. 2021, 100, 101323. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Chen, Y.; Yu, L.; Wang, J.; Huang, M.; Zhu, N. Effects of Lactobacillus plantarum on intestinal integrity and immune responses of egg-laying chickens infected with Clostridium perfringens under the free-range or the specific pathogen free environment. BMC Vet. Res. 2020, 16, 47. [Google Scholar] [CrossRef] [PubMed]
- Shokryazdan, P.; Faseleh Jahromi, M.; Liang, J.B.; Ramasamy, K.; Sieo, C.C.; Ho, Y.W. Effects of a Lactobacillus salivarius mixture on performance, intestinal health and serum lipids of broiler chickens. PLoS ONE 2017, 12, e0175959. [Google Scholar] [CrossRef] [PubMed]
- Fesseha, H.; Demlie, T.; Mathewos, M.; Eshetu, E. Effect of Lactobacillus species probiotics on growth performance of dual-purpose chicken. Vet. Med. 2021, 12, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Stergiou, O.S.; Tegopoulos, K.; Kiousi, D.E.; Tsifintaris, M.; Papageorgiou, A.C.; Tassou, C.C.; Chorianopoulos, N.; Kolovos, P.; Galanis, A. Whole-genome sequencing, phylogenetic and genomic analysis of Lactiplantibacillus pentosus L33, a potential probiotic strain isolated from fermented sausages. Front. Microbiol. 2021, 12, 746659. [Google Scholar] [CrossRef] [PubMed]
- Casey, P.G.; Gardiner, G.E.; Casey, G.; Bradshaw, B.; Lawlor, P.G.; Lynch, P.B.; Leonard, F.C.; Stanton, C.; Ross, R.P.; Fitzgerald, G.F.; et al. A five-strain probiotic combination reduces pathogen shedding and alleviates disease signs in pigs challenged with Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2007, 73, 1858–1863. [Google Scholar] [CrossRef] [PubMed]
- Altaher, Y.; Jahromi, M.; Ebrahim, R.; Zulkifli, I.; Liang, J. Lactobacillus pentosus Ita23 and L. acidipiscis Ita44 enhance feed conversion efficiency and beneficial gut microbiota in broiler chickens. Braz. J. Poult. Sci. 2015, 17, 159–164. [Google Scholar] [CrossRef]
- Faseleh Jahromi, M.; Wesam Altaher, Y.; Shokryazdan, P.; Ebrahimi, R.; Ebrahimi, M.; Idrus, Z.; Tufarelli, V.; Liang, J.B. Dietary supplementation of a mixture of Lactobacillus strains enhances performance of broiler chickens raised under heat stress conditions. Int. J. Biometeorol. 2016, 60, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Hu, C.; Yan, W.; Jiang, H.; Liu, G. Lactobacillus pentosus increases the abundance of Akkermansia and affects the serum metabolome to alleviate DSS-induced colitis in a murine model. Front. Cell Dev. Biol. 2020, 8, 591408. [Google Scholar] [CrossRef]
- García-Hernández, Y.; Pérez-Sánchez, T.; Boucourt, R.; Balcázar, J.L.; Nicoli, J.R.; Moreira-Silva, J.; Rodríguez, Z.; Fuertes, H.; Nuñez, O.; Albelo, N.; et al. Isolation, characterization and evaluation of probiotic lactic acid bacteria for potential use in animal production. Res. Vet. Sci. 2016, 108, 125–132. [Google Scholar] [CrossRef]
- Sika-Kadji, A.E.; Kéhi, T.B.; N’Guessan, F.K.; Koffi-Nevry, R.A. Probiotic potential of lactic acid bacteria isolated from broiler chickens in Côted’Ivoire. Int. J. Poult. Sci. 2022, 21, 119–128. [Google Scholar] [CrossRef]
- Shi, S.; Liu, J.; Dong, J.; Hu, J.; Liu, Y.; Feng, J.; Zhou, D. Research progress on the regulation mechanism of probiotics on the microecological flora of infected intestines in livestock and poultry. Lett. Appl. Microbiol. 2022, 74, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bu, Y.; Zhang, C.; Yi, H.; Liu, D.; Jiao, J. Development of a low-cost and high-efficiency culture medium for bacteriocin Lac-B23 production by Lactobacillus plantarum J23. Biology 2020, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Genta, M.L.; Heluane, H. Biochemical identification of most frequently encountered bacteria that cause food spoilage. In Food Microbiology Protocols. Methods in Biotechnology; Spencer, J.F.T., de Ragout Spencer, A.L., Eds.; Humana Press: Totowa, NJ, USA, 2001; Volume 14. [Google Scholar] [CrossRef]
- El-Hadedy, D.; El-Nour, S.A. Identification of Staphylococcus aureus and Escherichia coli isolated from Egyptian food by conventional and molecular methods. J. Genet. Eng. Biotechnol. 2012, 10, 129–135. [Google Scholar] [CrossRef]
- Chinese Pharmacopoeia commission. The Pharmacopoeia of the People’s Republic of China, 2020th ed.; China Medical Science and Technology Press: Beijing, China, 2020. [Google Scholar]
- Stachurska, X.; Roszak, M.; Jabłońska, J.; Mizielińska, M.; Nawrotek, P. Double-layer agar (DLA) modifications for the first step of the phage-antibiotic synergy (PAS) identification. Antibiotics 2021, 10, 1306. [Google Scholar] [CrossRef] [PubMed]
- Cappello, C.; Tlais, A.Z.A.; Acin-Albiac, M.; Lemos Junior, W.J.F.; Pinto, D.; Filannino, P.; Rinaldi, F.; Gobbetti, M.; Di Cagno, R. Identification and selection of prospective probiotics for enhancing gastrointestinal digestion: Application in pharmaceutical preparations and dietary supplements. Nutrients 2023, 15, 1306. [Google Scholar] [CrossRef] [PubMed]
- Moyes, R.B.; Reynolds, J.; Breakwell, D.P. Differential staining of bacteria: Gram stain. Curr. Protoc. Microbiol. 2009, 15, A.3C.1–A.3C.8. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Pang, H.; Wang, L.; Ma, C.; Wu, G.; Liu, Y.; Guan, Y.; Zhang, M.; Qin, G.; Tan, Z. Bacteriocin-producing lactic acid bacteria strains with antimicrobial activity screened from bamei pig feces. Foods 2022, 11, 709. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Zhang, L.; Liu, H.; Shen, J.; Zhang, Y.; Lu, L.; Zhang, X.; Ma, X. Bacillus subtilis M6 improves intestinal barrier, antioxidant capacity and gut microbial composition in AA broiler. Front. Nutr. 2022, 9, 965310. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis (Version 11). Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Sirisopapong, M.; Shimosato, T.; Okrathok, S.; Khempaka, S. Assessment of lactic acid bacteria isolated from the chicken digestive tract for potential use as poultry probiotics. Anim. Biosci. 2023, 36, 1209–1220. [Google Scholar] [CrossRef]
- Hu, G.; Jiang, H.; Zong, Y.; Datsomor, O.; Kou, L.; An, Y.; Zhao, J.; Miao, L. Characterization of lactic acid producing bacteria isolated from rumen: Growth, acid and bile salt toleranceand antimicrobial function. Fermentation 2022, 8, 385. [Google Scholar] [CrossRef]
- Sağlam, H.; Karahan, A.G. Plasmid stability of potential probiotic Lactobacillus plantarum strains in artificial gastric juice, at elevated temperature, and in the presence of novobiocin and acriflavine. Arch. Microbiol. 2021, 203, 183–191. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Lin, X.; Wu, C. Identification and evaluation of probiotic potential of Bifidobacterium breve AHC3 isolated from chicken intestines and its effect on necrotizing enterocolitis (NEC) in newborn SD rats. PLoS ONE 2023, 18, e0287799. [Google Scholar] [CrossRef]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken gut microbiota: Importance and detection technology. Front. Vet. Sci. 2018, 5, 254. [Google Scholar] [CrossRef]
- van Zyl, W.F.; Deane, S.M.; Dicks, L.M.T. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 2020, 12, 1831339. [Google Scholar] [CrossRef]
- Tang, H.; Huang, W.; Yao, Y.F. The metabolites of lactic acid bacteria: Classification, biosynthesis and modulation of gut microbiota. Microb. Cell 2023, 10, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Hai, D.; Lu, Z.; Huang, X.; Lv, F.; Bie, X. In vitro screening of chicken-derived lactobacillus strains that effectively inhibit salmonella colonization and adhesion. Foods 2021, 10, 569. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Li, P.; Gu, Q. Complete genome sequence analysis of a strain Lactobacillus pentosus ZFM94 and its probiotic characteristics. Genomics 2020, 112, 3142–3149. [Google Scholar] [CrossRef]
- Kumar, A.; Ruhal, R.; Kataria, R. Bacteriocins of lactic acid bacteria as a potential antimicrobial peptide. In Biomimicry Materials and Applications; Inamuddin, Altalhi, T., Alrogi, A., Eds.; Scrivener Publishing LLC.: Beverly, MA, USA, 2023; pp. 83–104. [Google Scholar] [CrossRef]
- Reuben, R.C.; Elghandour, M.M.M.Y.; Alqaisi, O.; Cone, J.W.; Márquez, O.; Salem, A.Z.M. Influence of microbial probiotics on ruminant health and nutrition: Sources, mode of action and implications. J. Sci. Food Agric. 2022, 102, 1319–1340. [Google Scholar] [CrossRef]
- Jung, J.Y.; Han, S.S.; Kim, Z.H.; Kim, M.H.; Kang, H.K.; Jin, H.M.; Lee, M.H. In-vitro characterization of growth inhibition against the gut pathogen of potentially probiotic lactic acid bacteria strains isolated from fermented products. Microorganisms 2021, 9, 2141. [Google Scholar] [CrossRef]
- Wang, C.; Cui, Y.; Qu, X. Mechanisms and improvement of acid resistance in lactic acid bacteria. Arch. Microbiol. 2018, 200, 195–201. [Google Scholar] [CrossRef]
- Shen, H.; Wang, T.; Dong, W.; Sun, G.; Liu, J.; Peng, N.; Zhao, S. Metagenome-assembled genome reveals species and functional composition of Jianghan chicken gut microbiota and isolation of Pediococcus acidilactic with probiotic properties. Microbiome 2024, 12, 25. [Google Scholar] [CrossRef]
- Ruiz, L.; Margolles, A.; Sánchez, B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 2013, 4, 396. [Google Scholar] [CrossRef]
- Bustos, A.Y.; Font de Valdez, G.; Fadda, S.; Taranto, M.P. New insights into bacterial bile resistance mechanisms: The role of bile salt hydrolase and its impact on human health. Food Res. Int. 2018, 112, 250–262. [Google Scholar] [CrossRef]
- Abedi, E.; Pourmohamadi, K.; Mousavifard, M.; Sayadi, M. Comparison between surface hydrophobicity of heated and thermosonicated cells to detoxify aflatoxin B1 by co-culture Lactobacillus plantarum and Lactobacillus rhamnosus in sourdough: Modeling studies. Food Sci. Technol. 2022, 154, 112616. [Google Scholar] [CrossRef]
- Ben Farhat, L.; Romeo, F.V.; Foti, P.; Russo, N.; Randazzo, C.L.; Caggia, C.; Abidi, F. Multi-functional potential of lactic acid bacteria strains and antimicrobial effects in minimally processed pomegranate (Punica granatum L. cv Jolly Red) arils. Microorganisms 2022, 10, 1876. [Google Scholar] [CrossRef]
- Wang, J.; Zeng, Y.; Wang, S.; Liu, H.; Zhang, D.; Zhang, W.; Wang, Y.; Ji, H. Swine-derived probiotic Lactobacillus plantarum inhibits growth and adhesion of enterotoxigenic Escherichia coli and mediates host defense. Front. Microbiol. 2018, 9, 1364. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, C.; Vanegas, C.; Klotz, B. Antagonistic effect of Lactobacillus strains against Escherichia coli and Listeria monocytogenes in milk. J. Dairy. Res. 2011, 78, 136–143. [Google Scholar] [CrossRef]
- Mizuno, K.; Mizuno, M.; Yamauchi, M.; Takemura, A.J.; Medrano Romero, V.; Morikawa, K. Adjacent-possible ecological niche: Growth of Lactobacillus species co-cultured with Escherichia coli in a synthetic minimal medium. Sci. Rep. 2017, 7, 12880. [Google Scholar] [CrossRef]
- Ma, K.; Chen, W.; Lin, X.Q.; Liu, Z.Z.; Wang, T.; Zhang, J.B.; Zhang, J.G.; Zhou, C.K.; Gao, Y.; Du, C.T.; et al. Culturing the chicken intestinal microbiota and potential application as probiotics development. Int. J. Mol. Sci. 2023, 24, 3045. [Google Scholar] [CrossRef]
| Strains | S. pullorum | E. coli | S. aureus |
|---|---|---|---|
| R6 | 23.37 ± 0.15 c | 19.58 ± 0.07 f | 20.33 ± 0.15 cd |
| R3 | 18.41 ± 0.0 l | 19.22 ± 0.03 g | 11.64 ± 0.05 j |
| R9 | 21.24 ± 0.08 e | 18.32 ± 0.25 i | 15.43 ± 0.08 i |
| R12 | 20.21 ± 0.02 h | 18.41 ± 0.16 ih | 18.25 ± 0.31 gh |
| R16 | 19.83 ± 0.10 i | 16.35 ± 0.05 k | 21.35 ± 0.05 b |
| R20 | 17.67 ± 0.12 m | 15.57 ± 0.08 e | 20.08 ± 0.03 cd |
| R22 | 20.70 ± 0.10 i | 19.92 ± 0.13 d | 22.32 ± 1.17 a |
| R25 | 21.44 ± 0.10 e | 19.30 ± 0.05 gf | 17.73 ± 0.12 h |
| R26 | 24.72 ± 0.11 a | 25.45 ± 0.05 a | 21.11 ± 0.10 b |
| R29 | 20.17 ± 0.12 h | 20.15 ± 0.05 e | 18.53 ± 0.06 g |
| R32 | 24.48 ± 0.03 b | 23.32 ± 0.02 b | 22.80 ± 0.70 a |
| R40 | 18.67 ± 0.06 k | 17.87 ± 0.06 j | 19.17 ± 0.15 f |
| R46 | 20.50 ± 0.10 fg | 18.66 ± 0.12 h | 20.13 ± 0.15 cd |
| R49 | 16.57 ± 0.12 n | 14.87 ± 0.31 m | 20.05 ± 0.13 cd |
| R53 | 18.50 ± 0.10 lk | 20.77 ± 0.25 d | 21.21 ± 0.19 b |
| R54 | 20.60 ± 0.10 f | 22.93 ± 0.32 c | 19.92 ± 0.13 cde |
| R64 | 20.43 ± 0.05 fg | 20.89 ± 0.53 d | 20.45 ± 0.05 c |
| R66 | 20.33 ± 0.29 gh | 17.97 ± 0.21 j | 19.37 ± 0.25 ef |
| R73 | 19.52 ± 0.06 j | 17.67 ± 0.21 j | 20.31 ± 0.09 cd |
| R75 | 22.53 ± 0.15 d | 20.38 ± 0.10 e | 19.70 ± 0.44 def |
| p Value | <0.001 | <0.001 | <0.001 |
| Strains (GenBank Accession No.) | The Highest Identity Strain (GenBank Accession No.) | Identity (%) |
|---|---|---|
| R6 (PP389395.1) | Lactiplantibacillus argentoratensis DKO 22 (NR 042254.1) | 99.96 |
| R22 (PP389397.1) | Lactiplantibacillus paraplantarum DSM 10667 (NR 025447.1) | 99.80 |
| R26 (PP389393.1) | Lactiplantibacillus pentosus 124-2 (NR 029133.1) | 100.00 |
| R32 (PP390060.1) | Lactiplantibacillus plantarum JCM 1149 (NR 117813.1) | 99.00 |
| R54 (PP390062.1) | Lactiplantibacillus fabifermentans DSM 21115 (NR113339.1) | 99.23 |
| R64 (PP390061.1) | Lactiplantibacillus plantarum LM 0901 (QQ569413.1) | 99.93 |
| R75 (PP389398.1) | Lactiplantibacillus plantarum CIP 103151 (NR 104573.1) | 100.00 |
| Strains | Items | pH Value of the Medium | p Value | |||||
|---|---|---|---|---|---|---|---|---|
| 6.5 | 3.5 | 2.5 | 1.5 | M | L | Q | ||
| L. pentosus R26 | Colony count (log CFU/mL) | 8.40 ± 0.03 a | 7.95 ± 0.02 b | 7.45 ± 0.04 c | 7.24 ± 0.02 d | <0.001 | <0.001 | <0.001 |
| Survival rate (%) | 100.00 ± 0.36 a | 94.72 ± 0.37 b | 88.69 ± 0.16 c | 86.29 ± 0.13 d | <0.001 | <0.001 | <0.001 | |
| L. plantarum R32 | Colony count (log CFU/mL) | 8.41 ± 0.03 a | 7.75 ± 0.09 b | 7.51 ± 0.05 b | 7.40 ± 0.03 c | <0.001 | <0.001 | 0.140 |
| Survival rate (%) | 100.00 ± 0.36 a | 92.23 ± 1.21 b | 89.33 ± 0.50 c | 87.99 ± 0.33 d | <0.001 | <0.001 | 0.151 | |
| Strains | Items | Concentrations of Bile Salt (%) | p Value | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 0.10 | 0.30 | 0.50 | M | L | Q | ||
| L. pentosus R26 | Colony count (log CFU/mL) | 8.37 ± 0.02 a | 8.33 ± 0.03 ab | 8.29 ± 0.02 b | 7.25 ± 0.03 c | <0.001 | <0.001 | <0.001 |
| Survival rate (%) | 100.00 ± 0.25 a | 99.56 ± 0.38 ab | 99.12 ± 0.30 b | 86.66 ± 0.52 c | <0.001 | <0.001 | <0.001 | |
| L. plantarum R32 | Colony count (log CFU/mL) | 8.33 ± 0.01 a | 8.24 ± 0.03 b | 8.22 ± 0.01 b | 7.13 ± 0.02 c | <0.001 | <0.001 | <0.001 |
| Survival rate (%) | 100.00 ± 0.07 a | 98.92 ± 0.41 b | 98.60 ± 0.14 b | 85.52 ± 0.24 c | <0.001 | <0.001 | <0.001 | |
| Strains | Items | Incubation Time (h) | p Value | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 2 | M | L | Q | ||
| L. pentosus R26 | Colony count (log CFU/mL) | 8.36 ± 0.03 a | 8.32 ± 0.05 a | 8.22 ± 0.05 b | 7.53 ± 0.01 c | <0.001 | <0.001 | <0.001 |
| Survival rate (%) | 100.00 ± 0.43 a | 99.48 ± 1.01 ab | 98.29 ± 0.89 b | 90.03 ± 0.33 c | <0.001 | <0.001 | <0.001 | |
| L. Plantarum R32 | Colony count (log CFU/mL) | 8.39 ± 0.04 a | 8.24 ± 0.01 b | 8.13 ± 0.03 c | 7.39 ± 0.03 d | <0.001 | <0.001 | <0.001 |
| Survival rate (%) | 100.00 ± 0.43 a | 98.17 ± 0.29 b | 96.86 ± 0.80 c | 88.16 ± 0.42 d | <0.001 | <0.001 | <0.001 | |
| Strains | Items | Incubation Time (h) | p Value | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 2 | M | L | Q | ||
| L. pentosus R26 | Colony count (log CFU/mL) | 8.37 ± 0.01 a | 8.35 ± 0.01 a | 8.30 ± 0.02 b | 8.28 ± 0.01 b | <0.001 | <0.001 | 0.276 |
| Survival rate (%) | 100.00 ± 0.12 a | 99.76 ± 0.07 a | 99.12 ± 0.36 b | 98.92 ± 0.24 b | 0.001 | <0.001 | 0.407 | |
| L. Plantarum R32 | Colony count (log CFU/mL) | 8.42 ± 0.01 a | 8.38 ± 0.04 a | 8.31 ± 0.02 b | 8.27 ± 0.05 b | 0.002 | <0.001 | 0.525 |
| Survival rate (%) | 100.00 ± 0.12 a | 99.56 ± 0.49 a | 98.65 ± 0.27 b | 98.22 ± 0.74 b | 0.006 | 0.001 | 0.590 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, C.; Wang, L.; Liu, M.; Liu, J.; Qiu, M.; Chen, Y. Isolation and Identification of Chicken-Derived Lactic Acid Bacteria: In Vitro Probiotic Properties and Antagonistic Effects against Salmonella pullorum, Staphylococcus aureus, and Escherichia coli. Microorganisms 2024, 12, 795. https://doi.org/10.3390/microorganisms12040795
Tian C, Wang L, Liu M, Liu J, Qiu M, Chen Y. Isolation and Identification of Chicken-Derived Lactic Acid Bacteria: In Vitro Probiotic Properties and Antagonistic Effects against Salmonella pullorum, Staphylococcus aureus, and Escherichia coli. Microorganisms. 2024; 12(4):795. https://doi.org/10.3390/microorganisms12040795
Chicago/Turabian StyleTian, Congcong, Lei Wang, Mengjian Liu, Jiancheng Liu, Mingxin Qiu, and Yong Chen. 2024. "Isolation and Identification of Chicken-Derived Lactic Acid Bacteria: In Vitro Probiotic Properties and Antagonistic Effects against Salmonella pullorum, Staphylococcus aureus, and Escherichia coli" Microorganisms 12, no. 4: 795. https://doi.org/10.3390/microorganisms12040795
APA StyleTian, C., Wang, L., Liu, M., Liu, J., Qiu, M., & Chen, Y. (2024). Isolation and Identification of Chicken-Derived Lactic Acid Bacteria: In Vitro Probiotic Properties and Antagonistic Effects against Salmonella pullorum, Staphylococcus aureus, and Escherichia coli. Microorganisms, 12(4), 795. https://doi.org/10.3390/microorganisms12040795






