Biological Traits and Comprehensive Genomic Analysis of Novel Enterococcus faecalis Bacteriophage EFP6
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Cultures and Culture Media
2.2. Isolation, Purification, and Initial Screening of the Lytic Bacteriophage
2.3. Assessment of Bacteriophage Host Spectrum
2.4. Biological Traits of EFP6
2.5. One-Step Growth Curve of EFP6
2.6. Concentration and Electron Microscopic Observation of EFP6
2.7. Whole Genome Sequencing of Phage EFP6
2.8. Phylogenetic Analysis and Collinearity of Phage EFP6
3. Results
3.1. Isolation, Purification, and Preliminary Screening of the Lytic Bacteriophage
3.2. Determination of Bacteriophage Host Range
3.3. Biological Characteristics of EFP6
3.4. One-Step Growth Curve of EFP6
3.5. Whole Genome Analysis of Phage EFP6
3.6. Functional Gene Analysis of Bacteriophage EFP6
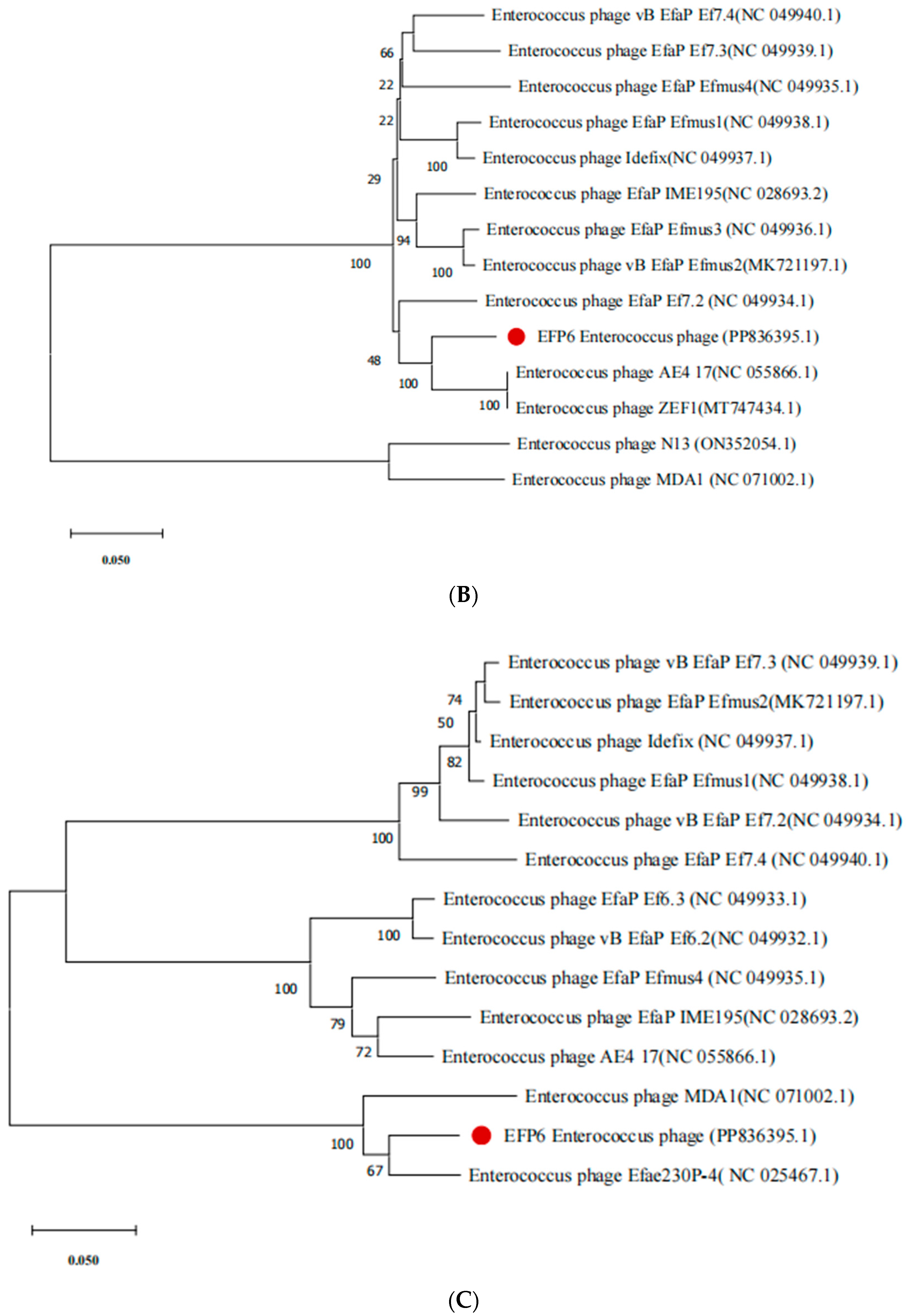
3.7. Evolutionary Analysis of Bacteriophage EFP6
3.8. Collinearity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lichtensteiger, A. Poultry veterinarians in health and production. Can. Vet. J. 2021, 62, 66. [Google Scholar] [PubMed]
- Shokryazdan, P.; Faseleh Jahromi, M.; Liang, J.B.; Ho, Y.W. Probiotics: From isolation to application. J. Am. Coll. Nutr. 2017, 36, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Falkovskaya, A.; Gowen, A. Literature review: Spectral imaging applied to poultry products. Poult. Sci. 2020, 99, 3709–3722. [Google Scholar] [CrossRef] [PubMed]
- Safonov, V. Comparison of LPO-AOS indices and biochemical composition of animal blood in biogeochemical provinces with different levels of selenium. Biol. Trace Elem. Res. 2022, 200, 2055–2061. [Google Scholar] [CrossRef] [PubMed]
- Trocino, A.; White, P.; Bordignon, F.; Ferrante, V.; Bertotto, D.; Birolo, M.; Pillan, G.; Xiccato, G. Effect of feed restriction on the behaviour and welfare of broiler chickens. Animals 2020, 10, 830. [Google Scholar] [CrossRef]
- Zhu, Q.; Sun, P.; Zhang, B.; Kong, L.; Xiao, C.; Song, Z. Progress on gut health maintenance and antibiotic alternatives in broiler chicken production. Front. Nutr. 2021, 8, 692839. [Google Scholar] [CrossRef] [PubMed]
- Averós, X.; Estevez, I. Meta-analysis of the effects of intensive rearing environments on the performance and welfare of broiler chickens. Poult. Sci. 2018, 97, 3767–3785. [Google Scholar] [CrossRef] [PubMed]
- Nhung, N.T.; Chansiripornchai, N.; Carrique-Mas, J.J. Antimicrobial resistance in bacterial poultry pathogens: A review. Front. Vet. Sci. 2017, 4, 126. [Google Scholar] [CrossRef] [PubMed]
- El Jeni, R.; Dittoe, D.K.; Olson, E.G.; Lourenco, J.; Corcionivoschi, N.; Ricke, S.C.; Callaway, T.R. Probiotics and potential applications for alternative poultry production systems. Poult. Sci. 2021, 100, 101156. [Google Scholar] [CrossRef]
- Krysiak, K.; Konkol, D.; Korczyński, M. Overview of the use of probiotics in poultry production. Animals 2021, 11, 1620. [Google Scholar] [CrossRef]
- Abd El-Hack, M.E.; El-Saadony, M.T.; Shafi, M.E.; Qattan, S.Y.; Batiha, G.E.; Khafaga, A.F.; Abdel-Moneim, A.M.E.; Alagawany, M. Probiotics in poultry feed: A comprehensive review. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1835–1850. [Google Scholar] [CrossRef] [PubMed]
- Yelubayeva, M.; Buralkhiyev, B.; Tyshchenko, V.; Terletskiy, V.; Ussenbekov, Y. Results of Camelus dromedarius and Camelus bactrianus genotyping by alpha-S 1-casein, kappa-casein loci, and DNA fingerprinting. Cytol. Genet. 2018, 52, 179–185. [Google Scholar] [CrossRef]
- Nami, Y.; Bakhshayesh, R.V.; Mohammadzadeh Jalaly, H.; Lotfi, H.; Eslami, S.; Hejazi, M.A. Probiotic properties of Enterococcus isolated from artisanal dairy products. Front. Microbiol. 2019, 10, 426946. [Google Scholar] [CrossRef]
- Wu, Y.; Zhen, W.; Geng, Y.; Wang, Z.; Guo, Y. Effects of dietary Enterococcus faecium NCIMB 11181 supplementation on growth performance and cellular and humoral immune responses in broiler chickens. Poult. Sci. 2019, 98, 150–163. [Google Scholar] [CrossRef]
- Holzapfel, W.; Arini, A.; Aeschbacher, M.; Coppolecchia, R.; Pot, B. Enterococcus faecium SF68 as a model for efficacy and safety evaluation of pharmaceutical probiotics. Benef. Microbes 2018, 9, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Olsen, R.; Schønheyder, H.; Christensen, H.; Bisgaard, M. Enterococcus faecalis of human and poultry origin share virulence genes supporting the zoonotic potential of E. faecalis. Zoonoses Public Health 2012, 59, 256–263. [Google Scholar] [CrossRef]
- Gregersen, R.H.; Petersen, A.; Christensen, H.; Bisgaard, M. Multilocus sequence typing of Enterococcus faecalis isolates demonstrating different lesion types in broiler breeders. Avian Pathol. 2010, 39, 435–440. [Google Scholar] [CrossRef]
- Devriese, L.; Hommez, J.; Wijfels, R.; Haesebrouck, F. Composition of the enterococcal and streptococcal intestinal flora of poultry. J. Appl. Microbiol. 1991, 71, 46–50. [Google Scholar] [CrossRef]
- Fertner, M.E.; Olsen, R.H.; Bisgaard, M.; Christensen, H. Transmission and genetic diversity of Enterococcus faecalis among layer chickens during hatch. Acta Vet. Scand. 2011, 53, 56. [Google Scholar] [CrossRef]
- Landman, W.J.M. Amyloid arthropathy in chickens: (Summary of thesis, Utrecht University, faculty of veterinary medicine, 1998). Vet. Q. 1999, 21, 78–82. [Google Scholar] [CrossRef]
- Jung, A.; Rautenschlein, S. Comprehensive report of an Enterococcus cecorum infection in a broiler flock in Northern Germany. BMC Vet. Res. 2014, 10, 311. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Nakamura, K.; Yamada, M.; Yamamoto, Y. Encephalomalacia with Enterococcus durans infection in the brain stem and cerebral hemisphere in chicks in Japan. Avian Dis. 2006, 50, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Moellering, R.C., Jr. Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. 1992, 14, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Noskin, G.A.; Peterson, L.R.; Warren, J.R. Enterococcus faecium and Enterococcus faecalis bacteremia: Acquisition and outcome. Clin. Infect. Dis. 1995, 20, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Hughes, E.R.; Winter, S.E. Enterococcus faecalis: E. coli’s siderophore-inducing sidekick. Cell Host Microbe 2016, 20, 411–412. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Fang, C.T.; Wang, H.K.; Hsue, P.; Chang, S.C.; Luh, K.T. Invasive infections due to vancomycin-resistant enterococci in adult patients. J. Microbiol. Immunol. Infect. 2001, 34, 281–286. [Google Scholar] [PubMed]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Li, W.; Song, F.; Yang, X.; Zhou, J.; Yu, H.; Ji, X.; Wei, Y. Biological characteristics and whole-genome analysis of the Enterococcus faecalis phage PEf771. Can. J. Microbiol. 2020, 66, 505–520. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, J.; Harbaugh, B.; Jones, J.; Somodi, G.; Jackson, L. H-mutant bacteriophages as a potential biocontrol of bacterial blight of geranium. HortScience 2001, 36, 98–100. [Google Scholar] [CrossRef]
- Guenther, S.; Huwyler, D.; Richard, S.; Loessner, M.J. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 2009, 75, 93–100. [Google Scholar] [CrossRef]
- Sulakvelidze, A. Using lytic bacteriophages to eliminate or significantly reduce contamination of food by foodborne bacterial pathogens. J. Sci. Food Agric. 2013, 93, 3137–3146. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Ruiz, M.; Vinés, E.D.; Montt, C.; Fernández, J.; Delgado, J.M.; Hormazábal, J.C.; Bittner, M. Isolation of a novel Aggregatibacter actinomycetemcomitans serotype b bacteriophage capable of lysing bacteria within a biofilm. Appl. Environ. Microbiol. 2011, 77, 3157–3159. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Brosh, Y.; Gelman, D.; Coppenhagen-Glazer, S.; Beyth, S.; Poradosu-Cohen, R.; Que, Y.-A.; Beyth, N.; Hazan, R. Targeting Enterococcus faecalis biofilms with phage therapy. Appl. Environ. Microbiol. 2015, 81, 2696–2705. [Google Scholar] [CrossRef] [PubMed]
- Shahin, K.; Bouzari, M.; Wang, R. Isolation, characterization and genomic analysis of a novel lytic bacteriophage vB_SsoS-ISF002 infecting Shigella sonnei and Shigella flexneri. J. Med. Microbiol. 2018, 67, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, X.; Wang, S.; Guan, L.; Li, X.; Hu, D.; Gao, D.; Song, J.; Chen, H.; Qian, P. A novel tail-associated O91-specific polysaccharide depolymerase from a podophage reveals lytic efficacy of Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 2020, 86, e00145-20. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. In Bacteriophages; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2009; Volume 501, pp. 69–76. [Google Scholar]
- Liu, J.; Zhu, Y.; Li, Y.; Lu, Y.; Xiong, K.; Zhong, Q.; Wang, J. Bacteriophage-resistant mutant of Enterococcus faecalis is impaired in biofilm formation. Front. Microbiol. 2022, 13, 913023. [Google Scholar] [CrossRef] [PubMed]
- Pajunen, M.; Kiljunen, S.; Skurnik, M. Bacteriophage φYeO3-12, specific for Yersinia enterocolitica serotype O: 3, is related to coliphages T3 and T7. J. Bacteriol. 2000, 182, 5114–5120. [Google Scholar] [CrossRef] [PubMed]
- Oduor, J.M.O.; Kadija, E.; Nyachieo, A.; Mureithi, M.W.; Skurnik, M. Bioprospecting Staphylococcus phages with therapeutic and bio-control potential. Viruses 2020, 12, 133. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Im, J.; Na, H.; Ryu, S.; Yun, C.-H.; Han, S.H. The novel Enterococcus phage vB_EfaS_HEf13 has broad lytic activity against clinical isolates of Enterococcus faecalis. Front. Microbiol. 2019, 10, 496990. [Google Scholar] [CrossRef]
- Rasool, M.H.; Yousaf, R.; Siddique, A.B.; Saqalein, M.; Khurshid, M. Isolation, characterization, and antibacterial activity of bacteriophages against methicillin-resistant Staphylococcus aureus in Pakistan. Jundishapur J. Microbiol. 2016, 9, e36135. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Krupovič, M.; Cvirkaitė-Krupovič, V.; Bamford, D.H. Identification and functional analysis of the Rz/Rz1-like accessory lysis genes in the membrane-containing bacteriophage PRD1. Mol. Microbiol. 2008, 68, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.; Savva, C.; Holzenburg, A.; Young, R. The lambda spanin components Rz and Rz1 undergo tertiary and quaternary rearrangements upon complex formation. Protein Sci. 2010, 19, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Hua, Y.; An, X.; Pei, G.; Li, S.; Wang, W.; Xu, X.; Fan, H.; Huang, Y.; Zhang, Z.; Mi, Z.; et al. Characterization of the morphology and genome of an Escherichia coli podovirus. Arch. Virol. 2014, 159, 3249–3256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, C.; Gao, R.; Yang, R. An ensemble method to distinguish bacteriophage virion from non-virion proteins based on protein sequence characteristics. Int. J. Mol. Sci. 2015, 16, 21734–21758. [Google Scholar] [CrossRef] [PubMed]
- Jurczak-Kurek, A.; Gąsior, T.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016, 6, 34338. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Coppenhagen-Glazer, S.; Shlezinger, M.; Kott-Gutkowski, M.; Adini, O.; Beyth, N.; Hazan, R. Complete genome sequence of Enterococcus bacteriophage EFLK1. Genome Announc. 2015, 3, e01308-15. [Google Scholar] [CrossRef]
- Wandro, S.; Oliver, A.; Gallagher, T.; Weihe, C.; England, W.; Martiny, J.B.; Whiteson, K. Predictable molecular adaptation of coevolving Enterococcus faecium and lytic phage EfV12-phi1. Front. Microbiol. 2019, 9, 428782. [Google Scholar] [CrossRef]
- Chapot, L.; Whatford, L.; Compston, P.; Tak, M.; Cuevas, S.; Garza, M.; Bennani, H.; Bin Aslam, H.; Hennessey, M.; Limon, G.J.S.; et al. A global media analysis of the impact of the COVID-19 pandemic on chicken meat food systems: Key vulnerabilities and opportunities for building resilience. Sustainability 2021, 13, 9435. [Google Scholar] [CrossRef]
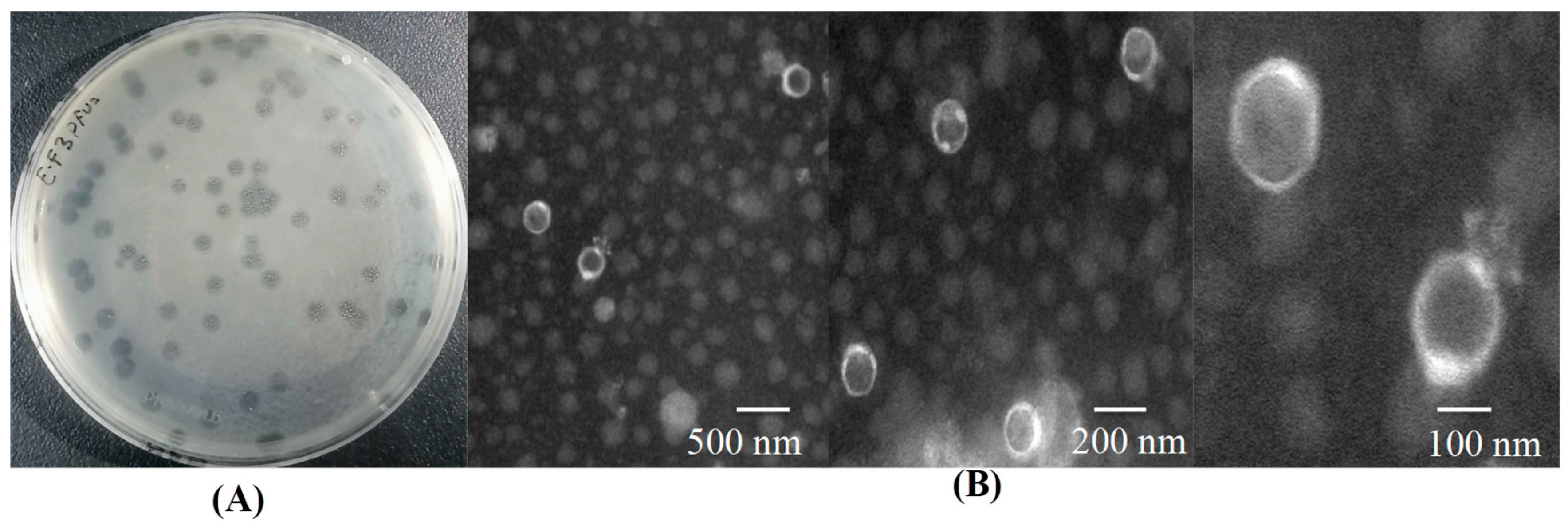
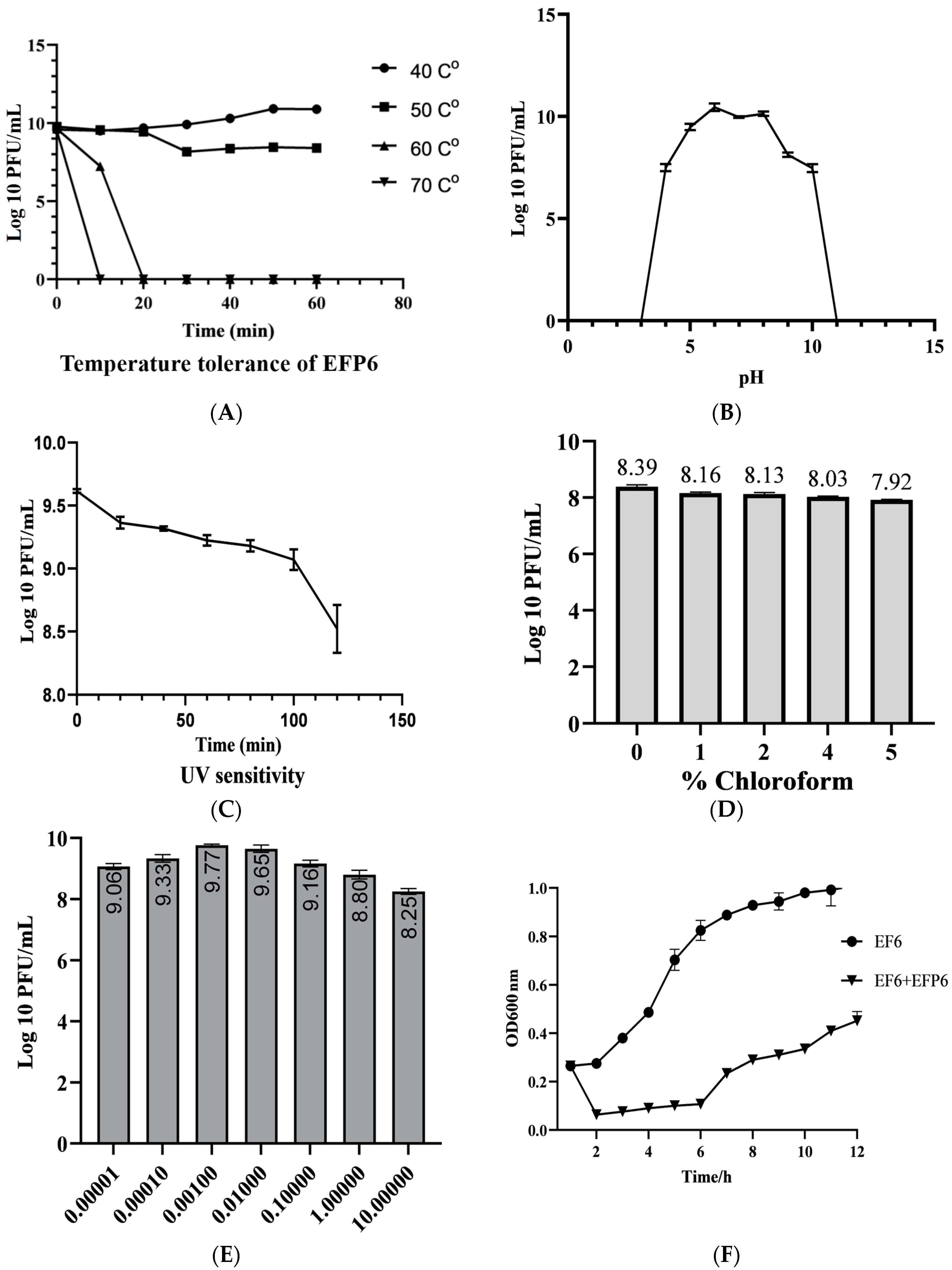
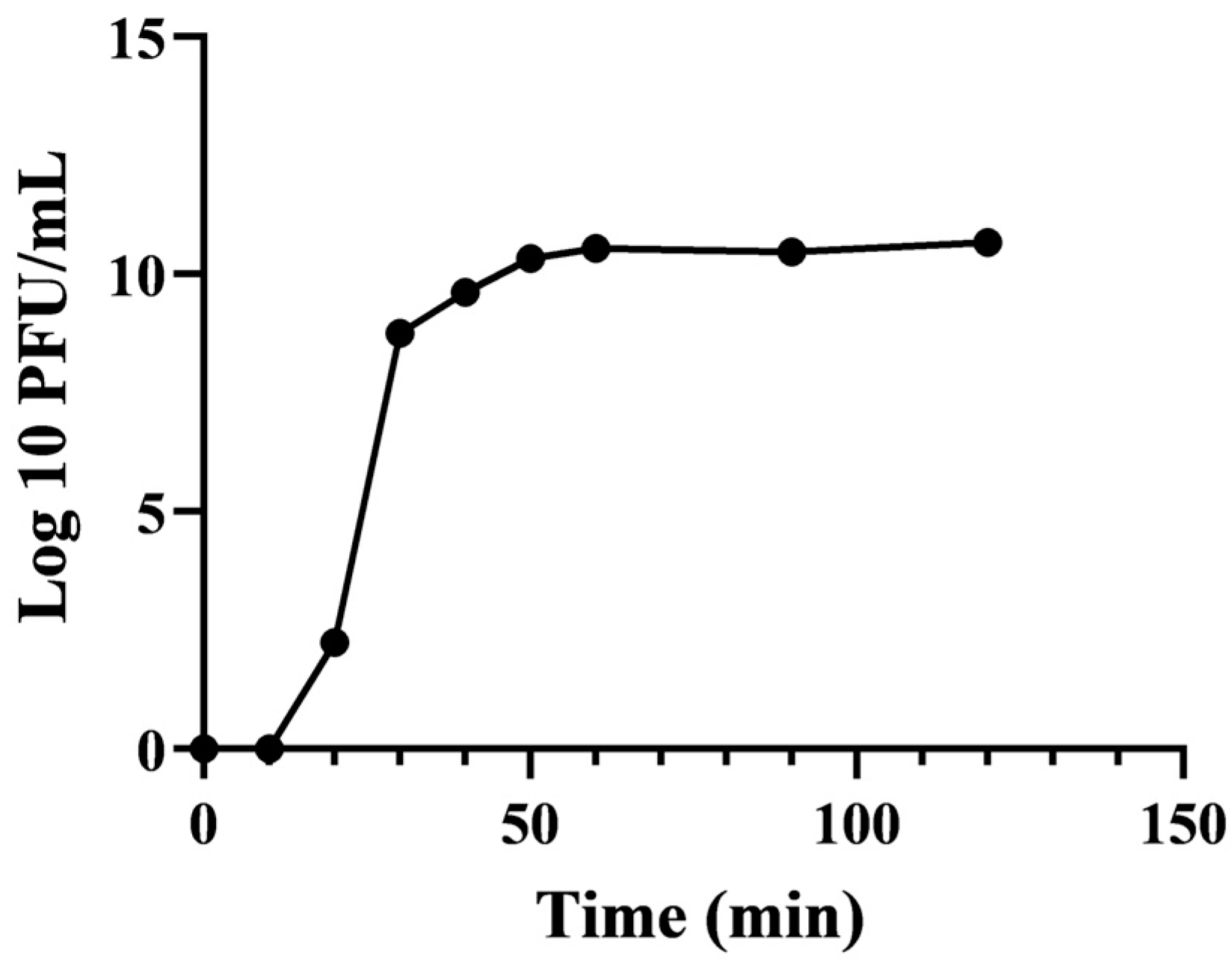
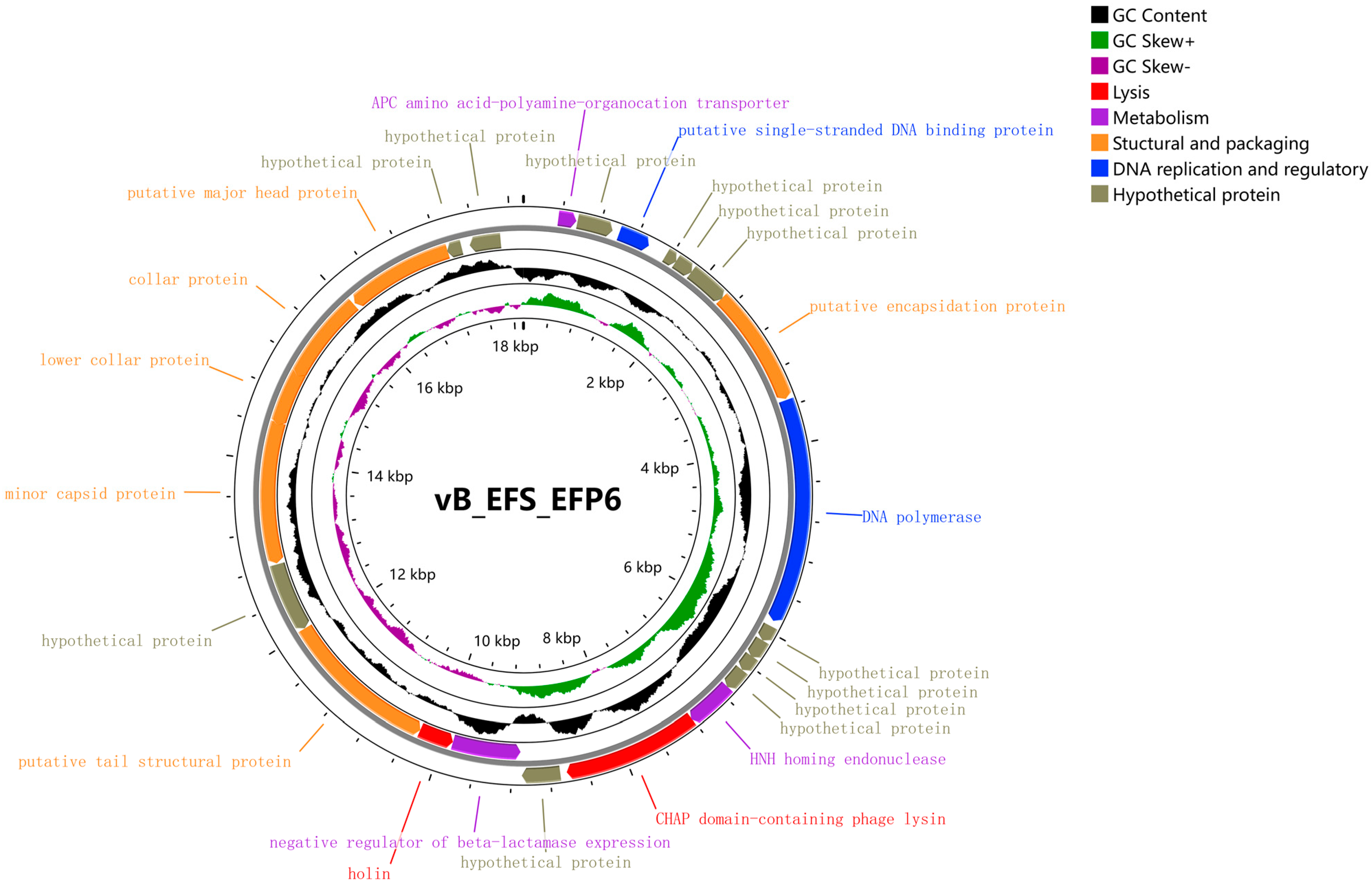



| NO. | Bacteria | Source | Phage Sensitivity | NO. | Bacteria | Source | Phage Sensitivity |
|---|---|---|---|---|---|---|---|
| 1 | Acidovorox | Dead embryo yolk | Resistant | 24 | E. coli 1 | Chicken liver | Resistant |
| 2 | B. bactirium | Dead embryo yolk | Resistant | 25 | E. coli 2 | Chicken liver | Resistant |
| 3 | A. baumannii 1 | Chicken liver | Resistant | 26 | E. coli 3 | Chicken liver | Resistant |
| 4 | A. baumannii 2 | Dead embryo yolk | Resistant | 27 | E. coli 4 | Chicken liver | Resistant |
| 5 | P. mirabilis 1 | Chicken blood | Resistant | 28 | E. coli 5 | Chicken liver | Resistant |
| 6 | P. mirabilis 2 | Chicken blood | Resistant | 29 | E. coli 6 | Chicken liver | Resistant |
| 7 | P. mirabilis 3 | Chicken blood | Resistant | 30 | E. coli 7 | Chicken liver | Resistant |
| 8 | P. mirabilis 4 | Dead embryo yolk | Resistant | 31 | E. coli 8 | Chicken liver | Resistant |
| 9 | P. mirabilis 5 | Dead embryo yolk | Resistant | 32 | E. coli 9 | Dead embryo yolk | Resistant |
| 10 | P. mirabilis 6 | Dead embryo yolk | Resistant | 33 | E. coli 10 | Dead embryo yolk | Resistant |
| 11 | P. mirabilis 7 | Dead embryo liver | Resistant | 34 | E. coli 11 | Dead embryo yolk | Resistant |
| 12 | P. mirabilis 8 | Dead embryo liver | Resistant | 35 | E. coli 12 | Dead embryo yolk | Resistant |
| 13 | S. aureus 1 | Chicken blood | Resistant | 36 | E. faecalis 1 | Dead embryo liver | Resistant |
| 14 | S. aureus 2 | Chicken blood | Resistant | 37 | E. faecalis 2 | Chicken blood | Resistant |
| 15 | Enterococcus spp. | Dead embryo yolk | Resistant | 38 | E. faecalis 3 | Dead embryo yolk | Resistant |
| 16 | S. argenteus 1 | Chicken blood | Resistant | 39 | E. faecalis 4 | Chicken blood | Resistant |
| 17 | S. argenteus 2 | Chicken blood | Resistant | 40 | E. faecalis 5 | Chicken blood | Susceptible |
| 18 | S. argenteus 3 | Chicken blood | Resistant | 41 | E. faecalis 6 | Chicken blood | Susceptible |
| 19 | S. argenteus 4 | Chicken blood | Resistant | 42 | E. faecalis 7 | Chicken liver | Susceptible |
| 20 | S. argenteus 5 | Chicken blood | Resistant | 43 | E. faecalis 8 | Dead embryo yolk | Susceptible |
| 21 | S. argenteus 6 | Chicken blood | Resistant | 44 | E. faecalis 9 | Chicken blood | Resistant |
| 22 | K. pneumoniae 1 | Chicken liver | Resistant | 45 | E. faecalis 10 | Chicken blood | Resistant |
| 23 | K. pneumoniae 2 | Chicken liver | Resistant |
| ORF | Start | Stop | F/R | Predicted Function | Best Match Phage | E Value | % Identity | Accession Number |
|---|---|---|---|---|---|---|---|---|
| EFP6_1 | 363 | 551 | F | APC amino acid–polyamine–organocation transporter | Enterococcus phage EFRM31 | 4 × 10−41 | 84.04% | NC_015270.1 |
| EFP6_2 | 564 | 938 | F | hypothetical protein | Enterococcus phage vB_EfaP_IME195 | 2 × 10−25 | 57.26% | YP_009191320.1 |
| EFP6_3 | 1012 | 1344 | F | putative single-stranded DNA binding protein | Enterococcus phage vB_Efae230P-4 | 5 × 10−72 | 98.18% | YP_009103979.1 |
| EFP6_4 | 1547 | 1672 | F | hypothetical protein | Enterococcus phage IME_EF3 | 1 × 10−19 | 85.85% | NC_023595.2 |
| EFP6_5 | 1669 | 1866 | F | hypothetical protein | Enterococcus phage vB_EfaP_IME195 | 5 × 10−8 | 39.34% | YP_009191325.1 |
| EFP6_6 | 1863 | 2285 | F | hypothetical protein | Enterococcus phage vB_EfaP_IME195 | 2 × 10−97 | 98.57% | YP_009191326.1 |
| EFP6_7 | 2278 | 3519 | F | putative encapsidation protein | Enterococcus phage vB_Efae230P-4 | 0.0 | 97.82% | YP_009103976.1 |
| EFP6_8 | 3532 | 5880 | F | DNA polymerase | Enterococcus phage vB_EfaP_IME195 | 0.0 | 97.44% | YP_009191328.1 |
| EFP6_9 | 5941 | 6111 | F | hypothetical protein | Enterococcus phage vB_EfaP_IME195 | 1 × 10−26 | 87.50% | YP_009191329.1 |
| EFP6_10 | 6111 | 6317 | F | hypothetical protein | Enterococcus phage vB_Efae230P-4 | 3 × 10−20 | 57.35% | YP_009103972.1 |
| EFP6_11 | 6308 | 8481 | F | hypothetical protein | Enterococcus phage vB_EfaP_IME195 | 2 × 10−24 | 84.21% | YP_009191331.1 |
| EFP6_12 | 6482 | 6715 | F | hypothetical protein | Enterococcus phage vB_EfaP_IME195 | 1 × 10−23 | 71.67% | YP_009191332.1 |
| EFP6_13 | 6712 | 7215 | F | HNH homing endonuclease | Enterococcus phage vB_EfaP_IME195 | 1 × 10−112 | 93.41% | YP_009191333.1 |
| EFP6_14 | 7208 | 8626 | F | CHAP domain-containing phage lysin | Enterococcus phage vB_EfaP_IME195 | 0.0 | 97.46% | YP_009191334.1 |
| EFP6_15 | 8694 | 9092 | F | hypothetical protein | Enterococcus phage vB_EfaP_IME195 | 4 × 10−28 | 87.93% | YP_009191335.1 |
| EFP6_16 | 9108 | 9881 | R | negative regulator of beta-lactamase expression | Enterococcus phage vB_EfaP_IME195 | 0.0 | 95.72% | YP_009191336.1 |
| EFP6_17 | 9882 | 10,274 | R | Holin | Enterococcus phage vB_EfaP_IME195 | 2 × 10−88 | 99.23% | YP_009191337.1 |
| EFP6_18 | 10,274 | 12,034 | R | putative tail structural protein | Enterococcus phage vB_Efae230P-4 | 0.0 | 95.90% | YP_009103964.1 |
| EFP6_19 | 12,036 | 12,824 | R | hypothetical protein | Enterococcus phage vB_EfaP_IME195 | 1 × 10−147 | 78.24% | YP_009191339.1 |
| EFP6_20 | 12,836 | 14,446 | R | minor capsid protein | Enterococcus phage vB_EfaP_IME195 | 0.0 | 91.17% | YP_009191340.1 |
| EFP6_21 | 14,415 | 15,056 | R | lower collar protein | Enterococcus phage vB_EfaP_IME195 | 8 × 10−124 | 97.75% | YP_009191341.1 |
| EFP6_22 | 15,013 | 16,050 | R | collar protein | Enterococcus phage vB_EfaP_IME195 | 0.0 | 95.38% | YP_009191342.1 |
| EFP6_23 | 16,067 | 17,278 | R | putative major head protein | Enterococcus phage vB_Efae230P-4 | 0.0 | 95.26% | YP_009103959.1 |
| EFP6_24 | 17,271 | 17,441 | R | hypothetical protein | Enterococcus phage vB_Efae230P-4.2 | 6 × 10−31 | 96.43% | YP_009103958.1 |
| EFP6_25 | 17,536 | 17,889 | R | hypothetical protein | Enterococcus phage vB_EfaP_IME195 | 9 × 10−14 | 85.48% | YP_009191345.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, S.; Leng, Q.; Hou, G.; Liang, Y.; Li, Y.; Qu, Y. Biological Traits and Comprehensive Genomic Analysis of Novel Enterococcus faecalis Bacteriophage EFP6. Microorganisms 2024, 12, 1202. https://doi.org/10.3390/microorganisms12061202
Ahmad S, Leng Q, Hou G, Liang Y, Li Y, Qu Y. Biological Traits and Comprehensive Genomic Analysis of Novel Enterococcus faecalis Bacteriophage EFP6. Microorganisms. 2024; 12(6):1202. https://doi.org/10.3390/microorganisms12061202
Chicago/Turabian StyleAhmad, Sajjad, Qingwen Leng, Gongmingzhu Hou, Yan Liang, Yanfang Li, and Yonggang Qu. 2024. "Biological Traits and Comprehensive Genomic Analysis of Novel Enterococcus faecalis Bacteriophage EFP6" Microorganisms 12, no. 6: 1202. https://doi.org/10.3390/microorganisms12061202
APA StyleAhmad, S., Leng, Q., Hou, G., Liang, Y., Li, Y., & Qu, Y. (2024). Biological Traits and Comprehensive Genomic Analysis of Novel Enterococcus faecalis Bacteriophage EFP6. Microorganisms, 12(6), 1202. https://doi.org/10.3390/microorganisms12061202





