Development of Multiplex RT qPCR Assays for Simultaneous Detection and Quantification of Faecal Indicator Bacteria in Bathing Recreational Waters
Abstract
1. Introduction
2. Materials and Methods
2.1. Target Selection and Design of Primers and TaqMan Probes
2.2. In Silico Specificity Validation
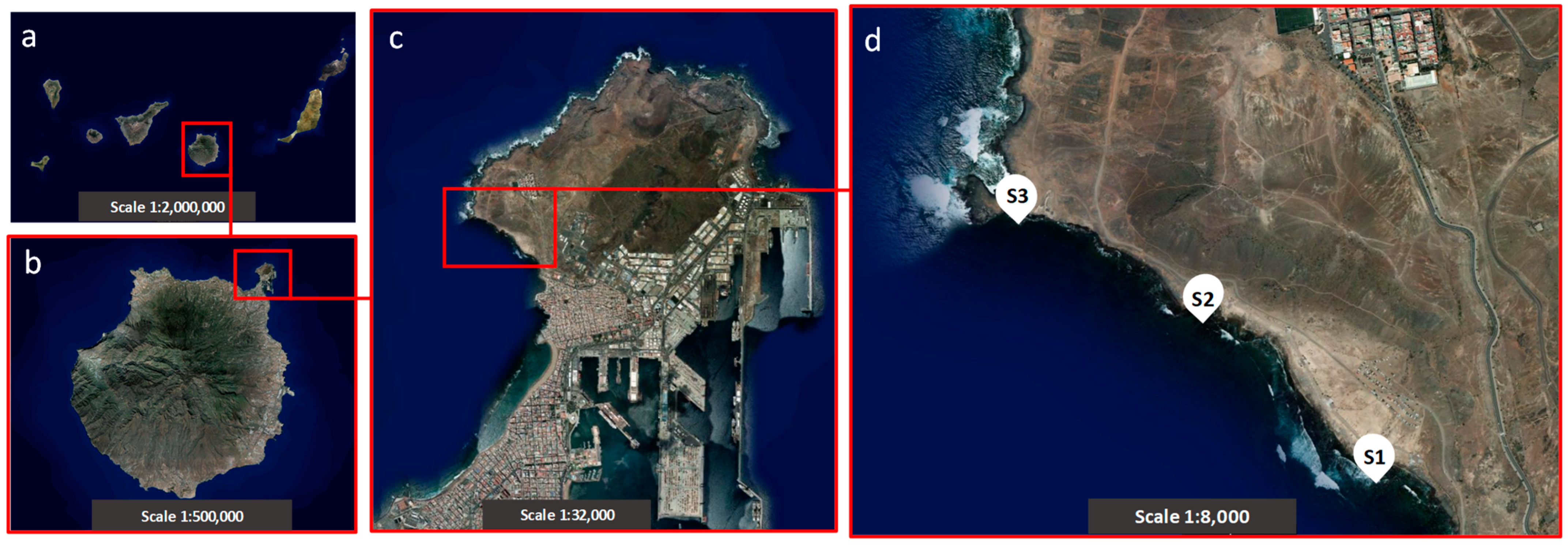
2.3. Study Area and Sample Collection and Preparation
2.4. DNA Extraction
2.5. Multiplex RT qPCR Amplification
2.5.1. Construction of Standard Curves for Gene Copy Number Determination
2.5.2. Quantification of Faecal Indicator Bacteria
2.6. Data Analysis
3. Results
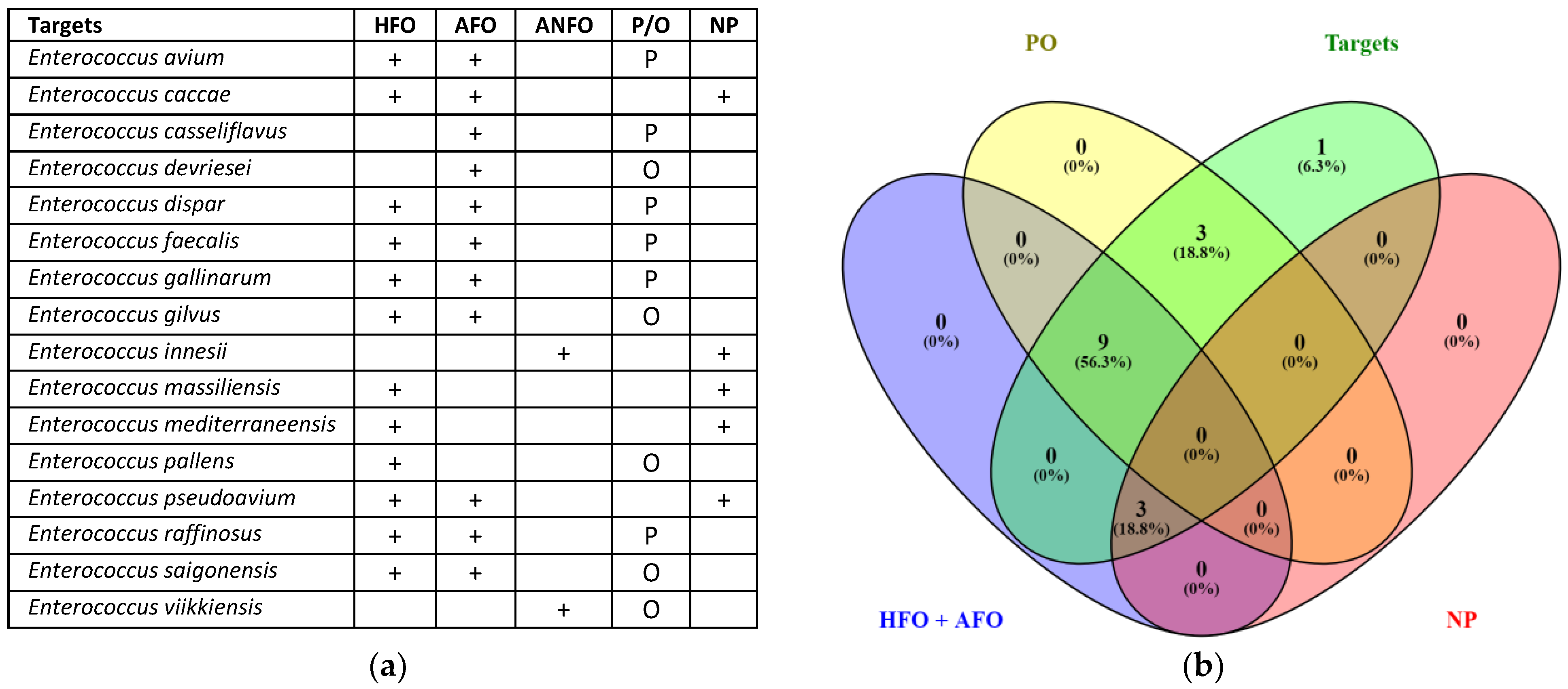
3.1. In Silico Validation for Group- and Species-Specific
3.2. Multiplex RT qPCR Amplification
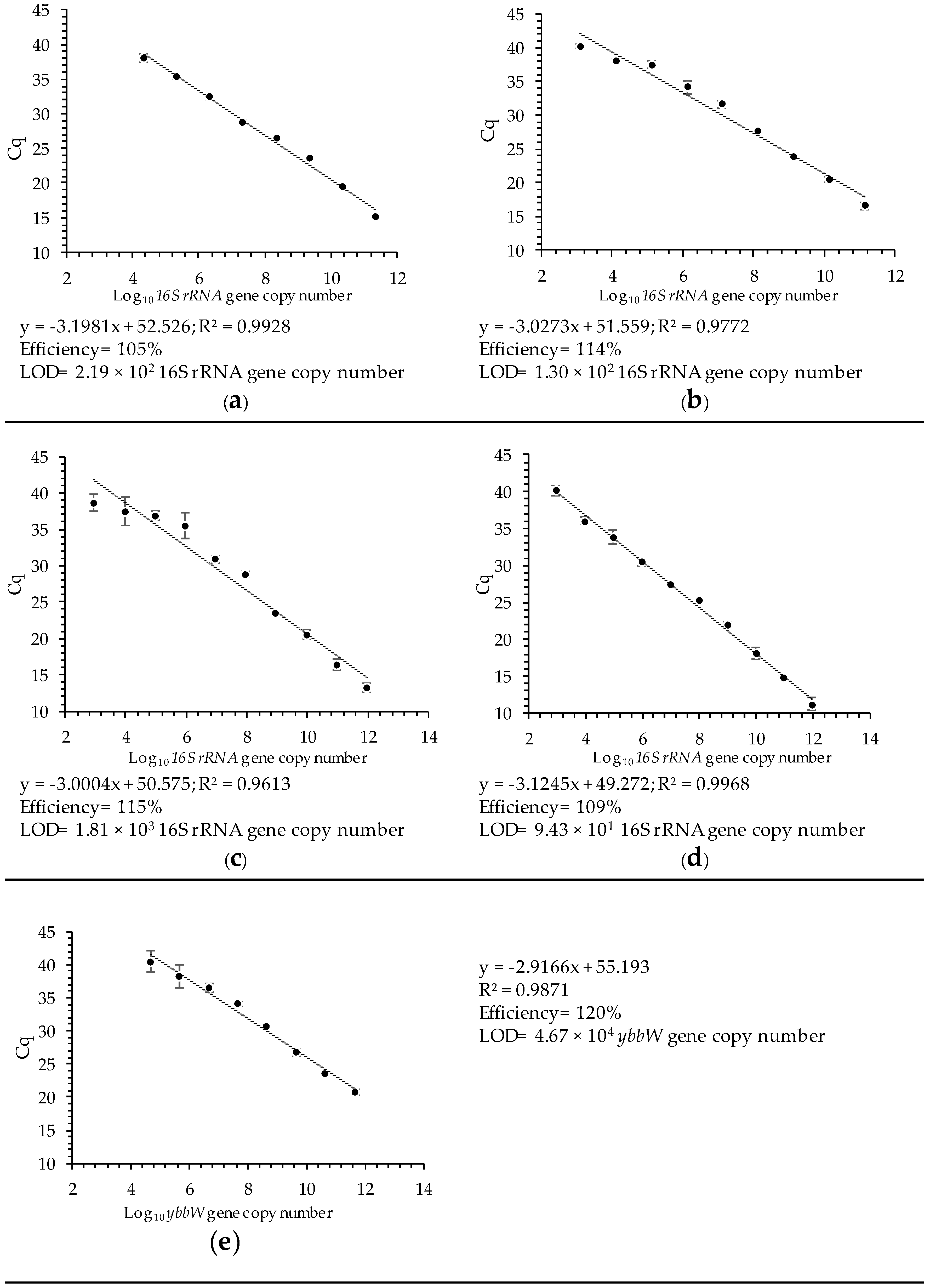
3.2.1. Linearity and Analytical Sensitivity of the Multiplex RT qPCR Assays
3.2.2. Quantification of Faecal Indicator Bacteria
4. Discussion
4.1. In Silico Validation for Group- and Species-Specific Data
4.2. Quantification of Faecal Indicator Bacteria
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levy, K.; Woster, A.P.; Goldstein, R.S.; Carlton, E.J. Untangling the impacts of climate change on waterborne diseases: A systematic review of relationships between diarrheal diseases and temperature, rainfall, flooding, and drought. Environ. Sci. Tech. 2016, 50, 4905–4922. [Google Scholar] [CrossRef]
- Kucuksezgin, F.; Gonul, L.T.; Pazi, I.; Kacar, A. Assessment of seasonal and spatial variation of surface water quality: Recognition of environmental variables and fecal indicator bacteria of the coastal zones of Izmir Bay, Eastern Aegean. Reg. Stud. Mar. Sci. 2020, 28, 100554. [Google Scholar] [CrossRef]
- Rafiee, M.; Hosseini, S.A.; Gholami-Borujeni, F.; Hesami Arani, M.; Niknejad, H. Health risk assessment of swimming beaches microbial contamination: A case study-Mahmoudabad, Iran. Int. J. Environ. Health Res. 2024, 34, 355–366. [Google Scholar] [CrossRef]
- Saleem, F.; Li, E.; Edge, T.A.; Tran, K.L.; Schellhorn, H.E. Identification of potential microbial risk factors associated with fecal indicator exceedances at recreational beaches. Environ. Microbiome 2024, 19, 4. [Google Scholar] [CrossRef]
- Stewart, J.R.; Fleming, L.E.; Fleisher, J.M.; Abdelzaher, A.M.; Lyons, M.M. Waterborne pathogens. In Marine Pollution and Human Health; Hester, R.E., Ed.; Royal Society of Chemistry: London, UK, 2011; pp. 25–67. [Google Scholar]
- Wade, T.J.; Pai, N.; Eisenberg, J.N.; Colford, J.M., Jr. Do US Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 2017, 111, 1102. [Google Scholar] [CrossRef]
- Kumar, N.; Hu, Y.; Singh, S.; Mizaikoff, B. Emerging biosensor platforms for the assessment of water-borne pathogens. Analyst 2018, 143, 359–373. [Google Scholar] [CrossRef]
- Archer, E.J.; Baker-Austin, C.; Osborn, T.J.; Jones, N.R.; Martínez-Urtaza, J.; Trinanes, J.; Oliver, J.D.; Colón-González, F.J.; Lake, I.R. Climate warming and increasing Vibrio vulnificus infections in North America. Sci. Rep. 2023, 13, 3893. [Google Scholar] [CrossRef]
- Yau, V.; Wade, T.J.; de Wilde, C.K.; Colford, J.M. Skin-related symptoms following exposure to recreational water: A systematic review and meta-analysis. Water Qual. Expo. Health 2009, 1, 79–103. [Google Scholar] [CrossRef]
- Su, Y.; Gao, R.; Huang, F.; Liang, B.; Guo, J.; Fan, L.; Wang, A.; Gao, S.H. Occurrence, transmission and risks assessment of pathogens in aquatic environments accessible to humans. J. Environ. Manag. 2024, 354, 120331. [Google Scholar] [CrossRef]
- McClung, R.P.; Roth, D.M.; Vigar, M.; Roberts, V.A.; Kahler, A.M.; Cooley, L.A.; Hilborn, E.D.; Wade, T.J.; Fullerton, K.E.; Yoder, J.S.; et al. Waterborne disease outbreaks associated with environmental and undetermined exposures to water—United States, 2013–2014. Morb. Mortal Wkly. Rep. 2017, 66, 1222. [Google Scholar] [CrossRef]
- United Nations. Transforming Our World: The 2030 Agenda for Sustainable Development. 2022. Available online: https://sdgs.un.org/2030agenda (accessed on 2 April 2024).
- United Nations. Decade of Ocean Science for Sustainable Development. 2023. Available online: https://oceandecade.org/actions/protection-and-sustainable-use-of-marine-areas-sustainmare (accessed on 6 April 2024).
- Issifu, I.; Dahmouni, I.; García-Lorenzo, I.; Sumaila, U.R. Economics in Marine Spatial Planning: A Review of Issues in British Columbia and Similar Jurisdictions. Sustainability 2024, 16, 1210. [Google Scholar] [CrossRef]
- European Parliament. Directive 2006/7/EC of the European Parliament, concerning the management of bathing water quality and repealing directive 76/160/EEC. Off. J. Eur. Commun. 2006, 64, 37–51. [Google Scholar]
- Ahmed, W.; Payyappat, S.; Cassidy, M.; Besley, C. A duplex PCR assay for the simultaneous quantification of Bacteroides HF183 and crAssphage CPQ_056 marker genes in untreated sewage and stormwater. Environ. Int. 2019, 126, 252–259. [Google Scholar] [CrossRef]
- Holcomb, D.A.; Knee, J.; Sumner, T.; Adriano, Z.; de Bruijn, E.; Nalá, R.; Cumming, O.; Brown, J.; Stewart, J.R. Human Fecal Contamination of Water, Soil, and Surfaces in Households Sharing Poor-Quality Sanitation Facilities in Maputo, Mozambique. Int. J. Hyg. Environ. Health 2020, 226, 113496. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Wang, Y.; Liu, F.; Chen, G. Establishment of a multiplex polymerase chain reaction assay for the detection of marine harmful algae. J. Appl. Phycol. 2024, 36, 243–258. [Google Scholar] [CrossRef]
- USEPA. Recreational Water Quality Criteria; Office of Water, United States Environmental Potection Agency: Washington, DC, USA, 2012.
- Shahraki, A.H.; Heath, D.; Chaganti, S.R. Recreational water monitoring: Nanofluidic qRT-PCR chip for assessing beach water safety. Environ. DNA 2019, 1, 305–315. [Google Scholar] [CrossRef]
- Alía, A.; Andrade, M.J.; Córdoba, J.J.; Martín, I.; Rodríguez, A. Development of a multiplex real-time PCR to differentiate the four major Listeria monocytogenes serotypes in isolates from meat processing plants. Food Microbiol. 2020, 87, 103367. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, L.; Brodsky, J.; Gablech, I.; Xu, F.; Li, Z.; Korabecna, M.; Neuzil, P. Quantitative or digital PCR? A comparative analysis for choosing the optimal one for biosensing applications. TrAC Trends Anal. Chem. 2024, 174, 117676. [Google Scholar] [CrossRef]
- Mekadim, C.; Killer, J.; Mrázek, J.; Bunešová, V.; Pechar, R.; Hroncová, Z.; Vlková, E. Evaluation of the infB and rpsB gene fragments as genetic markers intended for identification and phylogenetic analysis of particular representatives of the order Lactobacillales. Arch. Microbiol. 2018, 200, 1427–1437. [Google Scholar] [CrossRef]
- Deshmukh, R.; Prusty, A.K.; Roy, U.; Bhand, S. A capacitive DNA sensor for sensitive detection of Escherichia coli O157: H7 in potable water based on the z3276 genetic marker: Fabrication and analytical performance. Analyst 2020, 145, 2267–2278. [Google Scholar] [CrossRef]
- Wasiewska, L.A.; Juska, V.B.; Seymour, I.; Burgess, C.M.; Duffy, G.; O’Riordan, A. Electrochemical nucleic acid-based sensors for detection of Escherichia coli and Shiga toxin-producing E. coli—Review of the recent developments. Compr. Rev. Food Sci. Food Saf. 2023, 22, 1839–1863. [Google Scholar]
- Maheshwari, K.; Musyuni, P.; Moulick, A.; Mishra, H.; Ekielski, A.; Mishra, P.K.; Aggarwal, G. Unveiling the Microbial Symphony: Next-Gen Sequencing and Bioinformatics Insights into the Human Gut Microbiome. Health Sci. Rev. 2024, 11, 100173. [Google Scholar] [CrossRef]
- Silkie, S.S.; Tolcher, M.P.; Nelson, K.L. Reagent decontamination to eliminate false-positives in Escherichia coli qPCR. J. Microbiol. Methods 2008, 72, 275–282. [Google Scholar] [CrossRef]
- Walker, D.I.; McQuillan, J.; Taiwo, M.; Parks, R.; Stenton, C.A.; Morgan, H.; Lees, D.N. A highly specific Escherichia coli qPCR and its comparison with existing methods for environmental waters. Water Res. 2017, 126, 101–110. [Google Scholar] [CrossRef]
- Chevée, V.; Sachar, U.; Yadav, S.; Heryanto, C.; Eleftherianos, I. The peptidoglycan recognition protein PGRP-LE regulates the Drosophila immune response against the pathogen Photorhabdus. Microb. Pathog. 2019, 136, 103664. [Google Scholar] [CrossRef]
- McQuillan, J.S.; Wilson, M.W. ‘Ready Mixed’, improved nucleic acid amplification assays for the detection of Escherichia coli DNA and RNA. J. Microbiol. Methods 2019, 165, 105721. [Google Scholar] [CrossRef]
- McQuillan, J.S.; Wilson, M.W. Recombinase polymerase amplification for fast, selective, DNA-based detection of faecal indicator Escherichia coli. Lett. Appl. Microbiol. 2021, 72, 382–389. [Google Scholar] [CrossRef]
- Nallapareddy, S.R.; Wenxiang, H.; Weinstock, G.M.; Murray, B.E. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J. Bacteriol. 2005, 187, 5709–5718. [Google Scholar] [CrossRef]
- Palmer, K.L.; Godfrey, P.; Griggs, A.; Kos, V.N.; Zucker, J.; Desjardins, C.; Cerqueira, G.; Gevers, D.; Walker, S.; Wortman, J.; et al. Comparative genomics of enterococci: Variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 2012, 3, e00318-112012. [Google Scholar]
- Gotkowska-Płachta, A. The Prevalence of Virulent and Multidrug-Resistant Enterococci in River Water and in Treated and Untreated Municipal and Hospital Wastewater. Int. J. Environ. Res. Public Health 2021, 18, 563. [Google Scholar] [CrossRef]
- Mullally, C.A.; Fahriani, M.; Mowlaboccus, S.; Coombs, G.W. Non-faecium non-faecalis enterococci: A review of clinical manifestations, virulence factors, and antimicrobial resistance. CMR 2024, 37, e00121-23. [Google Scholar]
- de Kraker, M.E.A.; Jarlier, V.; Monen, J.C.M.; Heuer, O.E.; van de Sande, N.; Grundmann, H. The changing epidemiology of bacteraemias in Europe: Trends from the European Antimicrobial Resistance Surveillance System. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2013, 19, 860–868. [Google Scholar] [CrossRef]
- Sievert, D.M.; Ricks, P.; Edwards, J.R.; Schneider, A.; Patel, J.; Srinivasan, A.; Kallen, A.; Limbago, B.; Fridkin, S. Antimicrobial-resistant pathogens associated with health care associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 2013, 34, 1–14. [Google Scholar] [CrossRef]
- O’Mullan, G.D.; Juhl, A.R.; Reichert, R.; Schneider, E.; Martinez, N. Patterns of sediment-associated fecal indicator bacteria in an urban estuary: Benthic-pelagic coupling and implications for shoreline water quality. Sci. Total Environ. 2019, 656, 1168–1177. [Google Scholar] [CrossRef]
- Palmer, J.A.; Law, J.Y.; Soupir, M.L. Spatial and temporal distribution of E. coli contamination on three inland lake and recreational beach systems in the upper Midwestern United States. Sci. Total Environ. 2020, 722, 137846. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014, 42, D613–D616. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef]
- Gentilini, F.; Turba, M.E.; Calzolari, C.; Cinotti, S.; Forni, M.; Zannoni, A. Real time quantitative PCR using hairpin-shaped clone-specific primers for minimal residual disease assessment in an animal model of human non-Hodgkin lymphoma. Mol. Cel. Probes 2010, 24, 6–14. [Google Scholar] [CrossRef]
- Shen, Z.; Qu, W.; Wang, W.; Lu, Y.; Wu, Y.; Li, Z.; Hang, X.; Wang, X.; Zhao, D.; Zhang, C. MPprimer: A program for reliable multiplex PCR primer design. BMC Bioinform. 2010, 11, 143. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- San Millán, R.M.; Martínez-Ballesteros, I.; Rementeria, A.; Garaizar, J.; Bikandi, J. Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC Res. Notes 2013, 6, 513. [Google Scholar] [CrossRef] [PubMed]
- Luque-Escalona, A. Las áreas marinas protegidas en Canarias. Cartas Urbanas 2004, 10, 52–171. [Google Scholar]
- Ojeda, M.A.; Alonso, I.; Alcántara Carrió, J. Cartografía sedimentaria de la bahía de El Confital (Gran Canaria). Consideraciones sobre el transporte de sedimentos hacia la playa de Las Canteras. Geogaceta 1996, 20, 374–377. [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Acosta, M.; Garcia-Jimenez, P.; Herrera-Melián, J.A.; Peñate-Castellano, N.; Rivero-Rosales, A. The effects of plants on pollutant removal, clogging, and bacterial community structure in palm mulch-based vertical flow constructed wetlands. Sustainability 2019, 11, 632. [Google Scholar] [CrossRef]
- Dubin, K.; Pamer, E.G. Enterococci and their interactions with the intestinal microbiome. Microbiol. Spectr. 2017, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Wittwer, C.T. MIQE: A step toward more robust and reproducible quantitative PCR. Clin. Chem. 2017, 63, 1537–1538. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-Time PCR; Springer: Heidelberg/Berlin, Germany, 2001; pp. 21–34. [Google Scholar]
- Whelan, J.A.; Russell, N.B.; Whelan, M.A. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods 2003, 278, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. 2007–2015. Available online: https://biinfogp.cnb.csic.es/tools/venny/index.html (accessed on 10 February 2024).
- Orsi, R.H.; Stoppe, N.C.; Sato, M.I.Z.; Gomes, T.A.; Prado, P.I.; Manfio, G.P.; Ottoboni, L.M. Genetic variability and pathogenicity potential of Escherichia coli isolated from recreational water reservoirs. Res. Microbiol. 2007, 158, 420–427. [Google Scholar] [CrossRef]
- Maheux, A.F.; Picard, F.J.; Boissinot, M.; Bissonnette, L.; Paradis, S.; Bergeron, M.G. Analytical comparison of nine PCR primer sets designed to detect the presence of Escherichia coli/Shigella in water samples. Water Res. 2009, 43, 3019–3028. [Google Scholar] [CrossRef] [PubMed]
- Chattaway, M.A.; Schaefer, U.; Tewolde, R.; Dallman, T.J.; Jenkins, C. Identification of Escherichia coli and Shigella species from whole-genome sequences. J. Clin. Microbiol. 2017, 55, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Lan, R.; Reeves, P.R. Escherichia coli in disguise: Molecular origins of Shigella. Microb. Infect. 2002, 4, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Ragupathi, N.D.; Sethuvel, D.M.; Inbanathan, F.; Veeraraghavan, B. Accurate differentiation of Escherichia coli and Shigella serogroups: Challenges and strategies. New Microbes New Infect. 2018, 21, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Bej, A.K.; Mahbubani, M.H.; Atlas, R.M. Detection of viable Legionella pneumophila in water by polymerase chain reaction. Appl. Environ. Microbiol. 1991, 57, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Devane, M.L.; Moriarty, E.; Weaver, L.; Cookson, A.; Gilpin, B. Fecal indicator bacteria from environmental sources; strategies for identification to improve water quality monitoring. Water Res. 2020, 185, 116204. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Lee, J.E.; Lee, S.; Lee, H.T.; Hur, H.G.; Ko, G. Characterization of Enterococcus spp. from human and animal feces using 16S rRNA sequences, the esp gene, and PFGE for microbial source tracking in Korea. Environ. Sci. Tech. 2010, 44, 3423–3428. [Google Scholar] [CrossRef] [PubMed]
- Naphtali, P.; Mohiuddin, M.M.; Paschos, A.; Schellhorn, H.E. Application of high-throughput 16S rRNA sequencing to identify fecal contamination sources and to complement the detection of fecal indicator bacteria in rural groundwater. J. Water Health 2019, 17, 393–403. [Google Scholar] [CrossRef]
- Tamai, S.; Suzuki, Y. Diversity of fecal indicator enterococci among different hosts: Importance to water contamination source tracking. Microorganisms 2023, 11, 2981. [Google Scholar] [CrossRef]
- Karunakaran, E.; Battarbee, R.; Tait, S.; Brentan, B.M.; Berney, C.; Grinham, J.; Herrero, M.A.; Omolo, R.; Douterelo, I. Integrating molecular microbial methods to improve faecal pollution xmanagement in rivers with designated bathing waters. Sci. Total Environ. 2024, 912, 168565. [Google Scholar] [CrossRef]
- World Health Organization. Microbial aspects of beach sand quality. In Guidelines for Safe Recreational Water Environments, Coastal and Fresh Waters; World Health Organization, Ed.; World Health Organization: Geneva, Switzerland, 2003; Volume 1, pp. 118–126. [Google Scholar]
- Lane, M.J.; McNair, J.N.; Rediske, R.R.; Briggs, S.; Sivaganesan, M.; Haugland, R. Simplified analysis of measurement data from a rapid E. coli qPCR method (EPA Draft Method C) using a standardized Excel workbook. Water 2020, 12, 775. [Google Scholar] [CrossRef] [PubMed]
- Fratamico, P.M.; Strobaugh, T.P. Simultaneous detection of Salmonella spp. and Escherichia coli O157:H7 by multiplex PCR. J. Ind. Microbiol. Biotechnol. 1998, 21, 92–98. [Google Scholar] [CrossRef]
- Hadland, N.; Hamilton, C.W.; Duhamel, S. Young volcanic terrains are windows into early microbial colonization. Commun. Earth Environ. 2024, 5, 114. [Google Scholar] [CrossRef]
- Xiao, Z.; Goraya, M.U.; Ali, L.; Chen, X.; Yu, D. Nitrogen and phosphorus eutrophication enhance biofilm-related drug resistance in Enterococcus faecalis isolated from Water Sources. Microb. Pathog. 2024, 186, 106501. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Almeida, A.; Kumar, N.; Vervier, K.; Adoum, A.T.; Viciani, E.; Dawson, N.J.R.; Forster, S.C.; Cormie, C.; Goulding, D.; et al. Host adaptation in gut Firmicutes is associated with sporulation loss and altered transmission cycle. Genome Biol. 2021, 22, 204. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Pérez, M.C.; Anfuso, E.; Acevedo, A.; Perales-Vargas-Machuca, J.A. Microbial indicators of faecal contamination in waters and sediments of beach bathing zones. Int. J. Hyg. Environ. Health 2008, 211, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Weiskerger, C.J.; Brandão, J.; Ahmed, W.; Aslan, A.; Avolio, L.; Badgley, B.D.; Boehm, A.B.; Edge, T.A.; Fleisher, J.M.; Heaney, C.D.; et al. Impacts of a changing earth on microbial dynamics and human health risks in the continuum between beach water and sand. Water Res. 2019, 162, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Rumball, N.A.; Mayer, H.C.; McLellan, S.L. Selective survival of Escherichia coli phylotypes in freshwater beach sand. Appl. Environ. Microbiol. 2021, 87, e02473-20. [Google Scholar] [CrossRef]
- Koskey, A.M. Transcending Microbial Source Tracking Techniques Across Geographic Borders: An Examination of Human and Animal Microbiomes and the Integration of Molecular Approaches in Pathogen Surveillance in Brazil and the United States. Ph.D. Thesis, University of Wisconsin-Milwaukee, Milwaukee, WI, USA, 2013; p. 125. [Google Scholar]
- Kleinheinz, G.T.; McDermott, C.M.; Chomeau, V. Evaluation of avian waste and bird counts as predictors of Escherichia coli contamination at Door County, Wisconsin beaches. J. Great Lakes Res. 2006, 32, 117–123. [Google Scholar] [CrossRef]
| Target | FIB | Gene | Primers and Probes | Sequence (5′–3′) a | Product Size (bp) |
|---|---|---|---|---|---|
| Group-specific | Enterococci | 16S rRNA | EF399F EF484R ProbeEN | F: GCTCGTGTCGTGAGATGTT R: CGCTAGAGTGCCCAACTAAA P: [HEX]-AACAATAAG-[ZEN]-GGTTGCGCTCGTTGC-[IABkFQ] | 86 |
| Escherichia/Shigella | 16S rRNA | EC501F EC604R ProbeES | F: TCGGAATTACTGGGCGTAAAG R: GACTCAAGCTTGCCAGTATCA P: [6FAM]ACGCAGGCG-[ZEN]-GTTTGTTAAGTCAGA-[IABkFQ] | 104 | |
| Specie-specific | Enterococcus faecalis | 16S rRNA | EF1203F EF1305R ProbeEf | F: CTTATGACCTGGGCTACACAC R: CTGCAATCCGAACTGAGAGAA P: [Cy5]-ACCGCGAGG-[TAO]-TCATGCAAATCTCTT-[3IAbRQSp] | 103 |
| Enterococcus faecium | 16S rRNA | EFium96F EFium225R ProbeEFium | F: TGGCGAACGGGTGAGTAA R: TCCATCCATCAGCGACAC P: [6FAM]-CAAAACCGC-[ZEN]-ATGGTTTTGATTTGAAAGGCG-[3IABkFQ] | 140 | |
| E. coli | ybbW | ybbWF ybbWR ProbeybbW | F: GCAAAATCTGGCCGGGAT R: AATCGCCCAAATCGCCA P: [HEX]-CACTGCCAT-[ZEN]-TCTTAACCCGTGCATC-[3IABkFQ] | 203 |
| Percentage of Coverage in the Target Taxon (%) a % (Number of Sequences Matched/Total Number of Taxon Sequences Deposited) | ||||
|---|---|---|---|---|
| Target Assay Primers and Probes | Enterococci (Group-Specific) EF399F, EF484R, ProbeEN | Enterococcus faecalis (Species-Specific) EF1203F, EF1305R, ProbeEf | Enterococcus faecium (Species-Specific) EFium96F, Fium225R, ProbeEFium | |
| Target Taxon | ||||
| Bacteria Domain | 0.4 (1582/381,535) | 0.1 (302/381,535) | 0.0 (180/381,535) | |
| Desulfobacterota | 0.4 (1/7818) | 0.0 (0/7818) | 0.0 (0/7818) | |
| Proteobacteria | 0.0 (22/109,146) | 0.0 (1/109,146) | 0.0 (0/109,146) | |
| Firmicutes | 1.6 (1559/100,538) | 0.3 (301/100,538) | 0.2 (180/100,538) | |
| Dethiobacteria | 1.5 (2/137) | 0.0 (0/137) | 0.0 (0/137) | |
| Clostridia | 0.1 (32/50,639) | 0.0 (2/50,639) | 0.0 (0/50,639) | |
| Bacilli | 3.6 (1525/42,707) | 0.7 (299/42,707) | 0.7 (180/42,707) | |
| Lactobacillales | 12.0 (1525/13,176) | 19.0 (295/13,176) | 1.4 (180/13,176) | |
| Aerococcaceae | 0.8 (2/264) | 0.0 (0/264) | 0.0 (0/264) | |
| Streptococcaceae | 9.7 (520/5371) | 0.0 (1/5371) | 0.0 (1/5371) | |
| Lactobacillaceae | 0.2 (11/4739) | 0.0 (2/4739) | 0.0 (0/4739) | |
| Vagococcaceae | 78.3 (36/46) | 0.0 (0/46) | 0.0 (0/46) | |
| Enterococcaceae | 59.8 (956/1600) | 18.4 (292/1600) | 11.3 (179/1600) | |
| Melissococcus | 100.0 (9/9) | 0.0 (0/9) | 0.0 (0/9) | |
| Enterococcus | 61.1 (947/1551) | 19.0 (292/1551) | 11.7 (179/1551) | |
| Enterococcus faecalis | - | 80.2 (203/253) | 0.8 (3/253) | |
| Enterococcus faecium | - | 0.4 (1/261) | 49.4 (129/261) | |
| Enterococcus sp. | - | 16.0 (28/175) | 13.1 (23/175) | |
| Uncultured Enterococcus sp. | - | 23.1 (52/225) | 7.6 (17/225) | |
| Target Taxon | Percentage of Coverage in the Target Taxon (%) a % (Number of Sequences Matched/Total Number of Taxon Sequences Deposited) |
|---|---|
| Bacteria Domain | 0.6 (2153/381,535) |
| Firmicutes | 0.0 (5/100,538) |
| Proteobacteria | 2.0 (2147/109,146) |
| Burkholderiales | 0.0 (2/20,421) |
| Pseudomonadales | 0.0 (2/17,709) |
| Gammaproteobacteria | 2.9 (2147/75,026) |
| Enterobacterales | 9.3 (2143/23,028) |
| Erwiniaceae | 0.1 (1/1233) |
| Pectobacteriaceae | 0.2 (1/433) |
| Enterobacteriaceae | 27.0 (2141/7969) |
| Citrobacter | 0.2 (1/516) |
| Enterobacter | 0.2 (3/1670) |
| Klebsiella | 0.4 (5/1177) |
| Salmonella | 0.8 (8/952) |
| Escherichia/Shigella | 76.0 (2124/2807) |
| Escherichia coli | 98.2 (1019/1038) |
| Escherichia fergusonii | 95.7 (22/23) |
| Escherichia albertii | 27.3 (6/22) |
| Shigella flexneri | 100.0 (53/53) |
| Shigella boydii | 100.0 (24/24) |
| Shigella dysenteriae | 87.2 (34/39) |
| Shigella sonnei | 91.4 (32/35) |
| Escherichia sp. | 94.4 (34/36) |
| Shigella sp. | 90.9 (10/11) |
| Escherichia/Shigella b | 68.8 (242/352) |
| Uncultured Escherichia sp. | 28.6 (6/21) |
| Uncultured Shigella sp. | 70.0 (14/20) |
| Uncultured Escherichia/Shigella c | 59.6 (628/1054) |
| Sampling | Seawater (Gene Copy Number in 100 mL−1) | Marine Sediments (Gene Copy Number in 100 g−1) | |||
|---|---|---|---|---|---|
| Period | Site | Enterococci | Escherichia/Shigella | Enterococci | Escherichia/Shigella |
| Winter | S1 | 2.4 (1.7) × 107 c | 7.7 (3.4) × 105 c | 8.2 (0.2) × 108 | 2.4 (1.8) × 105 a;c |
| S2 | 1.2 (0.6) × 107 | 2.7 (0.9) × 106 a | 7.5 (0.3) × 109 | 2.1 (0.9) × 107 c | |
| S3 | 1.5 (0.1) × 107 c | 3.8 (3.1) × 104 a | 9.9 (0.7) × 105 a;c | 3.0 (2.2) × 107 c | |
| Spring | S1 | 1.8 (0.2) × 107 | 1.8 (0.6) × 106 | 4.5 (1.7) × 105 b | 1.2 (1.0) × 105 a |
| S2 | 6.3 (1.0) × 107 | 2.4 (0.8) × 106 | 1.8 (1.3) × 105 b | 9.2 (2.6) × 104 a;b | |
| S3 | 2.1 (0.5) × 107 | 2.0 (2.7) × 104 a;c | 1.3 (1.0) × 107 a;b | 2.4 (0.4) × 106 b | |
| Sampling | Seawater (Gene Copy Number in 100 mL−1) | Marine Sediments (Gene Copy Number in 100 g−1) | |||
|---|---|---|---|---|---|
| Period | Station | Enterococcus faecalis | Enterococcus faecium | Enterococcus faecalis | Enterococcus faecium |
| Winter | S1 | 4.7 (6.2) × 104 (0.20%) | No data | 5.4 (3.0) × 105 (0.07%) | No data |
| S2 | 4.3 (2.7) × 104 (0.36%) | 1.0 (1.7) × 103 | 3.6 (0.2) × 104 (0.001%) | No data | |
| S3 | 3.0 (3.2) × 104 (0.20%) | 2.3 (0.7) × 103 | 7.9 (2.6) × 105 (79.80%) c | 3.6 (0.8) × 102 b | |
| Spring | S1 | 1.5 (0.3) × 104 (0.08%) | No data | 1.9 (1.9) × 104 (4.20%) | 4.0 (1.6) × 103 b |
| S2 | 1.5 (0.1) × 106 (2.38%) a;b | No data | 4.0 (0.6) × 103 (2.20%) | 1.4 (0.1) × 102 b | |
| S3 | 1.1 (0.4) × 104 (0.05%) | 1.1 (1.8) × 103 | 8.5 (1.2) × 106 (65.39%) a | No data | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrasco-Acosta, M.; Garcia-Jimenez, P. Development of Multiplex RT qPCR Assays for Simultaneous Detection and Quantification of Faecal Indicator Bacteria in Bathing Recreational Waters. Microorganisms 2024, 12, 1223. https://doi.org/10.3390/microorganisms12061223
Carrasco-Acosta M, Garcia-Jimenez P. Development of Multiplex RT qPCR Assays for Simultaneous Detection and Quantification of Faecal Indicator Bacteria in Bathing Recreational Waters. Microorganisms. 2024; 12(6):1223. https://doi.org/10.3390/microorganisms12061223
Chicago/Turabian StyleCarrasco-Acosta, Marina, and Pilar Garcia-Jimenez. 2024. "Development of Multiplex RT qPCR Assays for Simultaneous Detection and Quantification of Faecal Indicator Bacteria in Bathing Recreational Waters" Microorganisms 12, no. 6: 1223. https://doi.org/10.3390/microorganisms12061223
APA StyleCarrasco-Acosta, M., & Garcia-Jimenez, P. (2024). Development of Multiplex RT qPCR Assays for Simultaneous Detection and Quantification of Faecal Indicator Bacteria in Bathing Recreational Waters. Microorganisms, 12(6), 1223. https://doi.org/10.3390/microorganisms12061223







