Comparative Analysis of Bacterial Information of Biofilms and Activated Sludge in Full-Scale MBBR-IFAS Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Treatment Process, Sewage Characteristic, and Sample Collection
2.2. High-Throughput Sequencing Methods
2.3. Potential Function Analysis
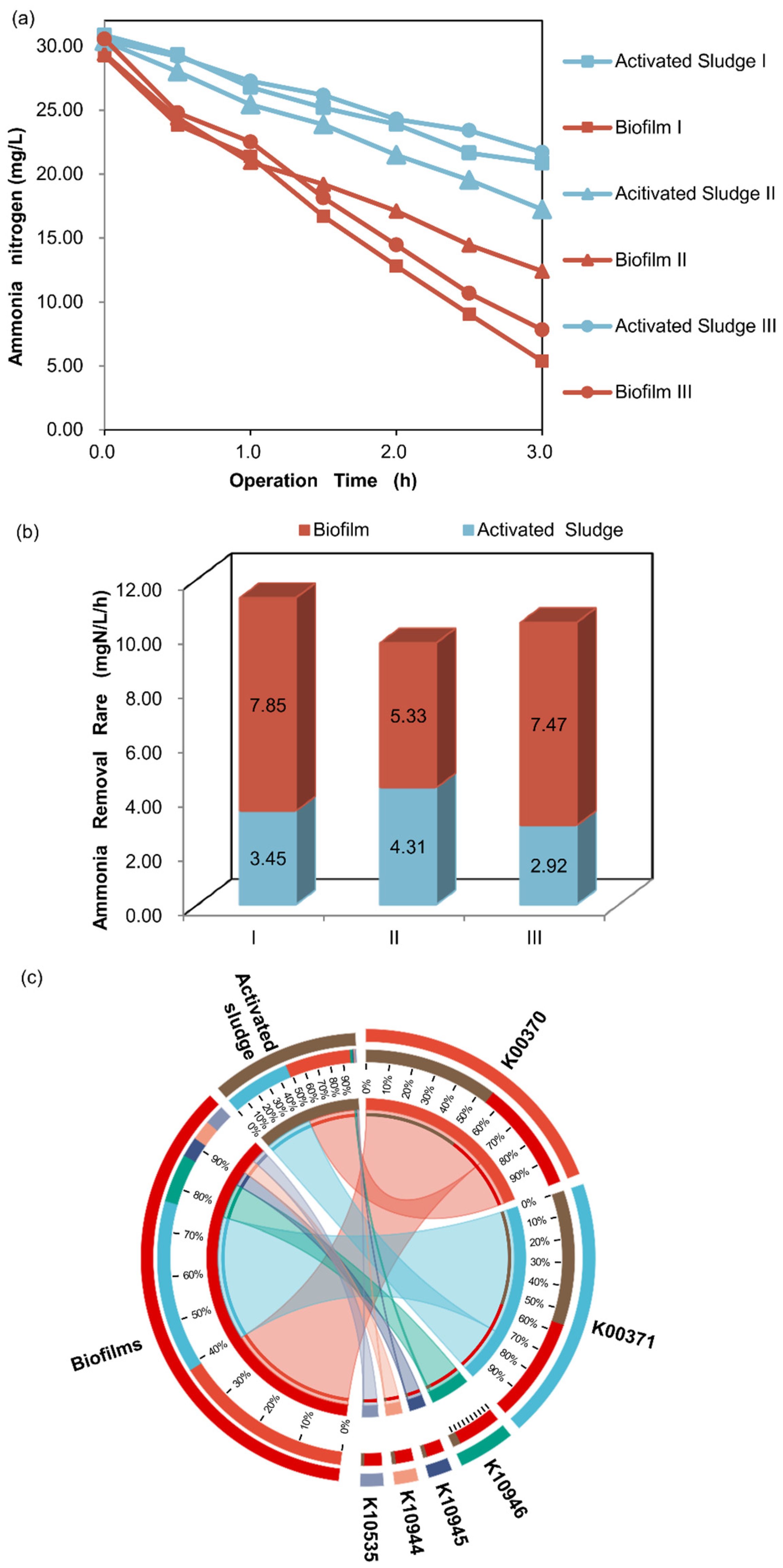
2.4. Ammonia Nitrogen Removal Capability Test
2.5. Statistical Analysis
3. Results and Discussion
3.1. Bacterial Community Diversity
3.1.1. Alpha Bacterial Community Diversity
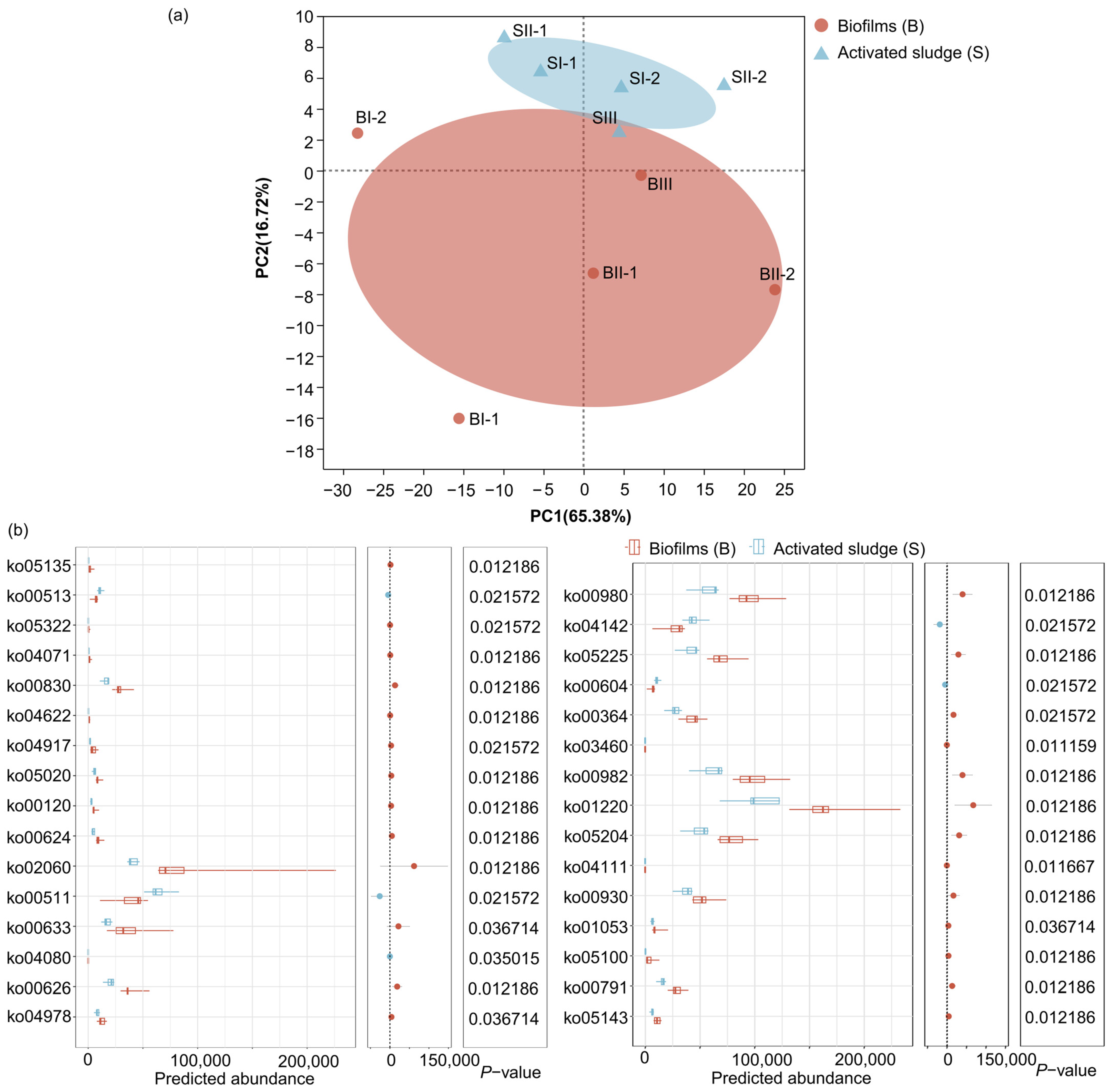
3.1.2. Beta Bacterial Community Diversity
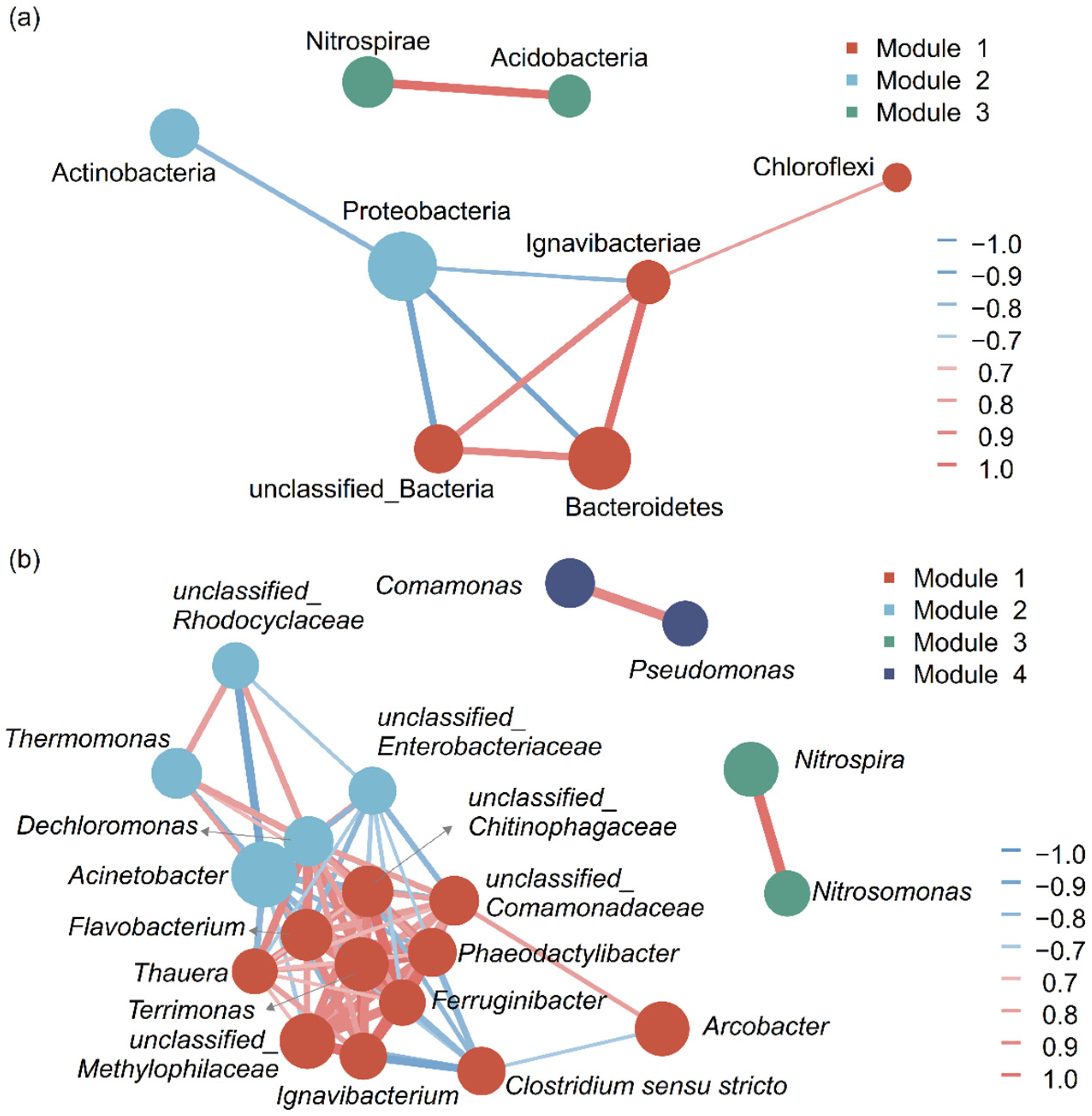
3.2. Bacterial Populations
3.2.1. Bacterial Populations at Phylum Level
3.2.2. Bacterial Populations at Genus Level
3.3. Bacterial Co-Occurrence Network
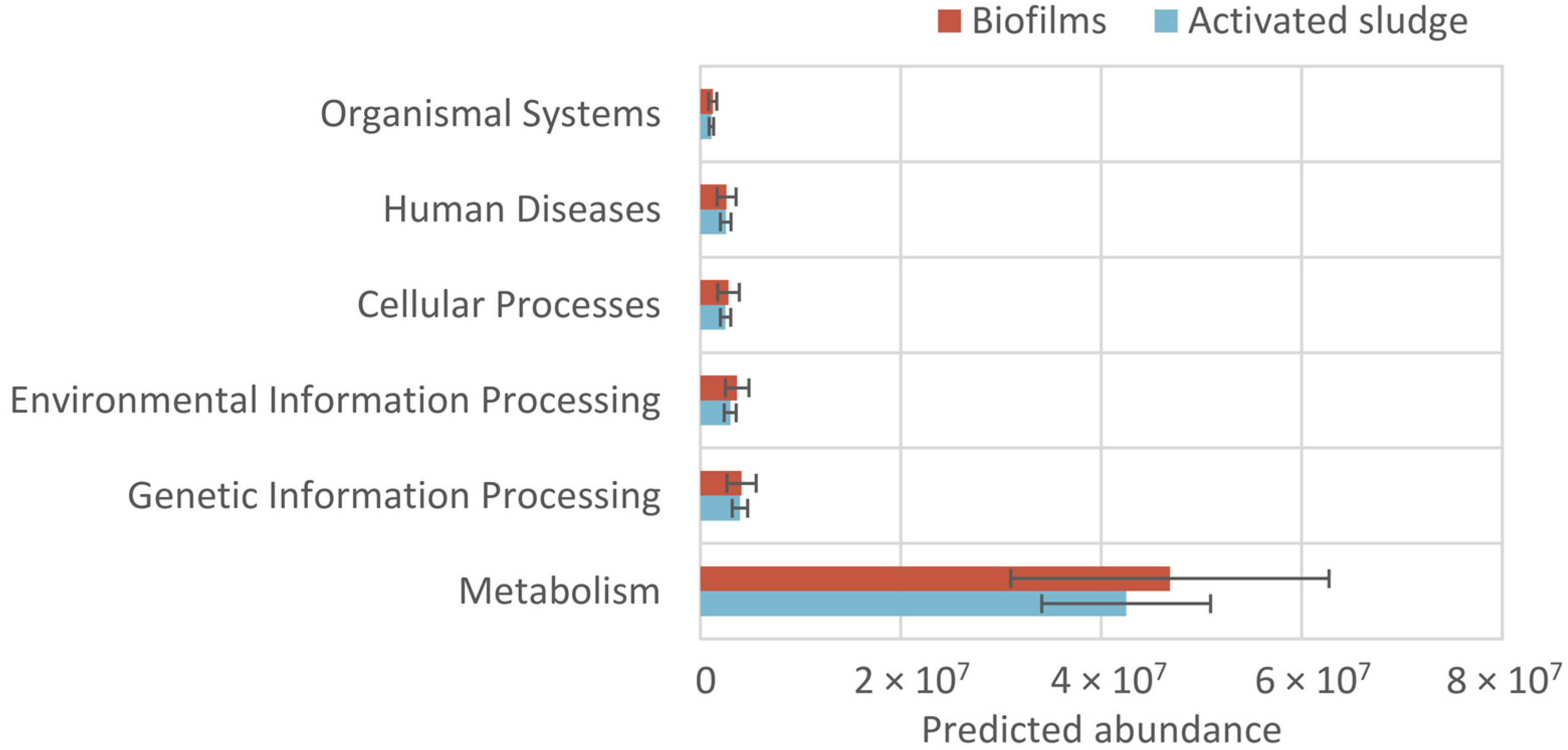
3.4. Potential Functions
3.4.1. Potential Functions of Pathways Based on KEGG
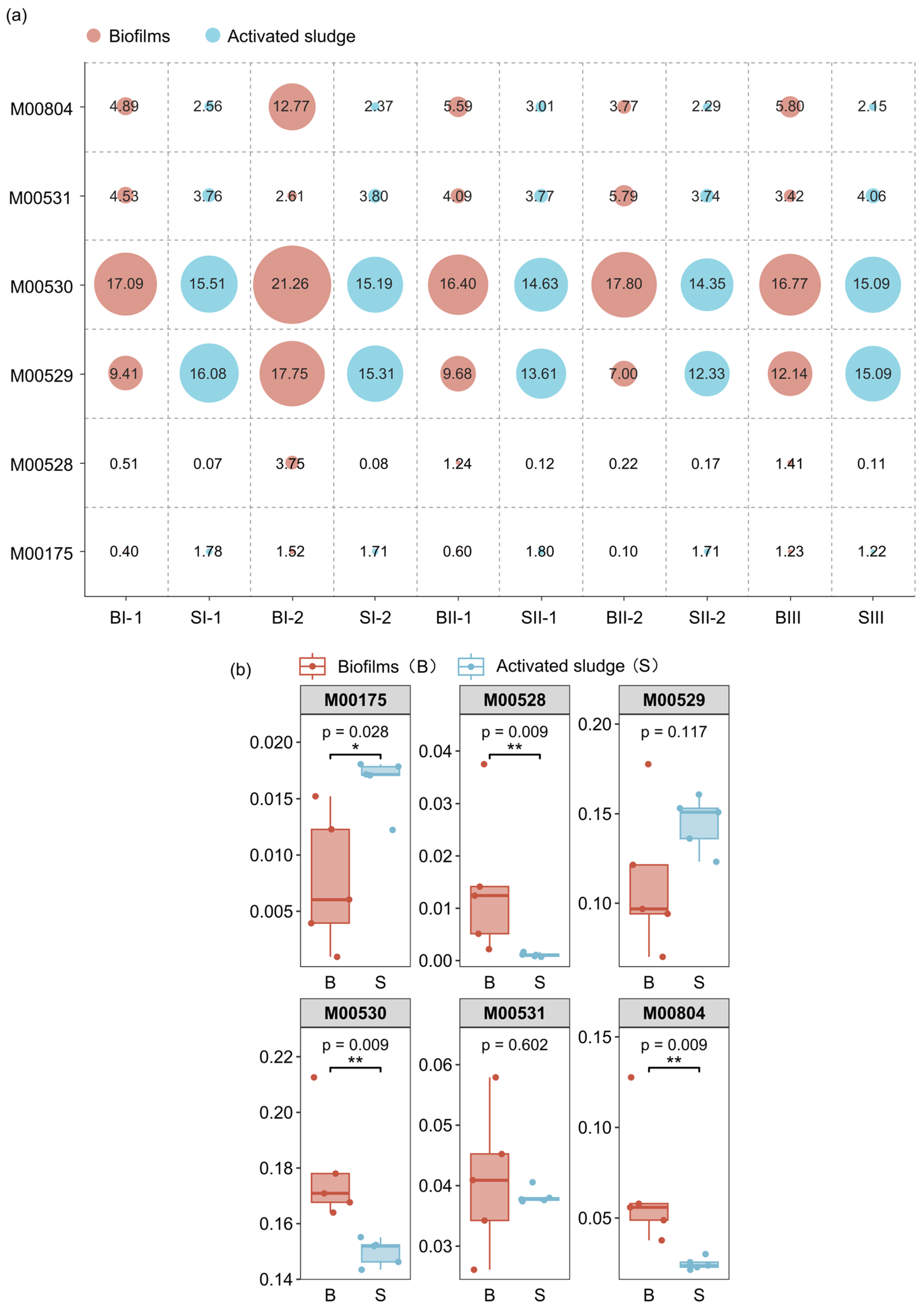
3.4.2. Potential Functions of Modules Involved in Nitrogen Metabolism
3.4.3. Potential Functions of KOs Involved in Nitrification
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Almomani, F. Prediction the performance of multistage moving bed biological process using artificial neural network (ANN). Sci. Total Environ. 2020, 744, 140854. [Google Scholar] [CrossRef] [PubMed]
- Mannina, G.; Capodici, M.; Cosenza, A.; Cinà, P.; Di Trapani, D.; Puglia, A.M.; Ekama, G.A. Bacterial community structure and removal performances in IFAS-MBRs: A pilot plant case study. J. Environ. Manag. 2017, 198, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, Y.; Xie, Q.; Shuo, C. Enhancing nitrogen removal efficiency and reducing nitrate liquor recirculation ratio by improving simultaneous nitrification and denitrification in integrated fixed-film activated sludge (IFAS) process. Water Sci. Technol. 2016, 73, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Ding, J.; Zhan, W.; Du, J.; Wang, G.; Pang, J.; Ren, N.; Yang, S. Effect of dissolved oxygen concentration on performance and mechanism of simultaneous nitrification and denitrification in integrated fixed-film activated sludge sequencing batch reactors. Bioresour. Technol. 2023, 387, 129616. [Google Scholar] [CrossRef]
- Sander, S.; Behnisch, J.; Wagner, M. Energy, cost and design aspects of coarse- and fine-bubble aeration systems in the MBBR IFAS process. Water Sci. Technol. 2017, 75, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Khudhair, D.N.; Hosseinzadeh, M.; Zwain, H.M.; Siadatmousavi, S.M.; Majdi, A.; Mojiri, A. Upgrading the MBBR Process to Reduce Excess Sludge Production in Activated Sludge System Treating Sewage. Water 2023, 15, 408. [Google Scholar] [CrossRef]
- Waqas, S.; Bilad, M.R.; Man, Z.; Wibisono, Y.; Jaafar, J.; Indra Mahlia, T.M.; Khan, A.L.; Aslam, M. Recent progress in integrated fixed-film activated sludge process for wastewater treatment: A review. J. Environ. Manag. 2020, 268, 110718. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Kim, T.-S.; Yu, G.H.; Jung, J.-H.; Park, H.-D. Bacterial community composition and diversity of a full-scale integrated fixed-film activated sludge system as investigated by pyrosequencing. J. Microbiol. Biotechnol. 2010, 20, 1717–1723. [Google Scholar] [PubMed]
- Meng, F.; Guo, S.; Zhang, L.; Lu, Y.; Li, M.; Tan, Y.; Zha, K.; Yuan, S. Ecological mechanisms of biofilm development in the hybrid sludge-biofilm process: Implications for process start-up and optimization. Water Res. 2023, 245, 120587. [Google Scholar] [CrossRef]
- Zhang, K.; Li, J.; Zheng, Z.; Zhang, J.; Sun, M.; Huang, S. Analyzing the sludge characteristics and microbial communities of biofilm and activated sludge in the partial nitrification/anammox process. J. Water Process Eng. 2022, 46, 102618. [Google Scholar] [CrossRef]
- Li, J.; Li, A.; Li, Y.; Cai, M.; Luo, G.; Wu, Y.; Tian, Y.; Xing, L.; Zhang, Q. PICRUSt2 functionally predicts organic compounds degradation and sulfate reduction pathways in an acidogenic bioreactor. Front. Environ. Sci. Eng. 2022, 16, 47. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Y.; Euverink, G.J.W. Effect of bioaugmentation combined with activated charcoal on the mitigation of volatile fatty acids inhibition during anaerobic digestion. Chem. Eng. J. 2022, 428, 131015. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Cayetano, R.D.A.; Park, J.; Kim, G.-B.; Jung, J.-H.; Kim, S.-H. Enhanced anaerobic digestion of waste-activated sludge via bioaugmentation strategy—Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt2) analysis through hydrolytic enzymes and possible linkage to system performance. Bioresour. Technol. 2021, 332, 125014. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Mai, J.; Cao, X.; Burberry, A.; Cominelli, F.; Zhang, L. ggpicrust2: An R package for PICRUSt2 predicted functional profile analysis and visualization. Bioinformatics 2023, 39, btad470. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, M.; Marco, C.; Alida, C.; Daniele, D.T.; Zhengyu, Z.; Yongmei, L. Integrated Fixed Film Activated Sludge (IFAS) membrane BioReactor: The influence of the operational parameters. Bioresour. Technol. 2020, 301, 122752. [Google Scholar]
- Huang, M.; Zhang, W.; Liu, C.; Hu, H. Fate of trace tetracycline with resistant bacteria and resistance genes in an improved AAO wastewater treatment plant. Process Saf. Environ. Prot. 2014, 93, 68–74. [Google Scholar] [CrossRef]
- Ashrafi, E.; Mehrabani Zeinabad, A.; Borghei, S.M.; Torresi, E.; Muñoz Sierra, J. Optimising nutrient removal of a hybrid five-stage Bardenpho and moving bed biofilm reactor process using response surface methodology. J. Environ. Chem. Eng. 2019, 7, 102861. [Google Scholar] [CrossRef]
- Water Environment Federation. Biofilm Reactors; McGraw-Hill Education: New York, NY, USA, 2010; pp. 212–234. [Google Scholar]
- Ping, L.; Bingyu, D.; Chunyan, F.; Shuwei, Y.; Ting, Z.; Xin, Q.; Huiying, L.; Xiaokui, G.; Ke, D.; Zhu, Y.; et al. Prevotella and Klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J. Affect. Disord. 2017, 207, 300–304. [Google Scholar]
- Yan, K.; Jian, Z.; Huijun, X.; Zizhang, G.; Ngo, H.H.; Wenshan, G.; Shuang, L. Enhanced nutrient removal and mechanisms study in benthic fauna added surface-flow constructed wetlands: The role of Tubifex tubifex. Bioresour. Technol. 2017, 224, 157–165. [Google Scholar]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Pang, H.L.; Xu, Y.M.; Ren, R.Y.; He, J.G.; Pan, X.L.; Wang, L. Enhanced anaerobic digestion of waste activated sludge by alkaline protease-catalyzing hydrolysis: Role and significance of initial pH adjustment. Chem. Eng. J. 2023, 467, 143323. [Google Scholar] [CrossRef]
- Walter, W.G. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1961. [Google Scholar]
- Xu, N.; Tan, G.C.; Wang, H.Y.; Gai, X.P. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.F.; He, Y.; Wu, J.Y.; Jiang, Y.X.; Tam, N.F.Y.; Zhou, H.W. Comparison of the Levels of Bacterial Diversity in Freshwater, Intertidal Wetland, and Marine Sediments by Using Millions of Illumina Tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef] [PubMed]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Herrera, A.M.; Riera, R.; Rodríguez, R.A. Alpha species diversity measured by Shannon’s H-index: Some misunderstandings and underexplored traits, and its key role in exploring the trophodynamic stability of dynamic multiscapes. Ecol. Indic. 2023, 156, 111118. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, H.; Xia, H.; Yang, X.; Yang, L.; Wang, S.; Wen, J.; Sun, G. Different effects of high-fat diets rich in different oils on lipids metabolism, oxidative stress and gut microbiota. Food Res. Int. 2021, 141, 110078. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, W.; Boyao, Z.; Yao, G.; Shi, X. Characteristics of Bacterial Community Structure in Wuliangsu Lake During an Irrigation Interval in Hetao Plain. Environ. Sci. 2022, 43, 1424–1433. [Google Scholar]
- Zhao, W.; Peng, Y.; Wang, M.; Huang, Y.; Li, X. Nutrient removal and microbial community structure variation in the two-sludge system treating low carbon/nitrogen domestic wastewater. Bioresour. Technol. 2019, 294, 122161. [Google Scholar] [CrossRef]
- Wang, X.; Bi, X.; Hem, L.J.; Ratnaweera, H. Microbial community composition of a multi-stage moving bed biofilm reactor and its interaction with kinetic model parameters estimation. J. Environ. Manag. 2018, 218, 340–347. [Google Scholar] [CrossRef]
- Fan, X.; Gao, J.; Pan, K.; Li, D.; Dai, H.; Li, X. Functional genera, potential pathogens and predicted antibiotic resistance genes in 16 full-scale wastewater treatment plants treating different types of wastewater. Bioresour. Technol. 2018, 268, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.S.; Parameswaran, P.; Rittmann, B.E. Effects of solids retention time on methanogenesis in anaerobic digestion of thickened mixed sludge. Bioresour. Technol. 2011, 102, 10266–10272. [Google Scholar] [CrossRef] [PubMed]
- Podosokorskaya, O.A.; Kadnikov, V.V.; Gavrilov, S.N.; Mardanov, A.V.; Merkel, A.Y.; Karnachuk, O.V.; Ravin, N.V.; Bonch-Osmolovskaya, E.A.; Kublanov, I.V. Characterization of Melioribacter roseus gen. nov., sp nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environ. Microbiol. 2013, 15, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Li, W.C.; Niu, Q.G.; Zhang, H.; Tian, Z.; Zhang, Y.; Gao, Y.X.; Li, Y.Y.; Nishimura, O.; Yang, M. UASB treatment of chemical synthesis-based pharmaceutical wastewater containing rich organic sulfur compounds and sulfate and associated microbial characteristics. Chem. Eng. J. 2015, 260, 55–63. [Google Scholar] [CrossRef]
- Song, J.Y.; Wu, H.Y.; Yuan, H.M.; Wu, J.; Qi, W.K.; Lu, J.B.; Li, W.C.; Li, Y.Y. Reclamation of pharmaceutical wastewater by UAF, contact oxidation, and auto brush-cleaning aiding nano ZrO2 coated ceramic membrane MBR—an industrial-scale practice and functional microbial diversity. J. Water Process Eng. 2022, 46, 102563. [Google Scholar] [CrossRef]
- Wang, K.M.; Zhou, L.X.; Ji, K.F.; Xu, S.N.; Wang, J.D. Evaluation of a modified internal circulation (MIC) anaerobic reactor for real antibiotic pharmaceutical wastewater treatment: Process performance, microbial community and antibiotic resistance genes evolutions. J. Water Process Eng. 2022, 48, 102914. [Google Scholar] [CrossRef]
- Guo, X.; Li, B.; Zhao, R.; Zhang, J.; Lin, L.; Zhang, G.; Li, R.-H.; Liu, J.; Li, P.; Li, Y.; et al. Performance and bacterial community of moving bed biofilm reactors with various biocarriers treating primary wastewater effluent with a low organic strength and low C/N ratio. Bioresour. Technol. 2019, 287, 121424. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Ye, W.; Wei, D.; Xu, W.; Du, B.; Wei, Q. Simultaneous nitrification-denitrification and membrane fouling alleviation in a submerged biofilm membrane bioreactor with coupling of sponge and biodegradable PBS carrier. Bioresour. Technol. 2018, 270, 156–165. [Google Scholar] [CrossRef]
- Zhao, B.; He, Y.L.; Hughes, J.; Zhang, X.F. Heterotrophic nitrogen removal by a newly isolated Acinetobacter calcoaceticus HNR. Bioresour. Technol. 2010, 101, 5194–5200. [Google Scholar] [CrossRef]
- Cao, L.; Ni, L.; Qi, L.; Wen, H.T.; Wang, Z.; Meng, J.Q.; Zhang, X.B.; Zhang, Y.F. The application of post-denitrification fixed biofilm reactor for polishing secondary effluent: Nitrate removal, soluble microbial products and micropollutants biotransformation. Bioresour. Technol. 2023, 369, 128511. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, X.; Cheng, Y.; Jiang, M.; Li, X.; Xue, G. Iron robustly stimulates simultaneous nitrification and denitrification under aerobic conditions. Environ. Sci. Technol. 2018, 52, 1404–1412. [Google Scholar] [CrossRef]
- Huang, C.; Yuan, L.; Niu, W.; Meng, Y. Effect of dosing suspended fillers on microbial community structure and investigation on variation in water quality. China Environ. Sci. 2021, 41, 207–213. [Google Scholar]
- Pishgar, R.; Dominic, J.A.; Sheng, Z.Y.; Tay, J.H. Denitrification performance and microbial versatility in response to different selection pressures. Bioresour. Technol. 2019, 281, 72–83. [Google Scholar] [CrossRef]
- Zhang, X.; Chi, Y.; Wang, Q.; Jin, X.; Shi, X.; Jin, P. Effects of aeration strategy on denitrifying performance of activated sludge processes in treating low-carbon-source municipal wastewater. Environ. Sci. 2020, 41, 3356–3364. [Google Scholar]
- Wang, D.P.; Meng, Y.B.; Meng, F.G. Genome-centric metagenomics insights into functional divergence and horizontal gene transfer of denitrifying bacteria in anammox consortia. Water Res. 2022, 224, 119062. [Google Scholar] [CrossRef]
- Wang, Z.C.; Yuan, S.Y.; Deng, Z.W.; Wang, Y.J.; Deng, S.L.; Song, Y.T.; Sun, C.T.; Bu, N.S.; Wang, X.R. Evaluating responses of nitrification and denitrification to the co-selective pressure of divalent zinc and tetracycline based on resistance genes changes. Bioresour. Technol. 2020, 314, 123769. [Google Scholar] [CrossRef] [PubMed]
- Roots, P.; Wang, Y.; Rosenthal, A.F.; Griffin, J.S.; Sabba, F.; Petrovich, M.; Yang, F.; Zhang, H.; Wells, G.F. Comammox Nitrospira are the dominant ammonia oxidizers in a mainstream low dissolved oxygen nitrification reactor. Water Res. 2019, 157, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Jousset, A.; Guo, S.; Karlsson, I.; Zhao, Q.Y.; Wu, H.S.; Kowalchuk, G.A.; Shen, Q.R.; Li, R.; Geisen, S. Soil protist communities form a dynamic hub in the soil microbiome. Isme J. 2018, 12, 634–638. [Google Scholar] [CrossRef]
- Luo, Y.L.; Jiang, Q.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I.; Price, W.E.; Wang, J.; Guo, W.S. Evaluation of micropollutant removal and fouling reduction in a hybrid moving bed biofilm reactor-membrane bioreactor system. Bioresour. Technol. 2015, 191, 355–359. [Google Scholar] [CrossRef]
- Zhou, X.; Bi, X.; Yang, T.; Fan, X.; Shi, X.; Wang, L.; Zhang, Y.; Cheng, L.; Zhao, F.; Maletskyi, Z. Metagenomic insights into microbial nitrogen metabolism in two-stage anoxic/oxic-moving bed biofilm reactor system with multiple chambers for municipal wastewater treatment. Bioresour. Technol. 2022, 361, 127729. [Google Scholar] [CrossRef]
- Preena, P.G.; Kumar, V.J.R.; Singh, I.S.B. Nitrification and denitrification in recirculating aquaculture systems: The processes and players. Rev. Aquac. 2021, 13, 2053–2075. [Google Scholar] [CrossRef]
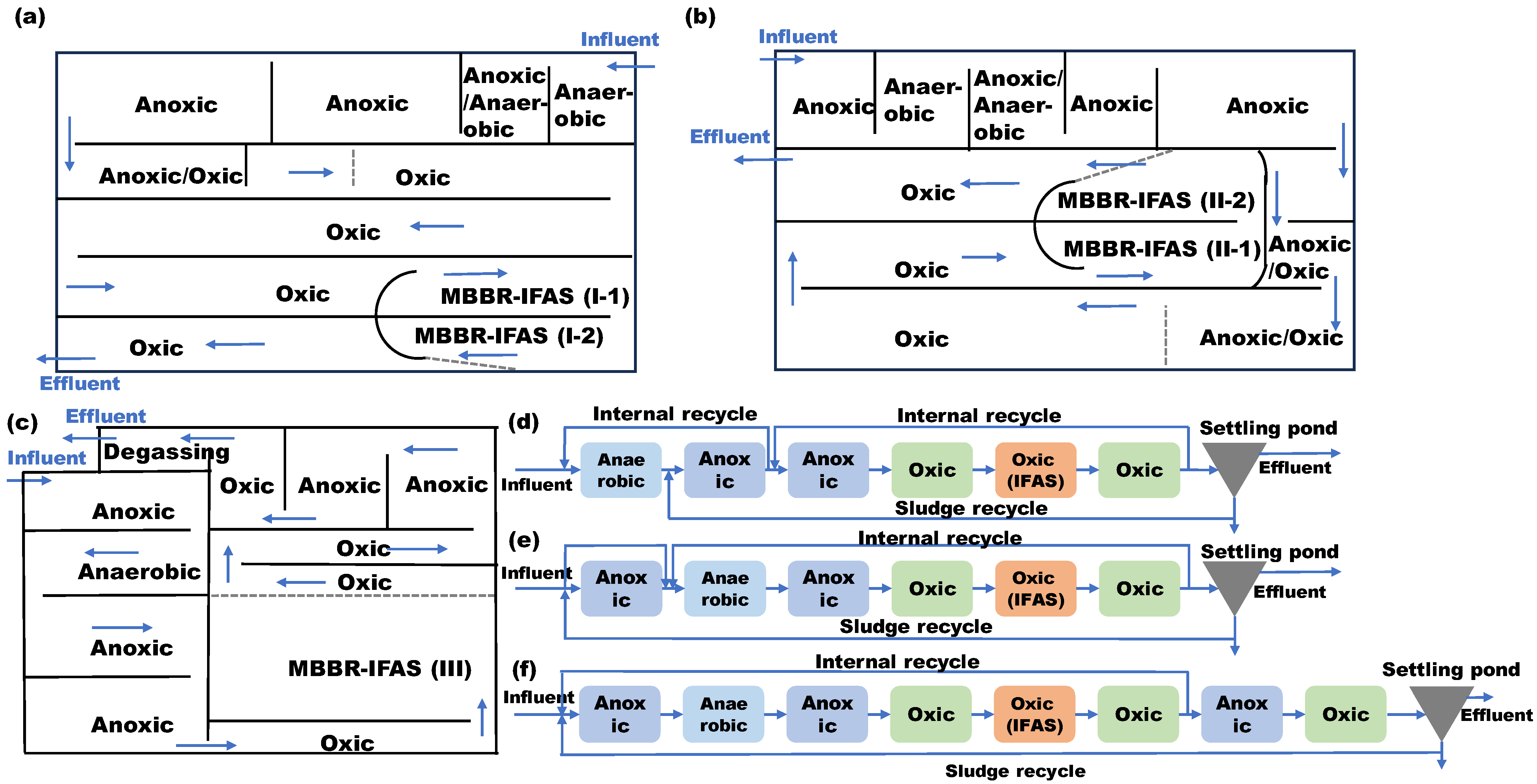




| Sample Number | Sample Type | System Source | Reactor Source |
|---|---|---|---|
| BI-1 | Biofilms | System I | The first MBBR-IFAS of System I |
| SI-1 | Activated sludge | System I | The first MBBR-IFAS of System I |
| BI-2 | Biofilms | System I | The second MBBR-IFAS of System I |
| SI-2 | Activated sludge | System I | The second MBBR-IFAS of System I |
| BII-1 | Biofilms | System II | The first MBBR-IFAS of System II |
| SII-1 | Activated sludge | System II | The first MBBR-IFAS of System II |
| BII-2 | Biofilms | System II | The second MBBR-IFAS of System II |
| SII-2 | Activated sludge | System II | The second MBBR-IFAS of System II |
| BIII | Biofilms | System III | The MBBR-IFAS of System III |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Liu, H.; Fan, X.; Wang, X.; Bi, X.; Cheng, L.; Huang, S.; Zhao, F.; Yang, T. Comparative Analysis of Bacterial Information of Biofilms and Activated Sludge in Full-Scale MBBR-IFAS Systems. Microorganisms 2024, 12, 1121. https://doi.org/10.3390/microorganisms12061121
Zhou X, Liu H, Fan X, Wang X, Bi X, Cheng L, Huang S, Zhao F, Yang T. Comparative Analysis of Bacterial Information of Biofilms and Activated Sludge in Full-Scale MBBR-IFAS Systems. Microorganisms. 2024; 12(6):1121. https://doi.org/10.3390/microorganisms12061121
Chicago/Turabian StyleZhou, Xiaolin, Haicheng Liu, Xing Fan, Xuyi Wang, Xuejun Bi, Lihua Cheng, Shujuan Huang, Fangchao Zhao, and Tang Yang. 2024. "Comparative Analysis of Bacterial Information of Biofilms and Activated Sludge in Full-Scale MBBR-IFAS Systems" Microorganisms 12, no. 6: 1121. https://doi.org/10.3390/microorganisms12061121






