Molecular Analysis of Tick-Borne Bacterial Pathogens from Ticks Infesting Animal Hosts in Kyrgyzstan, 2021
Abstract
:1. Introduction
2. Materials & Methods
2.1. Study Area and Tick Collection
2.2. DNA Extraction
2.3. Molecular Identification of Tick Species and Tick-Borne Pathogens
2.4. Nucleotide Sequencing and Phylogenetic Analysis
2.5. Statistical Analysis
3. Result
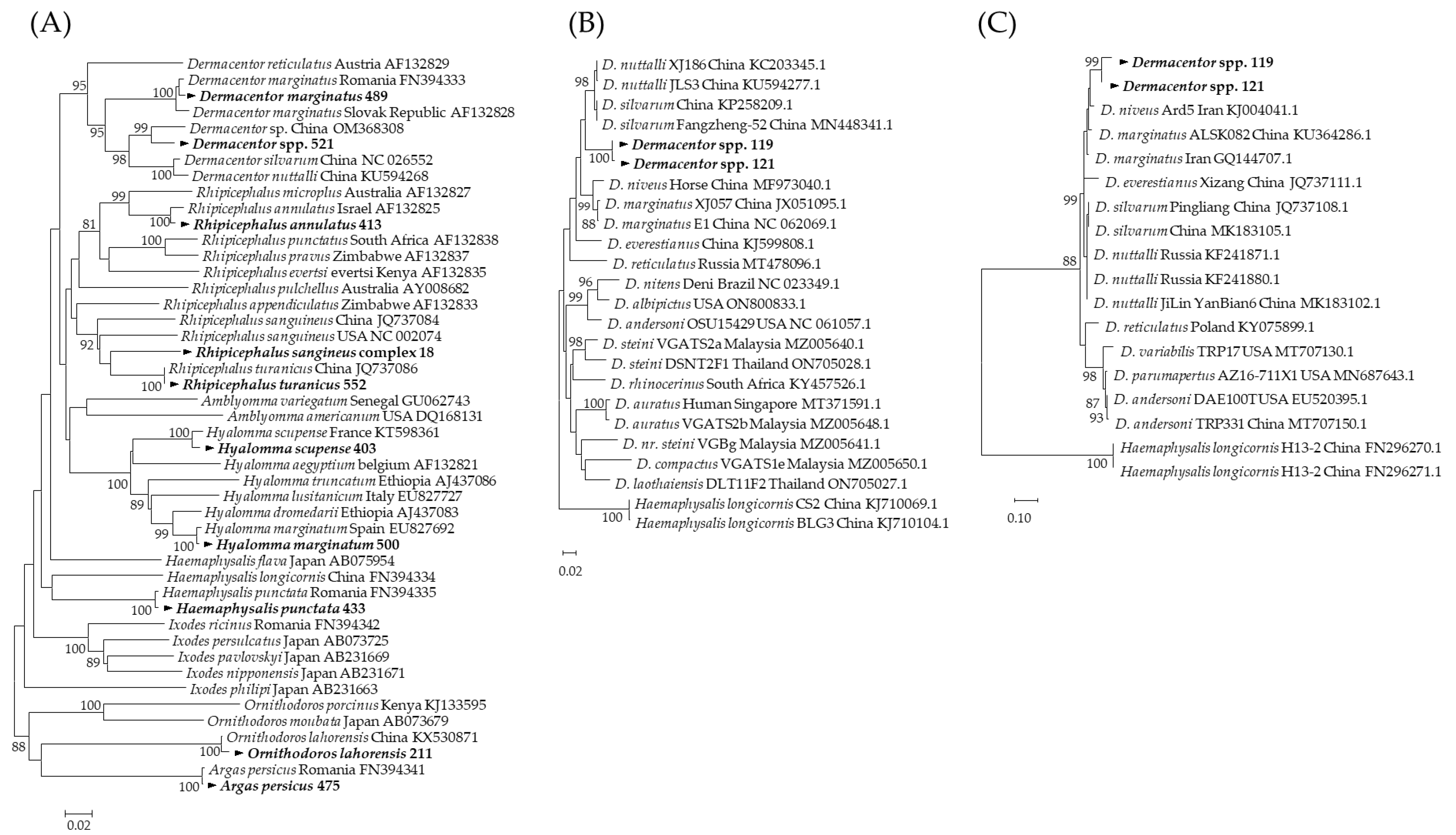
3.1. Molecular Identification of Tick Species
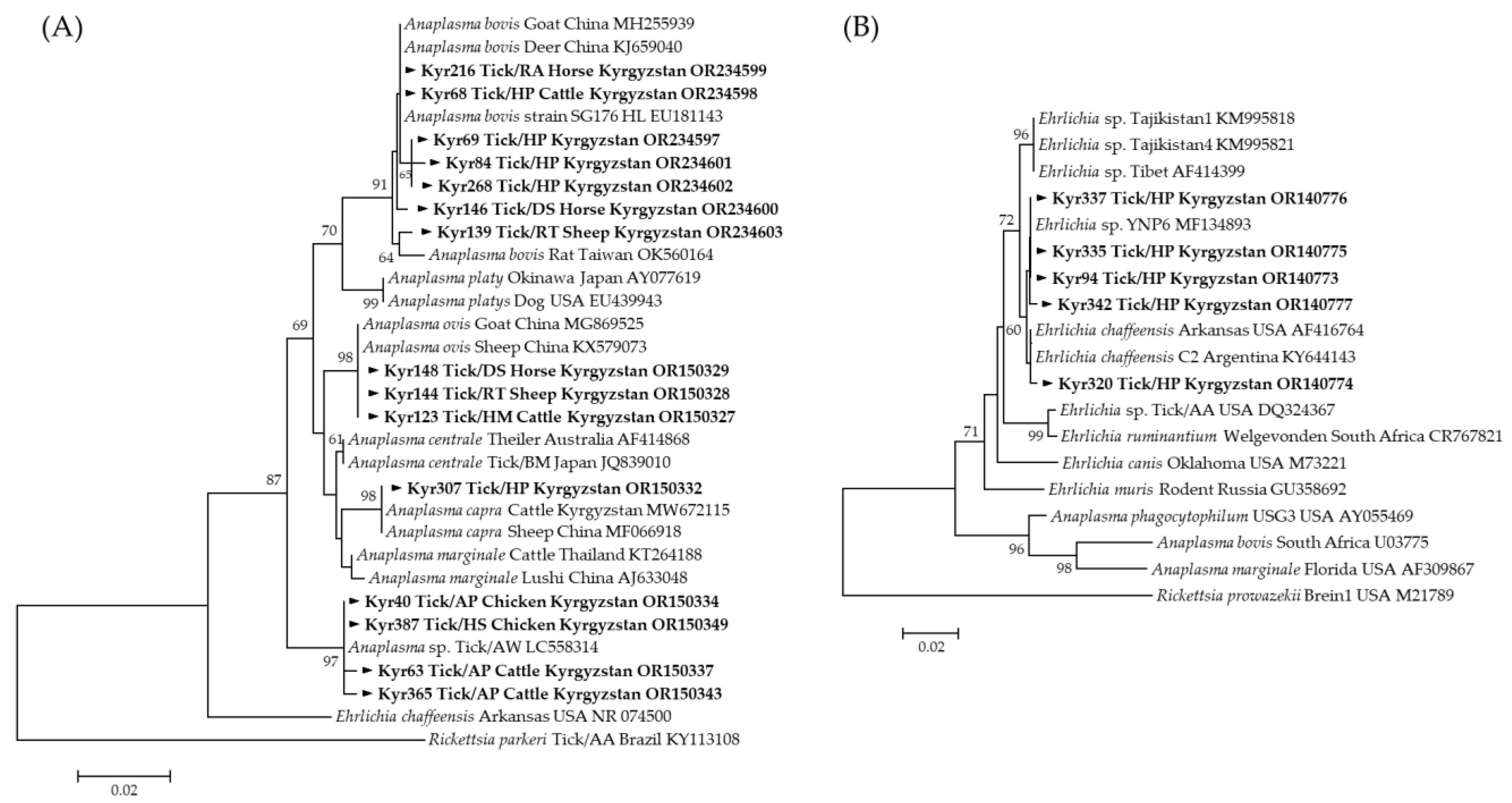
3.2. Detection of Tick-Borne Pathogens in Ticks
3.3. Molecular and Phylogenetic Analysis of Tick-Borne Pathogens
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhumanova, M. A Study on Livestock and Land Management in Kyrgyzstan; FAO: Rome, Italy, 2011. [Google Scholar]
- Kydyshov, K.; Usenbaev, N.; Sharshenbekov, A.; Aitkuluev, N.; Abdyraev, M.; Chegirov, S.; Kazybaeva, J.; Brangsch, H.; Melzer, F.; Neubauer, H.; et al. Brucellosis in humans and animals in Kyrgyzstan. Microorganisms 2022, 10, 1293. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, Y.; Zhang, H.; Su, Y.; Wu, X.; Yan, S.; Yang, S. Impact of socio-economic and environmental factors on livestock production in Kyrgyzstan. Front. Environ. Sci. 2022, 10, 1049187. [Google Scholar] [CrossRef]
- Sagynbekova, L. Environment, rural livelihoods, and labor migration: A case study in central Kyrgyzstan. Mt. Res. Dev. 2017, 37, 456–463. [Google Scholar] [CrossRef]
- Desta, B. Review on the impact of ticks on livestock health and productivity. J. Biogr. Agric. Health 2016, 6, 1–7. [Google Scholar]
- Hurtado, O.J.B.; Giraldo-Ríos, C. Economic and health impact of the ticks in production animals. In Ticks and Tick-Borne Pathogens; Abubakar, M., Perera, P.K., Eds.; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- Aktas, M.; Özübek, S.; Altay, K.; Ipek, N.D.; Balkaya, İ.; Utuk, A.E.; Kırbas, A.; Şimsek, S.; Dumanlı, N. Molecular detection of tick-borne rickettsial and protozoan pathogens in domestic dogs from Turkey. Parasites Vectors 2015, 8, 157. [Google Scholar] [CrossRef]
- Seo, J.Y.; Kim, Y.J.; Kim, S.Y.; Lee, H.I. Molecular detection of Anaplasma, Ehrlichia and Rickettsia pathogens in ticks collected from humans in the Republic of Korea, 2021. Pathogens 2023, 12, 802. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; He, B.; Huang, S.Y.; Wei, F.; Zhu, X.Q. Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. Lancet Infect. Dis. 2014, 14, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Briggs, B.J.; Atkinson, B.; Czechowski, D.M.; Larsen, P.A.; Meeks, H.N.; Carrera, J.P.; Duplechin, R.M.; Hewson, R.; Junushov, A.T.; Gavrilova, O.N.; et al. Tick-borne encephalitis virus, Kyrgyzstan. Emerg. Infect. Dis. 2011, 17, 876–879. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Choi, C.H.; Lee, M.; Kim, S.Y.; Aknazarov, B.; Nyrgaziev, R.; Atabekova, N.; Jetigenov, E.; Chung, Y.S.; Lee, H.I. Molecular detection and phylogenetic analysis of tick-borne encephalitis virus from ticks collected from cattle in Kyrgyzstan, 2023. Viruses 2024, 16, 107. [Google Scholar] [CrossRef]
- de la Fuente, J.; Estrada-Pena, A.; Venzal, J.M.; Kocan, K.M.; Sonenshine, D.E. Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front. Biosci. 2008, 13, 6938–6946. [Google Scholar] [CrossRef]
- de la Fuente, J.; Antunes, S.; Bonnet, S.; Cabezas-Cruz, A.; Domingos, A.G.; Estrada-Peña, A.; Johnson, N.; Kocan, K.M.; Mansfield, K.L.; Nijhof, A.M.; et al. Tick-pathogen interactions and vector competence: Identification of molecular drivers for tick-borne diseases. Front. Cell. Infect. Microbiol. 2017, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Kumar, S.; Sharma, A.K.; Jacob, S.S.; RamVerma, M.; Singh, N.K.; Shakya, M.; Sankar, M.; Ghosh, S. Economic impact of predominant ticks and tick-borne diseases on Indian dairy production systems. Exp. Parasitol. 2022, 243, 108408. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, C.; Dibernardo, A.; Koffi, J.; Wood, H.; Leighton, P.A.; Lindsay, L.R. N Increased risk of tick-borne diseases with climate and environmental changes. Can. Commun. Dis. Rep. 2019, 45, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Hussain, A.; Ho, J.; Li, J.; George, D.; Rehman, A.; Zeb, J.; Sparagano, O. An epidemiological survey regarding ticks and tick-borne diseases among livestock owners in Punjab, Pakistan: A one health context. Pathogens 2021, 10, 361. [Google Scholar] [CrossRef] [PubMed]
- Paguem, A.; Manchang, K.; Kamtsap, P.; Renz, A.; Schaper, S.; Dobler, G.; Bakkes, D.K.; Chitimia-Dobler, L. Ticks and Rickettsiae associated with wild animals sold in bush meat markets in Cameroon. Pathogens 2023, 12, 348. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Lv, J.; Wu, S.; Zhang, Y.; Chen, Y.; Feng, C.; Yuan, X.; Jia, G.; Deng, J.; Wang, C.; Wang, Q.; et al. Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida). Parasites Vectors 2014, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Abouelhassan, E.M.; El-Gawady, H.M.; Abdel-Aal, A.A.; El-Gayar, A.K.; Esteve-Gassent, M.D. Comparison of some molecular markers for tick species identification. J. Arthropod Borne Dis. 2019, 13, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, S.Z. Ixodidae ticks in Bishkek. Med. Parazitol. 2005, 4, 34–37. [Google Scholar]
- Aknazarov, B.; Jetigenov, E.; Atabekova, N.; Suerkulov, U.; Abdumanap, N. Spread of arthropod-borne infections in Kyrgyzstan. In E3S Web of Conferences; EDP Sciences: Les Ulis, France, 2023. [Google Scholar] [CrossRef]
- Orkun, Ö.; Karaer, Z.; Çakmak, A.; Nalbantoğlu, S. Identification of tick-borne pathogens in ticks feeding on humans in Turkey. PLoS Negl. Trop. Dis. 2014, 8, e3067. [Google Scholar] [CrossRef]
- Perveen, N.; Muzaffar, S.B.; Al-Deeb, M.A. Ticks and tick-borne diseases of livestock in the Middle East and North Africa: A review. Insects 2021, 12, 83. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Pan, Y.S.; Jiang, B.G.; Ye, R.Z.; Chang, Q.C.; Shao, H.Z.; Cui, X.M.; Xu, D.L.; Li, L.F.; Wei, W.; et al. Prevalence of multiple tick-borne pathogens in various tick vectors in Northeastern China. Vector Borne Zoonotic Dis. 2021, 21, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Jongejan, F.; Uilenberg, G. The global importance of ticks. Parasitology 2004, 129, S3–S14. [Google Scholar] [CrossRef] [PubMed]
- Aktaş, M.; Kısadere, İ.; Özübek, S.; Cihan, H.; Salıkov, R.; Cirak, V.Y. First molecular survey of piroplasm species in cattle from Kyrgyzstan. Parasitol. Res. 2019, 118, 2431–2435. [Google Scholar] [CrossRef] [PubMed]
- Ozubek, S.; Ulucesme, M.C.; Cirak, V.Y.; Aktas, M. Detection of Theileria orientalis genotypes from cattle in Kyrgyzstan. Pathogens 2022, 11, 1185. [Google Scholar] [CrossRef] [PubMed]
- Altay, K.; Erol, U.; Sahin, O.F.; Aytmirzakizi, A. First molecular detection of Anaplasma species in cattle from Kyrgyzstan; molecular identification of human pathogenic novel genotype Anaplasma capra and Anaplasma phagocytophilum related strain. Ticks Tick Borne Dis. 2022, 13, 101861. [Google Scholar] [CrossRef] [PubMed]
- Zhyldyz, A.; Aitakin, K.; Atabek, B.; Elmurat, J.; Rysbek, N.; Jailobek, O.; Ahedor, B.; Otgonsuren, D.; Mumbi, N.N.M.; Guswanto, A.; et al. An epidemiological survey of vector-borne pathogens infecting cattle in Kyrgyzstan. Parasitol. Int. 2023, 97, 102791. [Google Scholar] [CrossRef] [PubMed]
- Altay, K.; Erol, U.; Sahin, O.F.; Aytmirzakizi, A.; Temizel, E.M.; Aydin, M.F.; Dumanli, N.; Aktas, M. The detection and phylogenetic analysis of Anaplasma phagocytophilum-like 1, A. ovis and A. capra in sheep: A. capra divides into two genogroups. Vet. Res. Commun. 2022, 46, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Altay, K.; Erol, U.; Sahin, O.F.; Aydin, M.F.; Aytmirzakizi, A.; Dumanli, N. First molecular evidence of Babesia vogeli, Babesia vulpes, and Theileria ovis in dogs from Kyrgyzstan. Pathogens 2023, 12, 1046. [Google Scholar] [CrossRef]
- Lv, J.; Wu, S.; Zhang, Y.; Zhang, T.; Feng, C.; Jia, G.; Lin, X. Development of a DNA barcoding system for the Ixodida (Acari: Ixodida). Mitochondrial DNA 2014, 25, 142–149. [Google Scholar] [CrossRef]
- Wang, F.; Wang, D.; Guo, G.; Hu, Y.; Wei, J.; Liu, J. Species delimitation of the Dermacentor ticks based on phylogenetic clustering and niche modeling. PeerJ 2019, 7, e6911. [Google Scholar] [CrossRef] [PubMed]
- Barlough, J.E.; Madigan, J.E.; DeRock, E.; Bigornia, L. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus). Vet. Parasitol. 1996, 63, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Moon, B.; Bae, B.K.; Shin, E.; Ko, Y.H.; Kim, Y.; Park, Y.H.; Chae, J. Genetic identification and phylogenetic analysis of Anaplasma and Ehrlichia species in Haemaphysalis longicornis collected from Jeju Island, Korea. J. Bacteriol. Virol. 2009, 39, 257–267. [Google Scholar] [CrossRef]
- Lee, S.O.; Na, D.K.; Kim, C.M.; Li, Y.H.; Cho, Y.H.; Park, J.H.; Lee, J.H.; Eo, S.K.; Klein, T.A.; Chae, J.S. Identification and prevalence of Ehrlichia chaffeensis infection in Haemaphysalis longicornis ticks from Korea by PCR, sequencing and phylogenetic analysis based on 16S rRNA gene. J. Vet. Sci. 2005, 6, 151–155. [Google Scholar] [CrossRef]
- Coimbra-Dores, M.J.; Nunes, T.; Dias, D.; Rosa, F. Rhipicephalus sanguineus (Acari: Ixodidae) species complex: Morphometric and ultrastructural analyses. Exp. Appl. Acarol. 2016, 70, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Nava, S.; Estrada-Peña, A.; Petney, T.; Beati, L.; Labruna, M.B.; Szabó, M.P.; Venzal, J.M.; Mastropaolo, M.; Mangold, A.J.; Guglielmone, A.A. The taxonomic status of Rhipicephalus sanguineus (Latreille, 1806). Vet. Parasitol. 2015, 208, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Hekimoğlu, O.; Sağlam, İ.K.; Özer, N.; Estrada-Peña, A. New molecular data shed light on the global phylogeny and species limits of the Rhipicephalus sanguineus complex. Ticks Tick-Borne Dis. 2016, 7, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Camicas, J.L.; Hervy, J.P.; Adam, F.; Morel, P.C. The Ticks of the World (Acarida, Ixodida): Nomenclature, Described Stages, Hosts, Distribution; Orstom Editions: Paris, Italy, 1998. [Google Scholar]
- Gray, J.; Dantas-Torres, F.; Estrada-Peña, A.; Levin, M. Systematics and ecology of the brown dog tick, Rhipicephalus sanguineus. Ticks Tick-Borne Dis. 2013, 4, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.B.; Keirans, J.E.; Horak, I.G. The Genus Rhipicephalus (Acari, Ixodidae): A Guide to the Brown Ticks of the World; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Caetano, R.L.; Vizzoni, V.F.; Bitencourth, K.; Carriço, C.; Sato, T.P.; Pinto, Z.T.; De Oliveira, S.V.; Amorim, M.; Voloch, C.M.; Gazeta, G.S. Ultrastructural morphology and molecular analyses of tropical and temperate ‘species’ of Rhipicephalus sanguineus sensu lato (Acari: Ixodidae) in Brazil. J. Med. Entomol. 2017, 54, 1201–1212. [Google Scholar] [CrossRef] [PubMed]
- Sang, C.; Yang, M.; Xu, B.; Liu, G.; Yang, Y.; Kairullayev, K.; Bauyrzhan, O.; Hazihan, W.; Hornok, S.; Wang, Y. Tick distribution and detection of Babesia and Theileria species in Eastern and Southern Kazakhstan. Ticks Tick-Borne Dis. 2021, 12, 101817. [Google Scholar] [CrossRef]
- Kartashov, M.Y.; Kononova, Y.V.; Petrova, I.D.; Tupota, N.L.; Mikryukova, T.P.; Ternovoi, V.A.; Tishkova, F.H.; Loktev, V.B. Detection of Ehrlichia spp. and Theileria spp. in Hyalomma anatolicum ticks collected in Tajikistan. Vavilovskii Zhurnal Genet. Selektsii 2020, 24, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Tajedin, L.; Bakhshi, H.; Faghihi, F.; Telmadarraiy, Z. High infection of Anaplasma and Ehrlichia spp. among tick species collected from different geographical locations of Iran. Asian Pac. J. Trop. Dis. 2016, 6, 787–792. [Google Scholar] [CrossRef]
- Bowman, D.D. Introduction to the alpha-proteobacteria: Wolbachia and Bartonella, Rickettsia, Brucella, Ehrlichia, and Anaplasma. Top. Companion Anim. Med. 2011, 26, 173–177. [Google Scholar] [CrossRef]
- Dumler, J.S. Anaplasma and Ehrlichia infection. Ann. N. Y. Acad. Sci. 2005, 1063, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Palomar, A.M.; Portillo, A.; Santibáñez, P.; Mazuelas, D.; Roncero, L.; García-Álvarez, L.; Santibáñez, S.; Gutiérrez, Ó.; Oteo, J.A. Detection of tick-borne Anaplasma bovis, Anaplasma phagocytophilum and Anaplasma centrale in Spain. Med. Vet. Entomol. 2015, 29, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Belkahia, H.; Ben Said, M.; Ghribi, R.; Selmi, R.; Ben Asker, A.; Yahiaoui, M.; Bousrih, M.; Daaloul-Jedidi, M.; Messadi, L. Molecular detection, genotyping and phylogeny of Anaplasma spp. in Rhipicephalus ticks from Tunisia. Acta Trop. 2019, 191, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zheng, Y.C.; Ma, L.; Jia, N.; Jiang, B.G.; Jiang, R.R.; Huo, Q.B.; Wang, Y.W.; Liu, H.B.; Chu, Y.L.; et al. Human infection with a novel tick-borne Anaplasma species in China: A surveillance study. Lancet Infect. Dis. 2015, 15, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Tomassone, L.; Nuñez, P.; Gürtler, R.E.; Ceballos, L.A.; Orozco, M.M.; Kitron, U.D.; Farber, M. Molecular detection of Ehrlichia chaffeensis in Amblyomma parvum ticks, Argentina. Emerg. Infect. Dis. 2008, 14, 1953–1955. [Google Scholar] [CrossRef] [PubMed]
- Sparagano, O.A.E.; Allsopp, M.; Mank, R.; Rijpkema, S.; Figueroa, J.; Jongejan, F. Molecular detection of pathogen DNA in ticks (Acari: Ixodidae): A review. Exp. Appl. Acarol. 1999, 23, 929–960. [Google Scholar] [CrossRef]

| Target | Primers | Sequence (5′ to 3′) | Amplicon Length (bp) | References | |
|---|---|---|---|---|---|
| CO1 | LCO1490 HC02198 | GGTCAACAAATCATAAAGATATTGG TAAACTTCAGGGTGACCAAAAAATCA | 710 | [18] | |
| 16S rRNA | 16S_F 16S_R2 | TTAAATTGCTGTRGTATT CAACATCGAGGTCGCAAWCYA | 395 | [33] | |
| ITS2 | F3/1 R1/1 | GGGTCGATGAAGAACGCAGCCAGC TTCAGGGGGTTGTCTCGCCTGATG | ~1039 | [34] | |
| Anaplasma 16S rRNA | AE1-F AE1-R | 1st PCR | AAGCTTAACACATGCAAGTCGAA AGTCACTGACCCAACCTTAAATG | 1406 | [35] |
| EE3 EE4 | 2nd PCR | GTCGAACGGATTATTCTTTATAGCTTGC CCCTTCCGTTAAGAAGGATCTAATCTCC | 926 | ||
| Ehrlichia 16S rRNA | AE1-F AE1-R | 1st PCR | AAGCTTAACACATGCAAGTCGAA AGTCACTGACCCAACCTTAAATG | 1406 | [36] |
| HE1 HE3 | 2nd PCR | CAATTGCTTATAACCTTTTGGTTATAAAT TATAGGTACCGTCATTATCTTCCCTAT | 390 | [37] | |
| Host | Tick Species | Number of Tested Ticks | Detected Pathogens in Ticks | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A. bovis | A. capra | A. ovis | Anaplasma spp. | E. chaffeensis | Ehrlichia spp. | Total (%) | |||
| Cat | Rhipicephalus turanicus | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Subtotal | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0) | |
| Cattle | Argas persicus | 41 | 0 | 0 | 0 | 9 | 0 | 0 | 9 |
| Dermacentor marginatus | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dermacentor spp. | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Haemaphysalis punctata | 21 | 10 (1 *) | 0 | 0 | 0 | 1 (1 †) | 1 | 12 | |
| Hyalomma marginatum | 9 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| Hyalomma scupense | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Rhipicephalus annulatus | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 122 | 10 | 0 | 1 | 9 | 1 | 1 | 22 (18.0) | |
| Chicken | Argas persicus | 82 | 0 | 0 | 0 | 9 | 0 | 0 | 9 |
| Haemaphysalis punctata | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Hyalomma scupense | 19 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Ornithodoros lahorensis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 104 | 0 | 0 | 0 | 10 | 0 | 0 | 10 (9.6) | |
| Dog | Rhipicephalus annulatus | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rhipicephalus sangineus complex | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Rhipicephalus turanicus | 29 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | |
| Subtotal | 34 | 2 | 0 | 0 | 0 | 0 | 0 | 2 (5.9) | |
| Horse | Dermacentor spp. | 6 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| Haemaphysalis punctata | 20 | 1 | 1 (1 **) | 0 | 0 | 0 | 3 (1 ††) | 5 | |
| Rhipicephalus annulatus | 45 | 7 | 0 | 0 | 0 | 0 | 0 | 7 | |
| Subtotal | 71 | 9 | 1 | 1 | 0 | 0 | 3 | 14 (19.7) | |
| Sheep | Argas persicus | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Dermacentor marginatus | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dermacentor spp. | 61 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Haemaphysalis punctata | 46 | 22 | 1 | 0 | 0 | 0 | 0 | 23 | |
| Hyalomma marginatum | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Hyalomma scupense | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Rhipicephalus sangineus complex | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Rhipicephalus turanicus | 23 | 1 | 0 | 3 | 0 | 0 | 0 | 4 | |
| Subtotal | 161 | 23 | 1 | 3 | 1 | 0 | 0 | 28 (17.4) | |
| Total (%) | Argas persicus | 131 | 0 | 0 | 0 | 19 | 0 | 0 | 19 (14.5) |
| Dermacentor marginatus | 38 | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0.0) | |
| Dermacentor spp. | 79 | 1 | 0 | 1 | 0 | 0 | 0 | 2 (2.5) | |
| Haemaphysalis punctata | 89 | 33 | 2 | 0 | 0 | 1 | 4 | 40 (44.9) | |
| Hyalomma marginatum | 19 | 0 | 0 | 1 | 0 | 0 | 0 | 1 (5.3) | |
| Hyalomma scupense | 22 | 0 | 0 | 0 | 1 | 0 | 0 | 1 (4.5) | |
| Ornithodoros lahorensis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0.0) | |
| Rhipicephalus annulatus | 58 | 7 | 0 | 0 | 0 | 0 | 0 | 7 (12.1) | |
| Rhipicephalus sangineus complex | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0.0) | |
| Rhipicephalus turanicus | 54 | 3 | 0 | 3 | 0 | 0 | 0 | 6 (11.1) | |
| Total (%) | 494 | 44 (8.9) (CI 0.06–0.12) | 2 (0.4) (CI 0.0–0.01) | 5 (1.0) (CI 0.10–0.17) | 20 (4.0) (CI 0.02–0.06) | 1 (0.2) (CI 0.00–0.01) | 4 (0.8) (CI 0.00–0.02) | 76 (15.3) (CI 0.12–0.19) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.J.; Seo, J.Y.; Park, J.S.; Kim, S.Y.; Aknazarov, B.; Atabekova, N.; Lee, H.I. Molecular Analysis of Tick-Borne Bacterial Pathogens from Ticks Infesting Animal Hosts in Kyrgyzstan, 2021. Microorganisms 2024, 12, 1046. https://doi.org/10.3390/microorganisms12061046
Kim YJ, Seo JY, Park JS, Kim SY, Aknazarov B, Atabekova N, Lee HI. Molecular Analysis of Tick-Borne Bacterial Pathogens from Ticks Infesting Animal Hosts in Kyrgyzstan, 2021. Microorganisms. 2024; 12(6):1046. https://doi.org/10.3390/microorganisms12061046
Chicago/Turabian StyleKim, Yu Jung, Ji Ye Seo, Jin Seo Park, Seong Yoon Kim, Bekbolsun Aknazarov, Nurzina Atabekova, and Hee Il Lee. 2024. "Molecular Analysis of Tick-Borne Bacterial Pathogens from Ticks Infesting Animal Hosts in Kyrgyzstan, 2021" Microorganisms 12, no. 6: 1046. https://doi.org/10.3390/microorganisms12061046
APA StyleKim, Y. J., Seo, J. Y., Park, J. S., Kim, S. Y., Aknazarov, B., Atabekova, N., & Lee, H. I. (2024). Molecular Analysis of Tick-Borne Bacterial Pathogens from Ticks Infesting Animal Hosts in Kyrgyzstan, 2021. Microorganisms, 12(6), 1046. https://doi.org/10.3390/microorganisms12061046





