Development of a Plant-Expressed Subunit Vaccine against Brucellosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Constructs
2.2. Agrobacterium-Mediated Infiltration of N. benthamiana Plants
2.3. Protein Extraction and Small-Scale CLP Purification
2.4. Protein Confirmation Using LC-MS/MS-Based Peptide Sequencing
2.5. Transmission Electron Microscopy (TEM)
2.6. Antigen Preparation for Animal Trials
2.7. Efficacy Trials in Mice
2.8. Cellular Immune Response Assays
2.8.1. Spleen Homogenization
2.8.2. Interferon-Gamma Cytokine Secretion Assay
2.8.3. Flow Cytometry
2.8.4. Spleen Counts
3. Results
3.1. Expression and Assembly of Chimeric Orbivirus CLPs in Plants
3.2. Safety of Plant-Expressed Orbivirus CLPs in BALB/c Mice
3.3. Protective Efficacy of Plant-Expressed Orbivirus CLPs in BALB/c Mice
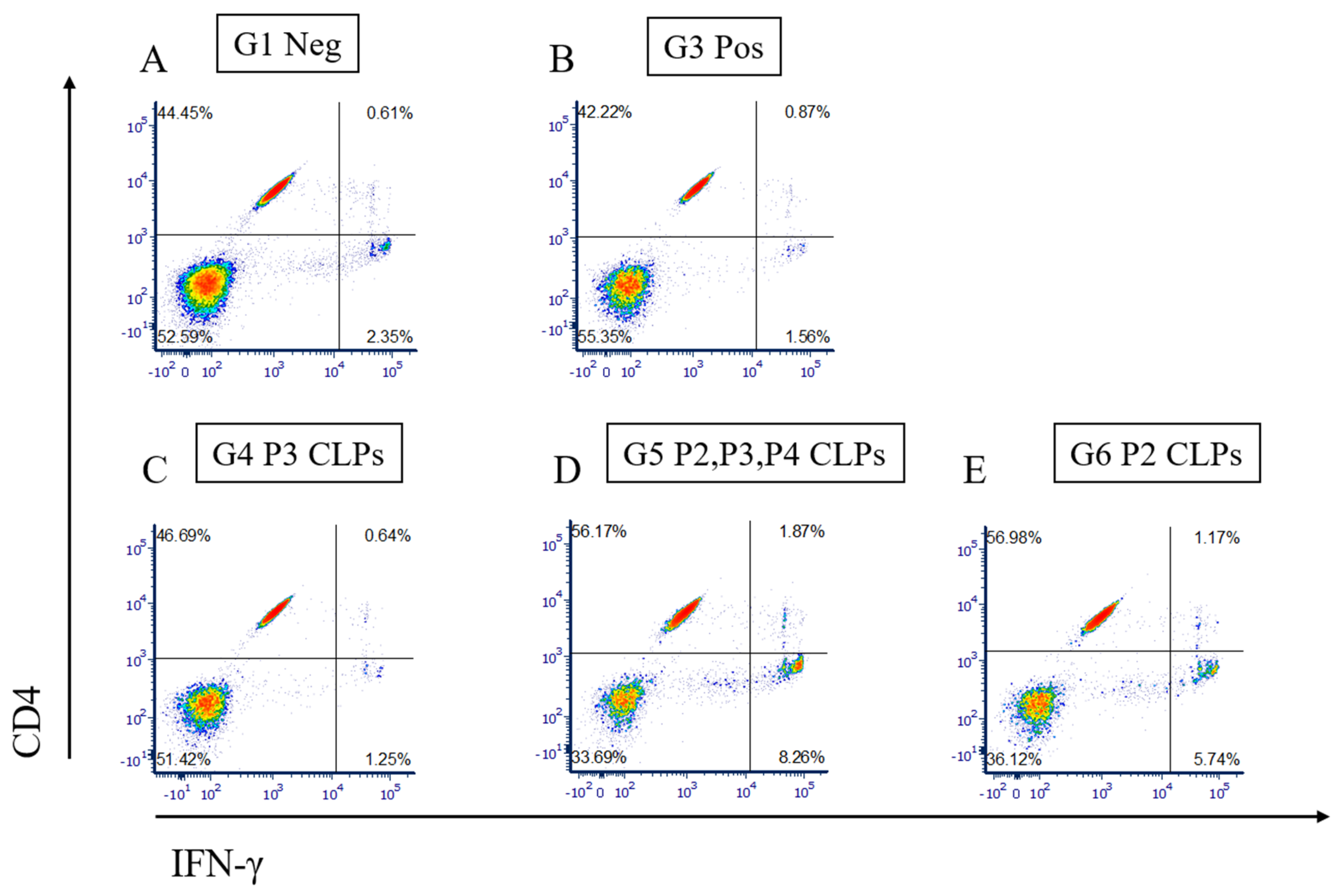
3.4. Measurement of Cellular Immune Response by Flow Cytometry and Cytokine Secretion Assay
3.5. Bacterial Spleen Count Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whatmore, A.M. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 2009, 9, 1168–1184. [Google Scholar] [CrossRef] [PubMed]
- Glynn, M.K.; Lynn, T.V. Brucellosis. J. Am. Vet. Med. Assoc. 2008, 233, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Pappas, G.; Papadimitriou, P.; Akritidis, N.; Christou, L.; Tsianos, E.V. The new global map of human brucellosis. Lancet Infect. Dis. 2006, 6, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Franco, M.P.; Mulder, M.; Gilman, R.H.; Smits, H.L. Human brucellosis. Lancet Infect. Dis. 2007, 7, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, Z.I.; Pascual, D.W. Brucellosis vaccines for livestock. Vet. Immunol. Immunopathol. 2016, 181, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J.; Savioli, L.; Fenwick, A. Neglected tropical diseases of the middle east and north africa: Review of their prevalence, distribution, and opportunities for control. PLoS Neglected Trop. Dis. 2012, 6, e1475. [Google Scholar] [CrossRef]
- Perkins, S.D.; Smither, S.J.; Atkins, H.S. Towards a Brucella vaccine for humans. FEMS Microbiol. Rev. 2010, 34, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Fuenmayor, J.; Gòdia, F.; Cervera, L. Production of virus-like particles for vaccines. New Biotechnol. 2017, 39, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Rutkowska, D.A.; Mokoena, N.B.; Tsekoa, T.L.; Dibakwane, V.S.; O’kennedy, M.M. Plant-produced chimeric virus-like particles—A new generation vaccine against African horse sickness. BMC Veter. Res. 2019, 15, 432. [Google Scholar] [CrossRef]
- Chackerian, B. Virus-like particles: Flexible platforms for vaccine development. Expert Rev. Vaccines 2007, 6, 381–390. [Google Scholar] [CrossRef]
- Grgacic, E.V.; Anderson, D.A. Virus-like particles: Passport to immune recognition. Methods 2006, 40, 60–65. [Google Scholar] [CrossRef]
- Frey, S.; Castro, A.; Arsiwala, A.; Kane, R.S. Bionanotechnology for vaccine design. Curr. Opin. Biotechnol. 2018, 52, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Moreno, N.; Mena, I.; Angulo, I.; Gómez, Y.; Crisci, E.; Montoya, M.; Castón, J.R.; Blanco, E.; Bárcena, J. Rabbit hemorrhagic disease virus capsid, a versatile platform for foreign B-cell epitope display inducing protective humoral immune responses. Sci. Rep. 2016, 6, 31844. [Google Scholar] [CrossRef] [PubMed]
- Pumpens, P.; Grens, E. The true story and advantages of the famous Hepatitis B virus core particles: Outlook 2016. Mol. Biol. 2016, 50, 489–509. [Google Scholar] [CrossRef]
- Adler, S.; Reay, P.; Roy, P.; Klenk, H.-D. Induction of T cell response by bluetongue virus core-like particles expressing a T cell epitope of the M1 protein of influenza A virus. Med Microbiol. Immunol. 1998, 187, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Roy, P. Genetically Engineered Particulate Virus-Like Structures and Their Use as Vaccine Delivery Systems. Intervirology 1996, 39, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Rutkowska, D.A.; Meyer, Q.C.; Maree, F.; Vosloo, W.; Fick, W.; Huismans, H. The use of soluble African horse sickness viral protein 7 as an antigen delivery and presentation system. Virus Res. 2011, 156, 35–48. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oliveira, S.; Harms, J.; Rech, E.; Rodarte, R.; Bocca, A.; Goes, A.; Splitter, G. The role of T cell subsets and cytokines in the regulation of intracellular bacterial infection. Braz. J. Med. Biol. Res. 1998, 31, 77–84. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Afley, P.; Dohre, S.K.; Prasad, G.B.K.S.; Kumar, S. Prediction of T cell epitopes of Brucella abortus and evaluation of their protective role in mice. Appl. Microbiol. Biotechnol. 2015, 99, 7625–7637. [Google Scholar] [CrossRef]
- Hisham, Y.; Ashhab, Y. Identification of Cross-Protective Potential Antigens against Pathogenic Brucella spp. through Combining Pan-Genome Analysis with Reverse Vaccinology. J. Immunol. Res. 2018, 2018, 1474517. [Google Scholar] [CrossRef]
- Belyaev, A.S.; Roy, P. Presentation of hepatitis B virus preS2 epitope on bluetongue virus core-like particles. Virology 1992, 190, 840–844. [Google Scholar] [CrossRef]
- Sainsbury, F.; Thuenemann, E.C.; Lomonossoff, G.P. pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 2009, 7, 682–693. [Google Scholar] [CrossRef]
- Fischer, R.; Buyel, J.F. Molecular farming—The slope of enlightenment. Biotechnol. Adv. 2020, 40, 107519. [Google Scholar] [CrossRef]
- Lomonossoff, G.P.; D’aoust, M.-A. Plant-produced biopharmaceuticals: A case of technical developments driving clinical deployment. Science 2016, 353, 1237–1240. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, M.A.; Couture, M.M.J.; Charland, N.; Trépanier, S.; Landry, N.; Ors, F.; Vézina, L.P. The production of hemagglutinin-based virus-like particles in plants: A rapid, efficient and safe response to pandemic influenza. Plant Biotechnol. J. 2010, 8, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Recent Developments in Deciphering the Biological Role of Plant Complex N-Glycans. Front. Plant Sci. 2022, 13, 897549. [Google Scholar] [CrossRef]
- Gomord, V.; Fitchette, A.; Menu-Bouaouiche, L.; Saint-Jore-Dupas, C.; Plasson, C.; Michaud, D.; Faye, L. Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol. J. 2010, 8, 564–587. [Google Scholar] [CrossRef]
- Landry, N.; Ward, B.J.; Trépanier, S.; Montomoli, E.; Dargis, M.; Lapini, G.; Vézina, L.-P. Preclinical and Clinical Development of Plant-Made Virus-Like Particle Vaccine against Avian H5N1 Influenza. PLoS ONE 2010, 5, e15559. [Google Scholar] [CrossRef] [PubMed]
- Pillet, S.; Couillard, J.; Trépanier, S.; Poulin, J.-F.; Yassine-Diab, B.; Guy, B.; Ward, B.J.; Landry, N. Immunogenicity and safety of a quadrivalent plant-derived virus like particle influenza vaccine candidate—Two randomized Phase II clinical trials in 18 to 49 and ≥50 years old adults. PLoS ONE 2019, 14, e0216533. [Google Scholar] [CrossRef]
- Ward, B.J.; Makarkov, A.; Séguin, A.; Pillet, S.; Trépanier, S.; Dhaliwall, J.; Libman, M.D.; Vesikari, T.; Landry, N. Efficacy, immunogenicity, and safety of a plant-derived, quadrivalent, virus-like particle influenza vaccine in adults (18–64 years) and older adults (≥65 years): Two multicentre, randomised phase 3 trials. Lancet 2020, 396, 1491–1503. [Google Scholar] [CrossRef]
- Ward, B.J.; Gobeil, P.; Seguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.-Y.; Couture, M.; D’Aoust, M.-A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Hager, K.J.; Marc, G.P.; Gobeil, P.; Diaz, R.S.; Heizer, G.; Llapur, C.; Makarkov, A.I.; Vasconcellos, E.; Pillet, S.; Riera, F.; et al. Efficacy and Safety of a Recombinant Plant-Based Adjuvanted COVID-19 Vaccine. N. Engl. J. Med. 2022, 386, 2084–2096. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, N.; Robinson, M.K.; Bernard, C.; Kawaguchi, Y.; Koujin, Y.; Koen, A.; Madhi, S.; Polasek, T.M.; McNeal, M.; Dargis, M.; et al. Safety and immunogenicity of a plant-derived rotavirus-like particle vaccine in adults, toddlers and infants. Vaccine 2021, 39, 5513–5523. [Google Scholar] [CrossRef] [PubMed]
- Pujol, M.; Ramírez, N.I.; Ayala, M.; Gavilondo, J.V.; Valdés, R.; Rodríguez, M.; Brito, J.; Padilla, S.; Gómez, L.; Reyes, B.; et al. An integral approach towards a practical application for a plant-made monoclonal antibody in vaccine purification. Vaccine 2005, 23, 1833–1837. [Google Scholar] [CrossRef]
- Liew, P.S.; Hair-Bejo, M. Farming of Plant-Based Veterinary Vaccines and Their Applications for Disease Prevention in Animals. Adv. Virol. 2015, 2015, 936940. [Google Scholar] [CrossRef] [PubMed]
- In brief: Taliglucerase (Elelyso) for Gaucher disease. Med. Lett. Drugs Ther. 2012, 54, 56.
- Ward, B.J.; Séguin, A.; Couillard, J.; Trépanier, S.; Landry, N. Phase III: Randomized observer-blind trial to evaluate lot-to-lot consistency of a new plant-derived quadrivalent virus like particle influenza vaccine in adults 18–49 years of age. Vaccine 2021, 39, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.H.; Church, D.; Koellhoffer, E.C.; Osota, E.; Shukla, S.; Rybicki, E.P.; Pokorski, J.K.; Steinmetz, N.F. Integrating plant molecular farming and materials research for next-generation vaccines. Nat. Rev. Mater. 2021, 7, 372–388. [Google Scholar] [CrossRef] [PubMed]
- Murad, S.; Fuller, S.; Menary, J.; Moore, C.; Pinneh, E.; Szeto, T.; Hitzeroth, I.; Freire, M.; Taychakhoonavudh, S.; Phoolcharoen, W.; et al. Molecular Pharming for low and middle income countries. Curr. Opin. Biotechnol. 2020, 61, 53–59. [Google Scholar] [CrossRef]
- Tsekoa, T.L.; Singh, A.A.; Buthelezi, S.G. Molecular farming for therapies and vaccines in Africa. Curr. Opin. Biotechnol. 2020, 61, 89–95. [Google Scholar] [CrossRef]
- Hodson, J. Centre for Epidemic Response and Innovation (CERI). Available online: https://www.csir.co.za/dr-patrick-soon-shiong-and-nantafrica-announce-launch-covid-19-and-cancer-vaccine-initiative (accessed on 11 February 2022).
- Chhiba-Govindjee, V.P.; Mathiba, K.; van der Westhuyzen, C.W.; Steenkamp, P.; Rashamuse, J.K.; Stoychev, S.; Bode, M.L.; Brady, D. Dimethylformamide is a novel nitrilase inducer in Rhodococcus rhodochrous. Appl. Microbiol. Biotechnol. 2018, 102, 10055–10065. [Google Scholar] [CrossRef] [PubMed]
- Mokoena, N.B.; Moetlhoa, B.; Rutkowska, D.A.; Mamputha, S.; Dibakwane, V.S.; Tsekoa, T.L.; O’Kennedy, M.M. Plant-produced Bluetongue chimaeric VLP vaccine candidates elicit serotype-specific immunity in sheep. Vaccine 2019, 37, 6068–6075. [Google Scholar] [CrossRef] [PubMed]
- Delpino, M.V.; Estein, S.M.; Fossati, C.A.; Baldi, P.C.; Cassataro, J. Vaccination with Brucella recombinant DnaK and SurA proteins induces protection against Brucella abortus infection in BALB/c mice. Vaccine 2007, 25, 6721–6729. [Google Scholar] [CrossRef] [PubMed]
- Pasquevich, K.A.; Estein, S.M.; Samartino, C.G.; Zwerdling, A.; Coria, L.M.; Barrionuevo, P.; Fossati, C.A.; Giambartolomei, G.H.; Cassataro, J. Immunization with Recombinant Brucella Species Outer Membrane Protein Omp16 or Omp19 in Adjuvant Induces Specific CD4+ and CD8+ T Cells as Well as Systemic and Oral Protection against Brucella abortus Infection. Infect. Immun. 2009, 77, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Radhakrishnan, G.; Harms, J.; Splitter, G. Invasive Escherichia coli vaccines expressing Brucella melitensis outer membrane proteins 31 or 16 or periplasmic protein BP26 confer protection in mice challenged with B. melitensis. Vaccine 2012, 30, 4017–4022. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Li, L.; Song, D.; Liu, Y.; Ju, W.; Song, X.; Wang, J.; Pang, B.; Xu, K.; Li, J. A novel recombinant multi-epitope protein against Brucella melitensis infection. Immunol. Lett. 2016, 175, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.J.; Andrews, N.; Ball, D.; Bellantuono, I.; Gray, J.; Hachoumi, L.; Holmes, A.; Latcham, J.; Petrie, A.; Potter, P.; et al. Does age matter? The impact of rodent age on study outcomes. Lab. Anim. 2017, 51, 160–169. [Google Scholar] [CrossRef] [PubMed]
- A Montaraz, J.; Winter, A.J. Comparison of living and nonliving vaccines for Brucella abortus in BALB/c mice. Infect. Immun. 1986, 53, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Barquero-Calvo, E.; Chacón-Díaz, C.; Chaves-Olarte, E.; Moreno, E. Bacterial Counts in Spleen. Bio-Protocol 2013, 3, e954. [Google Scholar] [CrossRef]
- Thuenemann, E.C.; Meyers, A.E.; Verwey, J.; Rybicki, E.P.; Lomonossoff, G.P. A method for rapid production of heteromultimeric protein complexes in plants: Assembly of protective bluetongue virus-like particles. Plant Biotechnol. J. 2013, 11, 839–846. [Google Scholar] [CrossRef]
- French, T.J. (NERC Institute of Virology and Environmental Microbiology, Oxford, United Kingdom), Roy P. Synthesis of bluetongue virus (BTV) corelike particles by a recombinant baculovirus expressing the two major structural core proteins of BTV. J. Virol. 1990, 64, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Goenka, R.; Parent, M.A.; Elzer, P.H.; Baldwin, C.L. B Cell–deficient Mice Display Markedly Enhanced Resistance to the Intracellular Bacterium Brucella abortus. J. Infect. Dis. 2011, 203, 1136–1146. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, Z.; Fasihi-Ramandi, M.; Bouzari, S. Nanoparticle-Based Vaccines for Brucellosis: Calcium Phosphate Nanoparticles-Adsorbed Antigens Induce Cross Protective Response in Mice. Int. J. Nanomed. 2020, 15, 3877–3886. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.; Mikhailov, M.; Bishop, D.H. Baculovirus multigene expression vectors and their use for understanding the assembly process of architecturally complex virus particles. Gene 1997, 190, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.; Sutton, G. New generation of African horse sickness virus vaccines based on structural and molecular studies of the virus particles. In African Horse Sickness; Springer: Vienna, Austria, 1998; Volume 14, pp. 177–202. [Google Scholar]
- Grimes, J.; Basak, A.K.; Roy, P.; Stuart, D. The crystal structure of bluetongue virus VP7. Nature 1995, 373, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Topp, E.; Irwin, R.; McAllister, T.; Lessard, M.; Joensuu, J.J.; Kolotilin, I.; Conrad, U.; Stöger, E.; Mor, T.; Warzecha, H.; et al. The case for plant-made veterinary immunotherapeutics. Biotechnol. Adv. 2016, 34, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Vishnu, U.S.; Sankarasubramanian, J.; Gunasekaran, P.; Rajendhran, J. Novel Vaccine Candidates against Brucella melitensis Identified through Reverse Vaccinology Approach. OMICS A J. Integr. Biol. 2015, 19, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Nazifi, N.; Tahmoorespur, M.; Sekhavati, M.H.; Haghparast, A.; Behroozikhah, A.M. In vivo immunogenicity assessment and vaccine efficacy evaluation of a chimeric tandem repeat of epitopic region of OMP31 antigen fused to interleukin 2 (IL-2) against Brucella melitensis in BALB/c mice. BMC Veter. Res. 2019, 15, 402. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sharma, A.; Tirpude, N.V.; Padwad, Y.; Hallan, V.; Kumar, S. Plant-derived immuno-adjuvants in vaccines formulation: A promising avenue for improving vaccines efficacy against SARS-CoV-2 virus. Pharmacol. Rep. 2022, 74, 1238–1254. [Google Scholar] [CrossRef]
- Jiang, X.; Baldwin, C.L. Effects of cytokines on intracellular growth of Brucella abortus. Infect. Immun. 1993, 61, 124–134. [Google Scholar] [CrossRef]
- Paranavitana, C.; Zelazowska, E.; Izadjoo, M.; Hoover, D. Interferon-γ associated cytokines and chemokines produced by spleen cells from Brucella-immune mice. Cytokine 2005, 30, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Resende, M.; Cardoso, M.S.; Ribeiro, A.R.; Flórido, M.; Borges, M.; Castro, A.G.; Alves, N.L.; Cooper, A.M.; Appelberg, R. Innate IFN-γ-Producing Cells Developing in the Absence of IL-2 Receptor Common γ-Chain. J. Immunol. 2017, 199, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Blasco, J. A review of the use of B. melitensis Rev 1 vaccine in adult sheep and goats. Prev. Veter. Med. 1997, 31, 275–283. [Google Scholar] [CrossRef] [PubMed]



| Group | Primary (i.p) | Boost (i.p) | Challenge with Virulent Brucella melitensis Strain i.p | Animals per Group (n) | |
|---|---|---|---|---|---|
| Day 1 | Day 15 | Day 45 | Day 75 | ||
| G1 Untouched control | 5 mice untouched | 5 mice untouched | 5 mice sacrificed for cellular immune assays | - | 5 |
| G2 Negative control | 10 mice injected with Bicine buffer + adj (100 μL/mouse) | 10 mice injected with Bicine buffer + adj (100 μL/mouse) | 5 mice sacrificed for cellular immune assays; 5 mice challenged with 5 × 105 B.melitensis | 5 mice sacrificed for spleen bacterial counts | 10 |
| G3 Positive control (Brucella melitensis vaccine Rev 1) | All 10 mice vaccinated with Rev1 5 × 105 | All 10 mice vaccinated with Rev1 5 × 105 | 5 mice sacrificed for cellular immune assays; 5 mice challenged with 5 × 105 B.melitensis | 5 mice sacrificed for spleen bacterial counts | 10 |
| G4 Test Group | All 10 mice inoculated with P3 CLPs + adj (2 μg/100 μL/mouse) | All 10 mice boosted with P3 CLPs + adj (2 μg/100 μL/mouse) | 5 mice sacrificed for cellular immune assays; 5 mice challenged with 5 × 105 B. melitensis | 5 mice sacrificed for spleen bacterial counts | 10 |
| G5 Test Group | All 10 mice inoculated with P2 + P3 + P4 CLPs + adj (2 μg/100 μL/mouse) | All 10 mice boosted with P2 + 3 + 4 CLPs + adj (2 μg/100 μL/mouse) | 5 mice sacrificed for cellular immune assays; 5 mice challenged with 5 × 105 B. melitensis | 5 mice sacrificed for spleen bacterial counts | 10 |
| G6 Test Group | All 10 mice inoculated with P2 CLPs+ adj (2 μg/100 μL/mouse) | All 10 mice boosted with P2 CLPs + adj (2 μg/100 μL/mouse) | 5 mice sacrificed for cellular immune assays; 5 mice challenged with 5 × 105 B. melitensis | 5 mice sacrificed for spleen bacterial counts | 10 |
| 55 |
| Average cfu */Spleen | |
|---|---|
| Group 2 (Negative control) | 6.05 × 105 (SD = 3.75 × 105) |
| Group 3 (Rev1 vaccine Positive control) | 0.7 × 105 (SD = 0.8 × 105) |
| Group 4 (P3 CLPs) | 11.01 × 105 (SD = 15.5 × 105) |
| Group 5 (P2, P3, P4 CLPs) | 0.625 × 105 (SD = 0.75 × 105) |
| Group 6 (P2 CLPs) | 0.7 × 105 (SD = 1.29 × 105) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rutkowska, D.A.; Du Plessis, L.H.; Suleman, E.; O’Kennedy, M.M.; Thimiri Govinda Raj, D.B.; Lemmer, Y. Development of a Plant-Expressed Subunit Vaccine against Brucellosis. Microorganisms 2024, 12, 1047. https://doi.org/10.3390/microorganisms12061047
Rutkowska DA, Du Plessis LH, Suleman E, O’Kennedy MM, Thimiri Govinda Raj DB, Lemmer Y. Development of a Plant-Expressed Subunit Vaccine against Brucellosis. Microorganisms. 2024; 12(6):1047. https://doi.org/10.3390/microorganisms12061047
Chicago/Turabian StyleRutkowska, Daria A., Lissinda H. Du Plessis, Essa Suleman, Martha M. O’Kennedy, Deepak B. Thimiri Govinda Raj, and Yolandy Lemmer. 2024. "Development of a Plant-Expressed Subunit Vaccine against Brucellosis" Microorganisms 12, no. 6: 1047. https://doi.org/10.3390/microorganisms12061047
APA StyleRutkowska, D. A., Du Plessis, L. H., Suleman, E., O’Kennedy, M. M., Thimiri Govinda Raj, D. B., & Lemmer, Y. (2024). Development of a Plant-Expressed Subunit Vaccine against Brucellosis. Microorganisms, 12(6), 1047. https://doi.org/10.3390/microorganisms12061047







