Evaluation of Probiotic Properties and Safety of Lactobacillus helveticus LH10 Derived from Vinegar through Comprehensive Analysis of Genotype and Phenotype
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
2.2. Extraction of Genome DNA
2.3. Genome Sequencing, Assembly, and Annotation
2.4. Evolutionary Position
2.5. Average Nucleotide Identity Analysis
2.6. Collinearity Analysis
2.7. Safety Evaluation of Lactobacillus Helveticus LH10
2.8. In Vitro Simulated Gastric and Intestinal Digestion
2.9. Binding Properties
2.10. Analysis of Bacteriostatic Properties
3. Results and Discussion
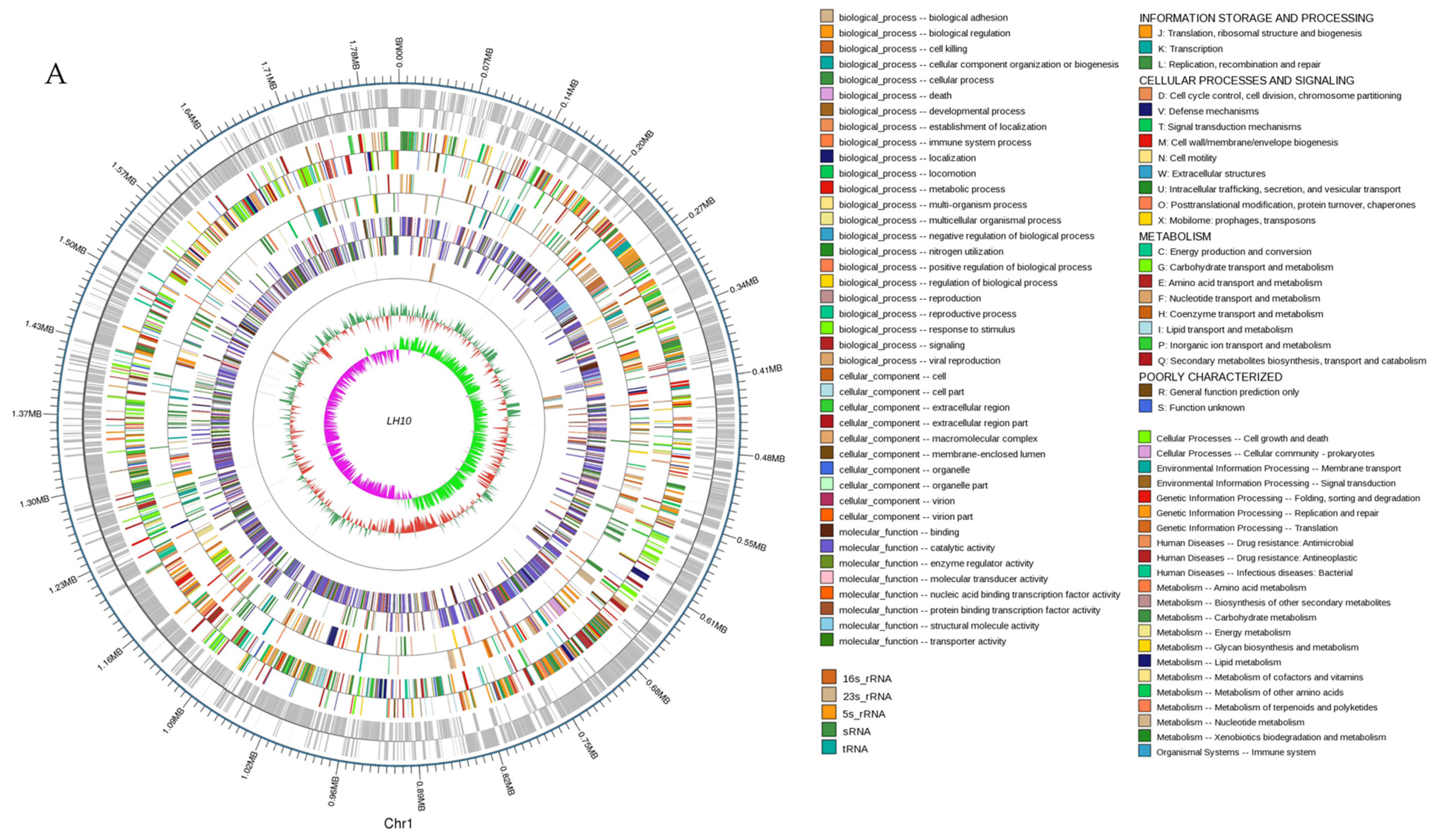
3.1. Genome Properties
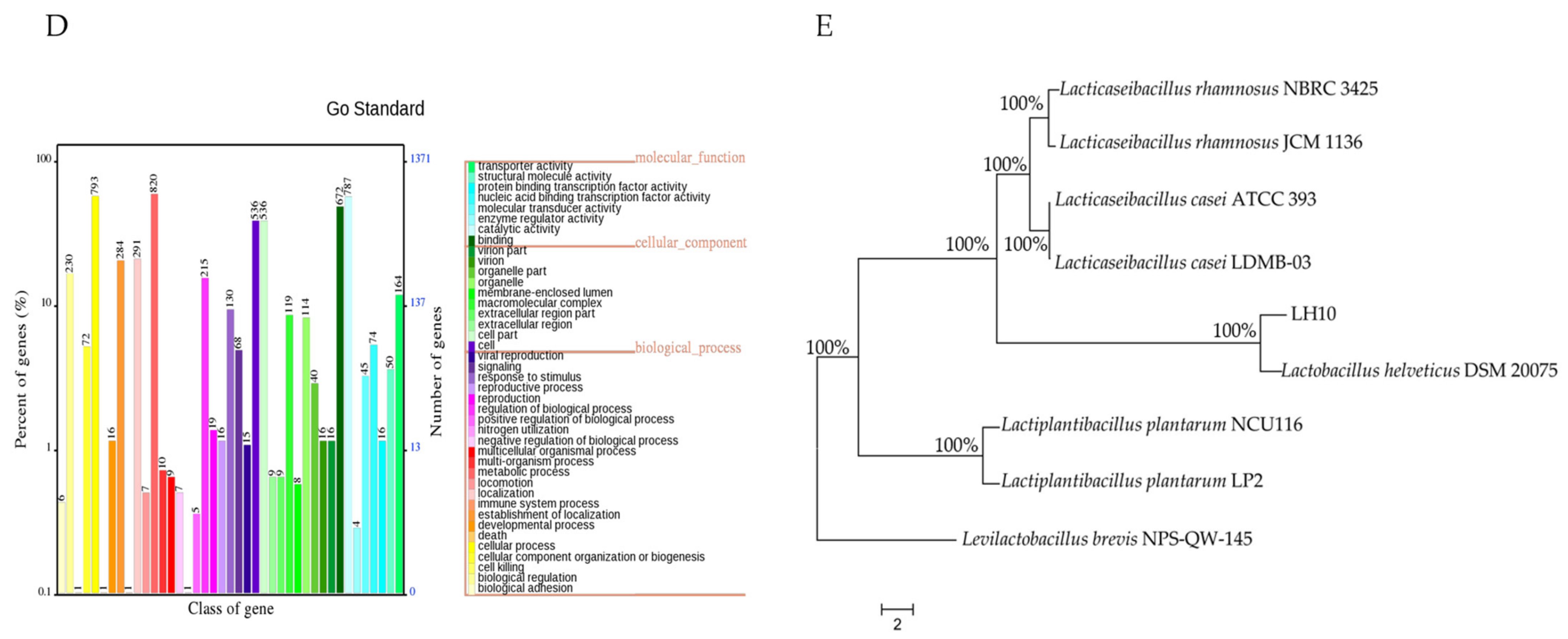
3.2. Phylogenetic Tree
3.3. Average Nucleotide Identity Analysis
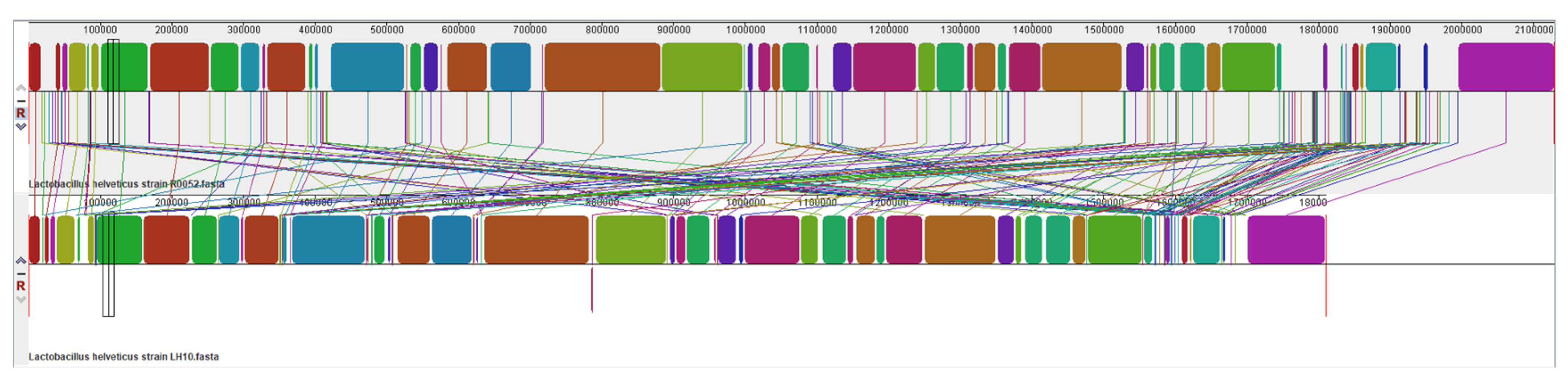
3.4. Collinearity Analysis
3.5. Antibiotic Susceptibility
3.6. Antibiotic Resistance Genotype
3.7. Hemolytic Activity Test
3.8. Virulence Factors of L. helveticus LH10
3.9. Binding Properties
3.10. Stress-Resistant Phenotype Analysis
3.10.1. Bile Salt Resistance
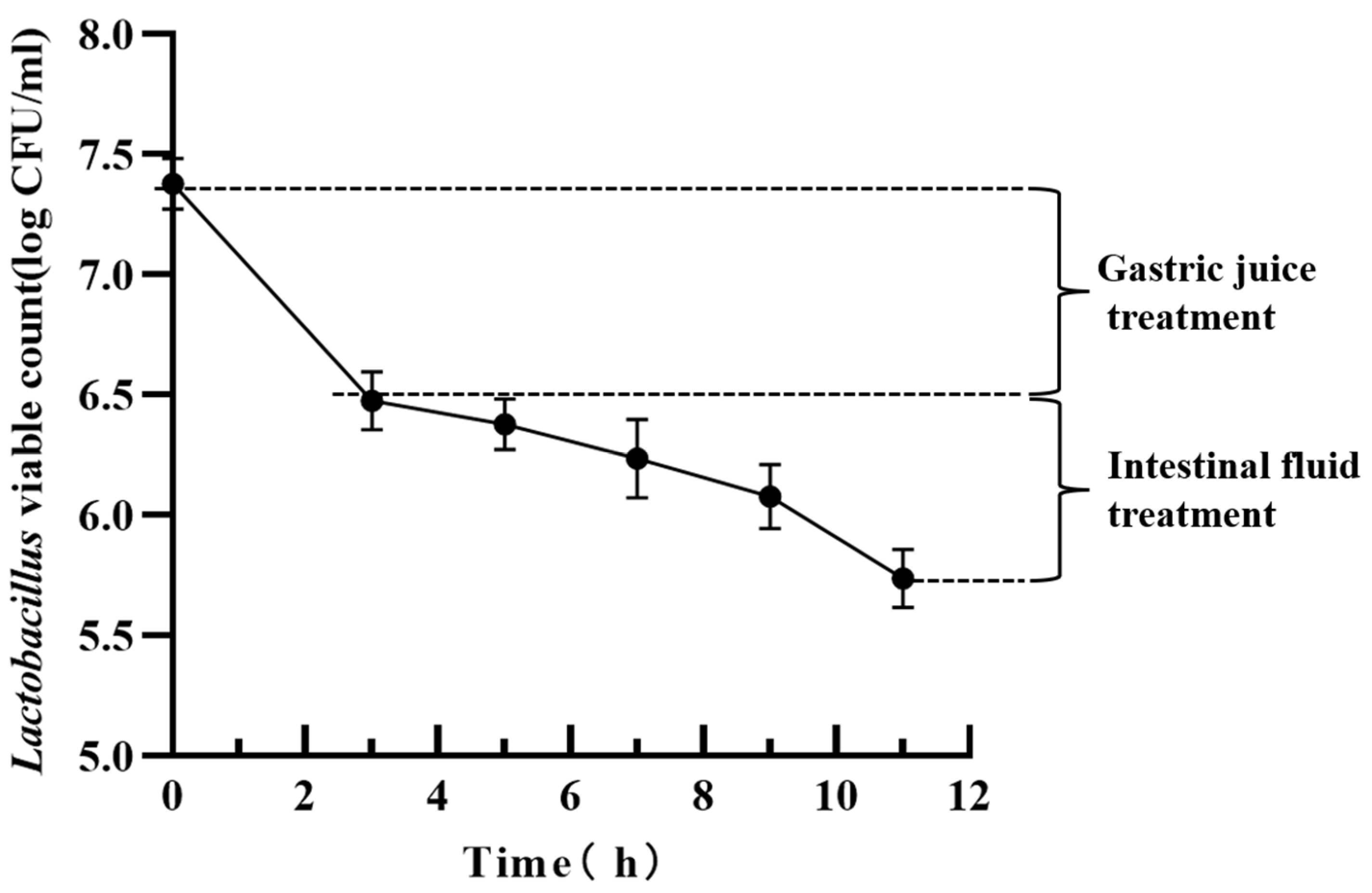
3.10.2. Gastrointestinal Fluid Resistance
3.11. Stress-Resistant Genotype Analysis
3.12. Bacteriostatic Ability
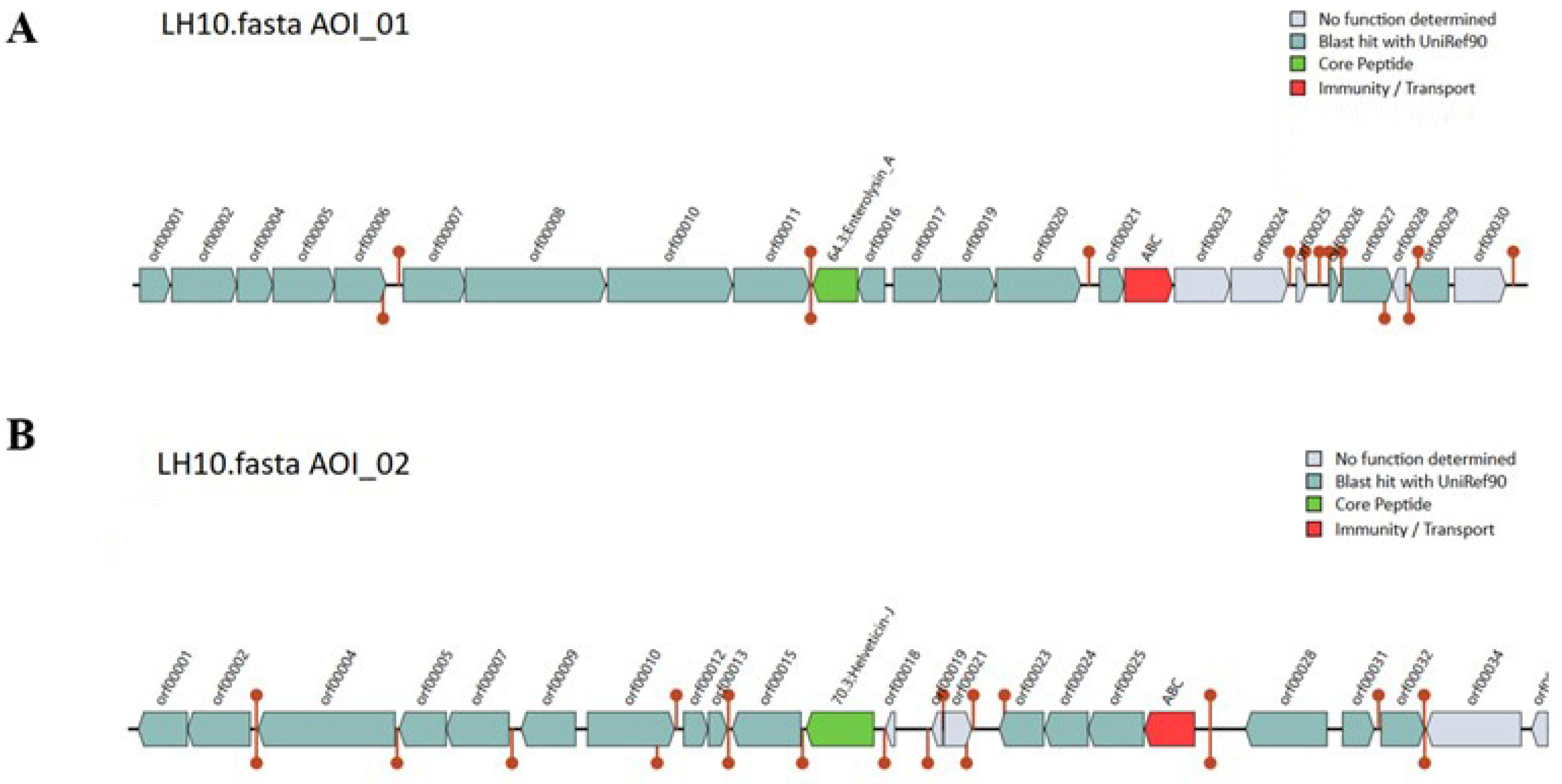
3.13. Bacteriocin Identification
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pimentel, T.C.; Cruz, A.G.; Pereira, E.; Almeida Da Costa, W.K.; Da Silva Rocha, R.; Targino De Souza Pedrosa, G.; Rocha, C.D.S.; Alves, J.M.; Alvarenga, V.O.; Sant’Ana, A.S.; et al. Postbiotics: An overview of concepts, inactivation technologies, health effects, and driver trends. Trends Food Sci. Technol. 2023, 138, 199–214. [Google Scholar] [CrossRef]
- Takeda, T.; Asaoka, D.; Nojiri, S.; Yanagisawa, N.; Nishizaki, Y.; Osada, T.; Koido, S.; Nagahara, A.; Katsumata, N.; Odamaki, T.; et al. Usefulness of Bifidobacterium longum BB536 in Elderly Individuals With Chronic Constipation: A Randomized Controlled Trial. Off. J. Am. Coll. Gastroenterol. ACG 2023, 118, 561. [Google Scholar] [CrossRef] [PubMed]
- Forsgård, R.A.; Rode, J.; Lobenius-Palmér, K.; Kamm, A.; Patil, S.; Tacken, M.G.J.; Lentjes, M.A.H.; Axelsson, J.; Grompone, G.; Montgomery, S.; et al. Limosilactobacillus reuteri DSM 17938 Supplementation and SARS-CoV-2 Specific Antibody Response in Healthy Adults: A Randomized, Triple-Blinded, Placebo-Controlled Trial. Gut Microbes 2023, 15, 2229938. [Google Scholar] [CrossRef]
- Fan, M.; Guo, T.; Li, W.; Chen, J.; Li, F.; Wang, C.; Shi, Y.; Li, D.X.; Zhang, S. Isolation and Identification of Novel Casein-Derived Bioactive Peptides and Potential Functions in Fermented Casein with Lactobacillus Helveticus. Food Sci. Hum. Wellness 2019, 8, 156–176. [Google Scholar] [CrossRef]
- Zago, M.; Massimiliano, L.; Bonvini, B.; Penna, G.; Giraffa, G.; Rescigno, M. Functional Characterization and Immunomodulatory Properties of Lactobacillus Helveticus Strains Isolated from Italian Hard Cheeses. PLoS ONE 2021, 16, e0245903. [Google Scholar] [CrossRef]
- Miao, Z.; Chen, L.; Zhang, Y.; Zhang, J.; Zhang, H. Bifidobacterium animalis subsp. Lactis Probio-M8 alleviates abnormal behavior and regulates gut microbiota in a mouse model suffering from autism. mSystems 2023, 9, e01013-23. [Google Scholar] [CrossRef]
- Yang, X.; Peng, Z.; He, M.; Li, Z.; Fu, G.; Li, S.; Zhang, J. Screening, probiotic properties, and inhibition mechanism of a Lactobacillus antagonistic to Listeria monocytogenes. Sci. Total Environ. 2024, 906, 167587. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, Y.; Duan, H.; Qiao, N.; Wang, G.; Zhao, J.; Zhai, Q.; Tian, F.; Chen, W. Latilactobacillus sakei: A candidate probiotic with a key role in food fermentations and health promotion. Crit. Rev. Food Sci. Nutr. 2022, 64, 978–995. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, J.; Xuan, J.; Chen, Y.; Tu, J.; Mu, H.; Wang, J.; Liu, G. Increase of γ-aminobutyric acid content and improvement of physicochemical characteristics of mulberry leaf powder by fermentation with a selected lactic acid bacteria strain. LWT 2023, 187, 115250. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, T.; Tang, H.; Li, X.; Chen, Y.; Zhang, L.; Zhang, J. Probiotic potential and amylolytic properties of lactic acid bacteria isolated from Chinese fermented cereal foods. Food Control 2020, 111, 107057. [Google Scholar] [CrossRef]
- Gong, M.; Zhou, Z.; Liu, S.; Zhu, S.; Li, G.; Zhong, F.; Mao, J. Dynamic changes in physico-chemical attributes and volatile compounds during fermentation of Zhenjiang vinegars made with glutinous and non-glutinous japonica rice. J. Cereal Sci. 2021, 100, 103246. [Google Scholar] [CrossRef]
- Wang, Z.; Lu, Z.; Shi, J.; Xu, Z. Exploring flavour-producing core microbiota in multispecies solid-state fermentation of traditional Chinese vinegar. Sci. Rep. 2016, 6, 26818. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.W.; Lazim, A.M.; Fazry, S.; Zaki, U.K.H.H.; Lim, S.J. Varieties, Production, Composition and Health Benefits of Vinegars: A Review. Food Chem. 2017, 221, 1621–1630. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, K.; Zhang, Y.; Li, Y.; Zhou, N.; Li, G. Probiotic characteristics and whole-genome sequence analysis of Pediococcus acidilactici isolated from the feces of adult beagles. Front. Microbiol. 2023, 14, 1179953. [Google Scholar] [CrossRef]
- Zhang, X.; Ruan, Y.; Liu, W.; Chen, Q.; Gu, L.; Guo, A. Whole genome sequencing and genome annotation of Dermacoccus abyssi strain HZAU 226 isolated from spoiled eggs. Genomics 2021, 113, 1199–1206. [Google Scholar] [CrossRef]
- Lim, H.J.; Lee, E.-H.; Yoon, Y.; Chua, B.; Son, A. Portable Lysis Apparatus for Rapid Single-step DNA Extraction of Bacillus Subtilis. J. Appl. Microbiol. 2016, 120, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A Self-Training Method for Prediction of Gene Starts in Microbial Genomes. Implications for Finding Sequence Motifs in Regulatory Regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Kolbe, D.L.; Eddy, S.R. Infernal 1.0: Inference of RNA Alignments. Bioinformatics 2009, 25, 1335–1337. [Google Scholar] [CrossRef]
- Hsiao, W.; Wan, I.; Jones, S.J.; Brinkman, F.S.L. IslandPath: Aiding Detection of Genomic Islands in Prokaryotes. Bioinformatics 2003, 19, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, H.; Mu, G.; Wu, X. Safety evaluation and complete genome analysis emphasis on extracellular polysaccharide of two strains of Limosilactobacillus fermentum MWLf-4 and Lactipiantibacillus plantarum MWLp-12 from human milk. Food Biosci. 2023, 51, 102356. [Google Scholar] [CrossRef]
- De Oliveira, F.L.; Salgaço, M.K.; de Oliveira, M.T.; Mesa, V.; Sartoratto, A.; Peregrino, A.M.; Ramos, W.S.; Sivieri, K. Exploring the potential of Lactobacillus helveticus R0052 and Bifidobacterium longum R0175 as promising psychobiotics Using SHIME. Nutrients 2023, 15, 1521. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Chen, L. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2004, 33, D325–D328. [Google Scholar] [CrossRef]
- Margalho, L.P.; Van Schalkwijk, S.; Bachmann, H.; Sant’Ana, A.S. Enterococcus spp. in Brazilian artisanal cheeses: Occurrence and assessment of phenotypic and safety properties of a large set of strains through the use of high throughput tools combined with multivariate statistics. Food Control 2020, 118, 107425. [Google Scholar] [CrossRef]
- Mulaw, G.; Sisay Tessema, T.; Muleta, D.; Tesfaye, A. In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented Ethiopian food products. Int. J. Microbiol. 2019, 2019, 7179514. [Google Scholar] [CrossRef]
- Chen, M.; Ke, W.; Zhang, J.; Tang, J.; Wang, L.; Ding, W. Ex vivo and in vivo probiotic properties of antioxidant-active lactic acid bacteria from yak yogurt on the Tibetan Plateau. Food Sci. 2017, 38, 178–183. [Google Scholar]
- Topçu, K.C.; Kaya, M.; Kaban, G. Probiotic properties of lactic acid bacteria strains isolated from pastırma. LWT 2020, 134, 110216. [Google Scholar] [CrossRef]
- Sharma, S.; Kandasamy, S.; Kavitake, D.; Shetty, P.H. Probiotic characterization and antioxidant properties of Weissella confusa KR780676, isolated from an Indian fermented food. LWT 2018, 97, 53–60. [Google Scholar] [CrossRef]
- Liu, X.; Lv, X.; Sun, Y.; Liu, C.; Wang, R.; Liu, R.; Ma, Y.; Li, Q. Probiotic properties of Lacticaseibacillus rhamnosus grx10 revolved with complete genome. Food Biosci. 2023, 51, 102219. [Google Scholar] [CrossRef]
- Hinc, K.; Kabała, M.; Iwanicki, A.; Martirosian, G.; Negri, A.; Obuchowski, M. Complete Genome Sequence of the Newly Discovered Temperate Clostridioides Difficile Bacteriophage phiCDKH01 of the Family Siphoviridae. Arch. Virol. 2021, 166, 2305–2310. [Google Scholar] [CrossRef]
- Fontana, A.; Falasconi, I.; Molinari, P.; Treu, L.; Basile, A.; Vezzi, A.; Campanaro, S.; Morelli, L. Genomic comparison of Lactobacillus helveticus strains highlights probiotic potential. Front. Microbiol. 2019, 10, 1380. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.-A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Xia, A.-N.; Meng, X.-S.; Tang, X.-J.; Zhang, Y.-Z.; Lei, S.-M.; Liu, Y.-G. Probiotic and Related Properties of a Novel Lactic Acid Bacteria Strain Isolated from Fermented Rose Jam. LWT 2021, 136, 110327. [Google Scholar] [CrossRef]
- Ouwehand, A.C.; Forssten, S.; Hibberd, A.A.; Lyra, A.; Stahl, B. Probiotic approach to prevent antibiotic resistance. Ann. Med. 2016, 48, 246–255. [Google Scholar] [CrossRef]
- Saroj, D.B.; Gupta, A.K. Genome based safety assessment for Bacillus coagulans strain LBSC (DSM 17654) for probiotic application. Int. J. Food Microbiol. 2020, 318, 108523. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Mao, Y.; Ma, T.; Liu, X.; Cheng, X.; Bai, Y.; Li, S. Screening and genome analysis of lactic acid bacteria with high exopolysaccharide production and good probiotic properties. Food Biosci. 2023, 56, 103211. [Google Scholar] [CrossRef]
- Shangpliang HN, J.; Sharma, S.; Rai, R.; Tamang, J.P. Some Technological Properties of Lactic Acid Bacteria Isolated from Dahi and Datshi, Naturally Fermented Milk Products of Bhutan. Front. Microbiol. 2017, 8, 216942. [Google Scholar] [CrossRef]
- Aslim, B.; Onal, D.; Beyatli, Y. Factors influencing autoaggregation and aggregation of Lactobacillus delbrueckii subsp. Bulgaricus isolated from handmade yogurt. J. Food Prot. 2007, 70, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Montoro, B.P.; Benomar, N.; Lavilla Lerma, L.; Castillo Gutiérrez, S.; Gálvez, A.; Abriouel, H. Fermented aloreña Table Olives as a source of potential probiotic Lactobacillus pentosus Strains. Front. Microbiol. 2016, 7, 219927. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Prajapati, J.B.; Holst, O.; Ljungh, A. Determining probiotic potential of exopolysaccharide producing lactic acid bacteria isolated from vegetables and traditional Indian fermented food products. Food Biosci. 2014, 5, 27–33. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.M.; Bongers, R.S.; de Vos, W.M.; Kleerebezem, M. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl. Environ. Microbiol. 2008, 74, 4719–4726. [Google Scholar] [CrossRef] [PubMed]
- Pfeiler, E.A.; Klaenhammer, T.R. Role of Transporter Proteins in Bile Tolerance of Lactobacillus acidophilus. Appl. Environ. Microbiol. 2009, 75, 6013–6016. [Google Scholar] [CrossRef]
- Ye, K.; Li, P.; Gu, Q. Complete genome sequence analysis of a strain Lactobacillus pentosus ZFM94 and its probiotic characteristics. Genomics 2020, 112, 3142–3149. [Google Scholar] [CrossRef]
- Kommineni, S.; Kristich, C.J.; Salzman, N.H. Harnessing bacteriocin biology as targeted therapy in the GI tract. Gut Microbes 2016, 7, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Joerger, M.C.; Klaenhammer, T.R. Characterization and purification of helveticin J and evidence for a chromosomally determined bacteriocin produced by Lactobacillus helveticus 481. J. Bacteriol. 1986, 167, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.; Nes, I.F.; Holo, H. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2003, 69, 2975–2984. [Google Scholar] [CrossRef] [PubMed]




| Strains | Lactobacillus helveticus | |||
|---|---|---|---|---|
| LH10 | D75 | LH5 | R0052 | |
| Number of scaffolds | 1 | 1 | 3 | 1 |
| Genome size (Mb) | 1.81 | 2.05 | 2.16 | 2.13 |
| GC (%) | 36.6 | 37.03 | 36.92 | 36.80 |
| Coding gene number | 2044 | 2187 | 2193 | 2288 |
| rRNA | 12 | 15 | 12 | 12 |
| tRNA | 63 | 64 | 63 | 61 |
| GenBank | CP149445.1 | CP020029.1 | CP019581.1 | CP003799.1 |
| Category of Drug Sensitivity Test Paper | Name of Drug Sensitivity Test Paper | Judgment Standard of Bacteriostatic Circle Diameter (mm) | Result | ||
|---|---|---|---|---|---|
| R | I | S | |||
| Lincosamide antibiotics | clindamycin | ≤14 | 15–20 | ≥21 | 28 ± 0.7 (S) |
| β Lactam antibiotics | penicillin | ≤19 | 20–23 | ≥24 | 25 ± 0.5 (S) |
| ampicillin | ≤19 | 20–23 | ≥24 | 22 ± 0.3 (I) | |
| cefamezin | ≤19 | 20–22 | ≥23 | /(R) | |
| Tetracyclines antibiotics | tetracycline | ≤14 | 15–18 | ≥19 | 24 ± 0.3 (S) |
| Glycopeptides antibiotics | vancomycin | — | — | ≥15 | /(R) |
| Sulfonamides antibiotics | sulfamethoxazole | ≤10 | 11–15 | ≥16 | 7 ± 0.2 (R) |
| Aminoglycoside antibiotics | gentamicin | ≤12 | 13–14 | ≥15 | 12 ± 0.5 (R) |
| Macrolide antibiotics | erythromycin | ≤13 | 14–22 | ≥23 | 32 ± 0.3 (S) |
| Quinolone antibiotics | ciprofloxacin | ≤15 | 16–20 | ≥21 | 12 ± 0.6 (R) |
| Rifamycin antibiotics | rifampicin | ≤16 | 17–19 | ≥20 | 25 ± 0.4 (S) |
| Miscellaneous agents | chloramphenicol | ≤12 | 13–17 | ≥18 | 22 ± 0.7 (S) |
| Drug Class | Gene Name | Locus_Tag | Definition |
|---|---|---|---|
| Macrolide | macB | LH10_GM001871 | ATP-binding cassette (ABC) transporter that exports macrolides with 14- or 15- membered lactones |
| carA | LH10_GM000399 | efflux pump complex or subunit conferring antibiotic resistance | |
| msr(B) | LH10_GM000751 | efflux pump complex or subunit conferring antibiotic resistance | |
| Glycopeptides | vanHO | LH10_GM000065 | antibiotic resistance gene cluster, cassette, or operon, determinant of resistance to glycopeptide antibiotics, protein(s) conferring antibiotic resistance via molecular bypass |
| vanSI | LH10_GM000102 | complex or subunit conferring antibiotic resistance, protein(s) and two-component regulatory system modulating antibiotic efflux | |
| vanRM | LH10_GM001853 | antibiotic resistance gene cluster, cassette, or operon, determinant of aminoglycoside resistance, determinant of resistance to glycopeptide antibiotics, efflux pump complex or subunit conferring antibiotic resistance | |
| vanB | LH10_GM000161 | a D-Ala-D-Ala ligase homolog that synthesizes D-Ala-D-Lac, an alternative substrate for peptidoglycan synthesis that reduces vancomycin binding affinity. | |
| vanA | LH10_GM000161 | a D-Ala-D-Ala ligase homolog that synthesizes D-Ala-D-Lac, an alternative substrate for peptidoglycan synthesis that reduces vancomycin binding affinity. | |
| vanC | LH10_GM000161 | a D-Ala-D-Ala ligase homolog that synthesizes D-Ala-D-Lac, an alternative substrate for peptidoglycan synthesis that reduces vancomycin binding affinity. | |
| Tetracyclines | Tet(Q) | LH10_GM000061 | antibiotic target protection protein, determinant of tetracycline resistance |
| tetA(60) | LH10_GM000817 | efflux pump complex or subunit conferring antibiotic resistance | |
| tcmA | LH10_GM000470 | efflux pump complex or subunit conferring antibiotic resistance | |
| adeR | LH10_GM001410 | positive regulator of AdeABC efflux system | |
| tetA (58) | LH10_GM000159 | Tetracycline efflux pump | |
| Peptide | arnA | LH10_GM000779 | determinant of polymyxin resistance, gene altering cell wall charge |
| Phosphonic | mdtG | LH10_GM001887 | efflux pump complex or subunit conferring antibiotic resistance |
| Diaminopyrimidine | dfrA26 | LH10_GM001252 | antibiotic target replacement protein, determinant of diaminopyrimidine resistance |
| dfrE | LH10_GM001253 | antibiotic target replacement protein, determinant of diaminopyrimidine resistance | |
| Aminoglycoside | kdpE | LH10_GM000607 | transcriptional activator that is part of the two-component system KdpD/KdpE |
| Lincosamide | lmrD | LH10_GM001591 | efflux pump complex or subunit conferring antibiotic resistance |
| Elfamycin | Scin_EFTu_ELF | LH10_GM001297 | sequence variants of Streptomyces cinnamoneus elongation factor Tu that confer resistance to elfamycin antibiotics |
| Stress Response | Gene Locus | Gene Name | Definition |
|---|---|---|---|
| Temperature | LH10_GM000282 | clpC | ATP-dependent Clp protease ATP-binding subunit ClpC |
| LH10_GM001466 | clpP | ATP-dependent Clp protease, protease subunit | |
| LH10_GM001524 | clpE | ATP-dependent Clp protease ATP-binding subunit ClpE | |
| LH10_GM001967 | clpE | ATP-dependent Clp protease ATP-binding subunit ClpE | |
| LH10_GM001295 | clpX | ATP-dependent Clp protease ATP-binding subunit ClpX | |
| LH10_GM001156 | hslV, clpQ | ATP-dependent HslUV protease, peptidase subunit HslV | |
| LH10_GM001305 | — | Lon-like protease | |
| LH10_GM000217 | HSP20 | HSP20 family protein | |
| LH10_GM000276 | hslO | molecular chaperone Hsp33 | |
| LH10_GM000417 | groES, | HSPE1 chaperonin GroES | |
| LH10_GM000418 | groEL, | HSPD1 chaperonin GroEL | |
| LH10_GM000839 | hrcA | heat-inducible transcriptional repressor | |
| LH10_GM000840 | GRPE | molecular chaperone GrpE | |
| LH10_GM000841 | dnaK, | HSPA9 molecular chaperone DnaK | |
| LH10_GM000842 | dnaJ | molecular chaperone DnaJ | |
| LH10_GM001327 | cspA | cold shock protein (beta-ribbon, CspA family) | |
| LH10_GM001618 | cspA | cold shock protein (beta-ribbon, CspA family) | |
| LH10_GM000122 | htpX | heat shock protein HtpX | |
| Acid | LH10_GM001377 | ATPF1E, atpC | F-type H+-transporting ATPase subunit epsilon |
| LH10_GM001378 | ATPF1B, atpD | F-type H+-transporting ATPase subunit beta | |
| LH10_GM001379 | ATPF1G, atpG | F-type H+-transporting ATPase subunit gamma | |
| LH10_GM001380 | ATPF1A, atpA | F-type H+-transporting ATPase subunit alpha | |
| LH10_GM001381 | ATPF1D, atpH | F-type H+-transporting ATPase subunit delta | |
| LH10_GM001382 | ATPF0B, atpF | F-type H+-transporting ATPase subunit b | |
| LH10_GM001383 | ATPF0C, atpE | F-type H+-transporting ATPase subunit c | |
| LH10_GM001384 | ATPF0A, atpB | F-type H+-transporting ATPase subunit a | |
| LH10_GM001621 | nhaC | Na+:H+ antiporter, NhaC family Alkaline LH10_GM001206 aspS aspartyl-tRNA synthetase | |
| Bile salt | LH10_GM000642 | lmrB | MFS transporter, DHA2 family, lincomycin resistance protein |
| LH10_GM001601 | mdtG | MFS transporter, DHA1 family, multidrug resistance protein | |
| LH10_GM001887 | mdtG | MFS transporter, DHA1 family, multidrug resistance protein | |
| LH10_GM002020 | pbuG | putative MFS transporter, AGZA family, xanthine/uracil permease | |
| LH10_GM002024 | pbuG | putative MFS transporter, AGZA family, xanthine/uracil permease | |
| LH10_GM002029 | pbuG | putative MFS transporter, AGZA family, xanthine/uracil permease | |
| LH10_GM002035 | yaaU | MFS transporter, putative metabolite transport protein | |
| LH10_GM000027 | ABC.X2.A | putative ABC transport system ATP-binding protein | |
| LH10_GM000028 | ABC.X2.P | putative ABC transport system permease protein | |
| LH10_GM000052 | ABC.X4.A | putative ABC transport system ATP-binding protein | |
| LH10_GM000053 | ABC.X4.P | putative ABC transport system permease protein | |
| LH10_GM000054 | ABC.X4.S | putative ABC transport system substrate-binding protein | |
| LH10_GM001761 | ABC.CD.P | putative ABC transport system permease protein | |
| LH10_GM001762 | ABC.CD.A | putative ABC transport system ATP-binding protein | |
| LH10_GM001871 | ABC.CD.A | putative ABC transport system ATP-binding protein | |
| LH10_GM001872 | ABC.CD.P | putative ABC transport system permease protein | |
| LH10_GM001970 | ABC.CD.A | putative ABC transport system ATP-binding protein | |
| LH10_GM001972 | ABC.CD.P | putative ABC transport system permease protein | |
| LH10_GM002007 LH10_GM001256 | ABC.CD.A Bsh | putative ABC transport system ATP-binding protein bile salt hydrolase | |
| Osmotic stress | LH10_GM000804 | TC.APA | basic amino acid/polyamine antiporter, APA family |
| Oxidative stress | LH10_GM001866 | nfr2 | flavin reductase (NADH) subunit 2 |
| LH10_GM000437 | trxA | thioredoxin 1 | |
| LH10_GM000492 | trxA | thioredoxin 1 | |
| LH10_GM000494 | trxB, | TRR thioredoxin reductase (NADPH) | |
| LH10_GM001520 | spxA | regulatory protein spx |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Y.; Xu, J.; Li, J.; Wu, R. Evaluation of Probiotic Properties and Safety of Lactobacillus helveticus LH10 Derived from Vinegar through Comprehensive Analysis of Genotype and Phenotype. Microorganisms 2024, 12, 831. https://doi.org/10.3390/microorganisms12040831
Du Y, Xu J, Li J, Wu R. Evaluation of Probiotic Properties and Safety of Lactobacillus helveticus LH10 Derived from Vinegar through Comprehensive Analysis of Genotype and Phenotype. Microorganisms. 2024; 12(4):831. https://doi.org/10.3390/microorganisms12040831
Chicago/Turabian StyleDu, Yang, Jingru Xu, Jinquan Li, and Renwei Wu. 2024. "Evaluation of Probiotic Properties and Safety of Lactobacillus helveticus LH10 Derived from Vinegar through Comprehensive Analysis of Genotype and Phenotype" Microorganisms 12, no. 4: 831. https://doi.org/10.3390/microorganisms12040831
APA StyleDu, Y., Xu, J., Li, J., & Wu, R. (2024). Evaluation of Probiotic Properties and Safety of Lactobacillus helveticus LH10 Derived from Vinegar through Comprehensive Analysis of Genotype and Phenotype. Microorganisms, 12(4), 831. https://doi.org/10.3390/microorganisms12040831





