Caldanaerobacter subterraneus subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Cultivation
2.2. Microscopy
2.3. Physiological Characterization
2.4. 16S rRNA Gene Sequence Analysis
2.5. Genome Sequencing and Phylogenomics
2.6. Feather Degradation Test
3. Results
3.1. Isolation and Morphology
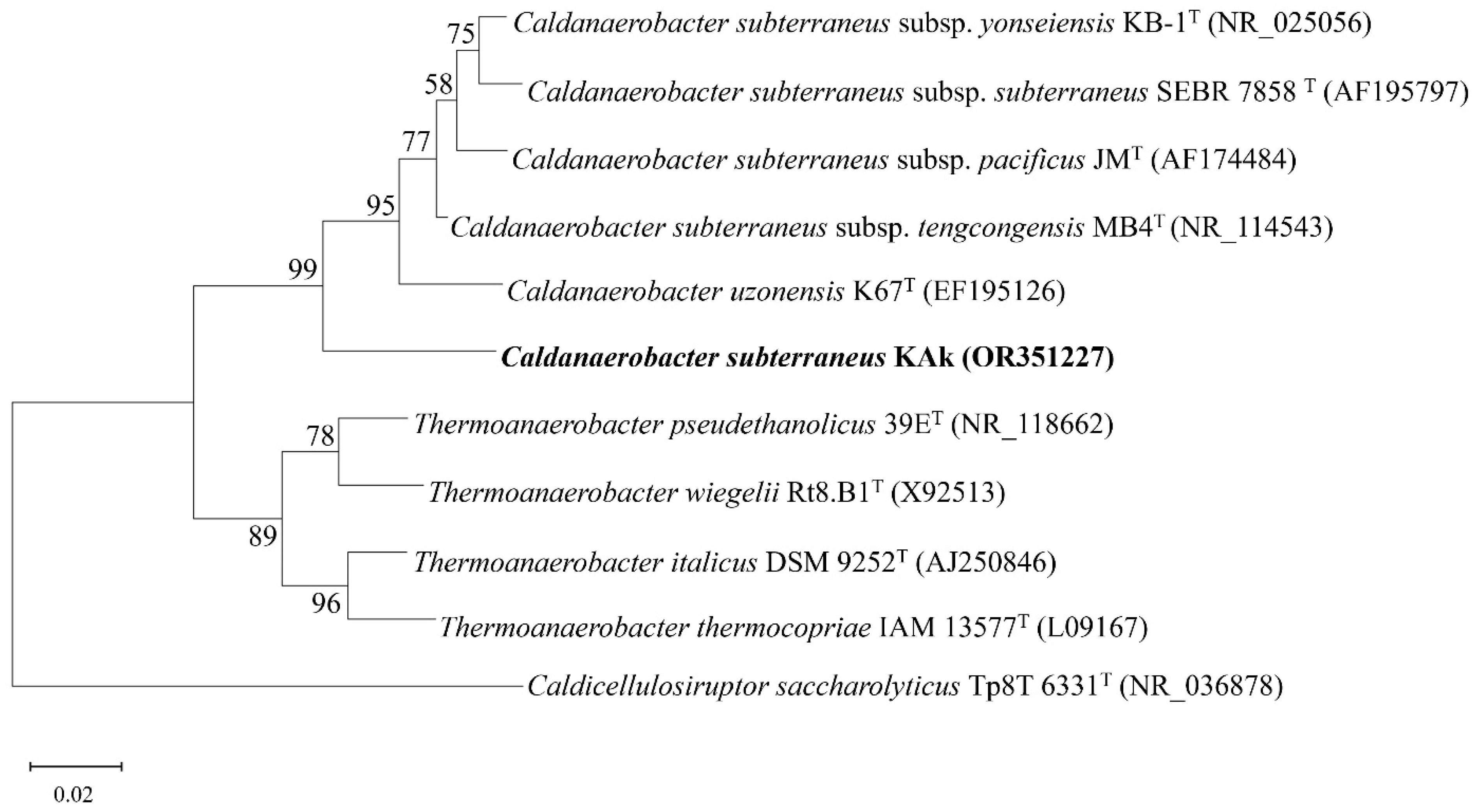
3.2. Phylogenetic Identification
3.3. Physiology
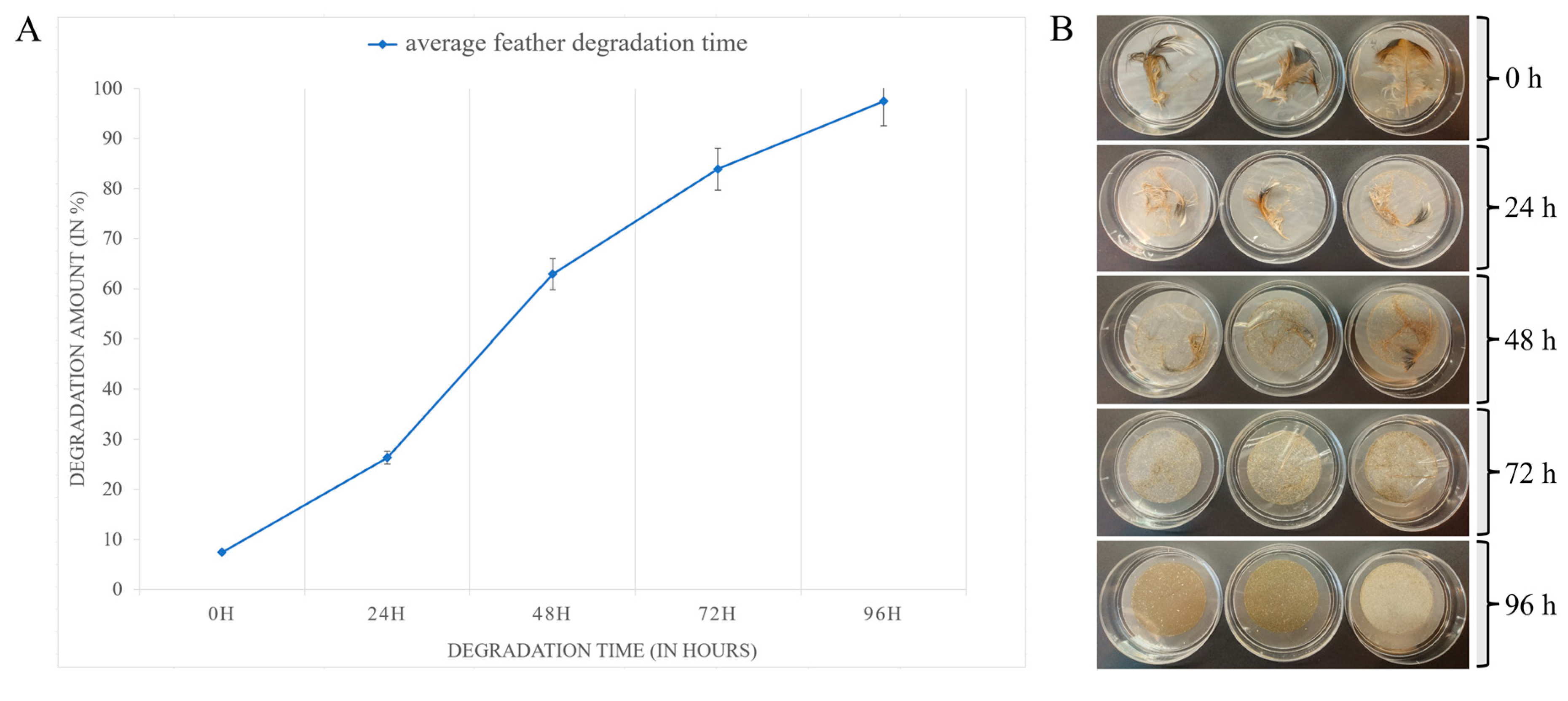
3.4. Feather Degradation
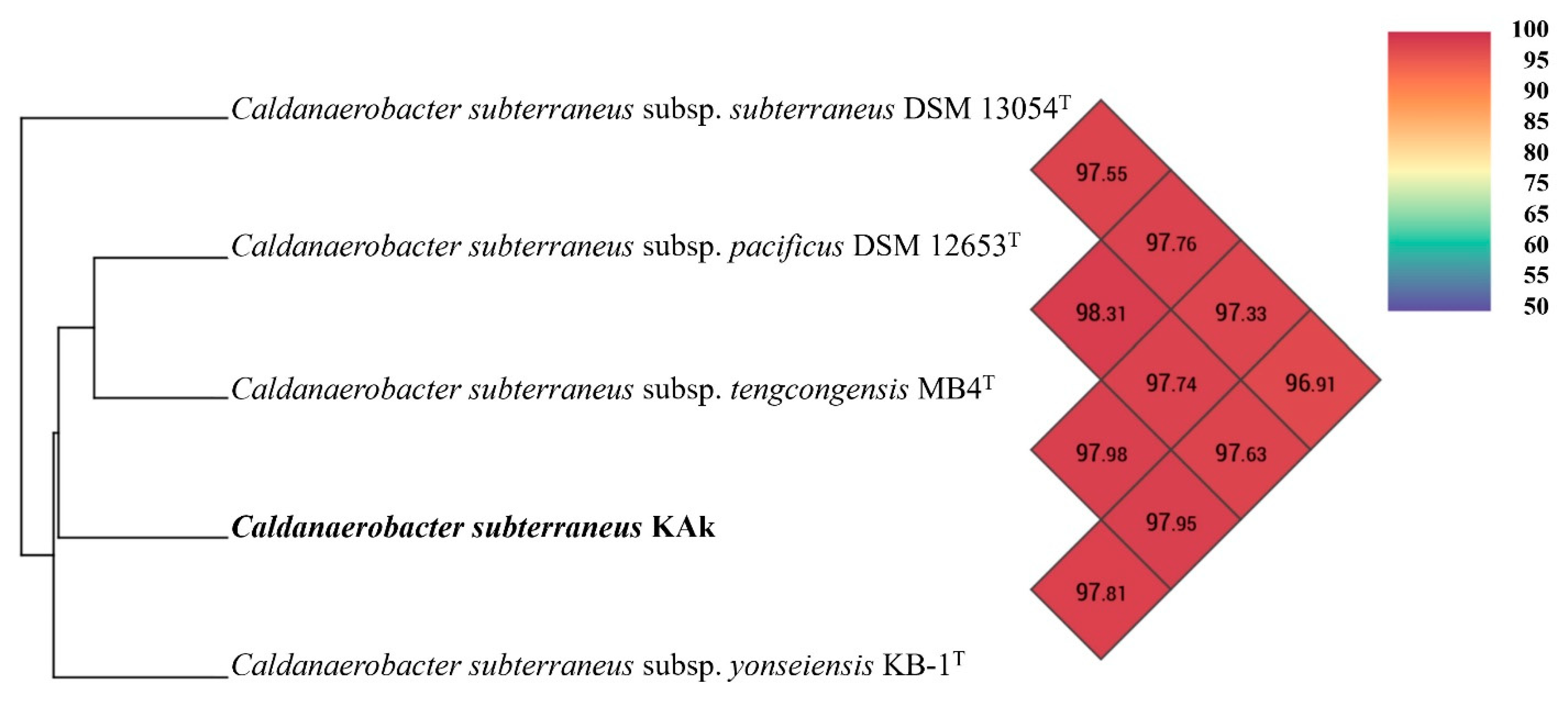
3.5. Genome Characteristics and Phylogenomics
4. Discussion
5. Conclusions
6. Caldanaerobacter subterraneus subsp. keratinolyticus subsp. nov.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Kozina, I.V.; Kublanov, I.V.; Kolganova, T.V.; Chernyh, N.A.; Bonch-Osmolovskaya, E.A. Caldanaerobacter uzonensis sp. nov., an anaerobic, thermophilic, heterotrophic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2010, 60 Pt 6, 1372–1375. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fardeau, M.L.; Magot, M.; Patel, B.K.; Thomas, P.; Garcia, J.L.; Ollivier, B. Thermoanaerobacter subterraneus sp. nov., a novel thermophile isolated from oilfield water. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 6, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Fardeau, M.L.; Salinas, M.B.; L’Haridon, S.; Jeanthon, C.; Verhé, F.; Cayol, J.L.; Patel, B.K.C.; Garcia, J.L.; Ollivier, B. Isolation from oil reservoirs of novel thermophilic anaerobes phylogenetically related to Thermoanaerobacter subterraneus: Reassignment of T. subterraneus, Thermoanaerobacter yonseiensis, Thermoanaerobacter tengcongensis and Carboxydibrachium pacificum to Caldanaerobacter subterraneus gen. nov., sp. nov., comb. nov. as four novel subspecies. Int. J. Syst. Evol. Microbiol. 2004, 54 Pt 2, 467–474. [Google Scholar] [PubMed]
- Sokolova, T.G.; González, J.M.; Kostrikina, N.A.; Chernyh, N.A.; Tourova, T.P.; Kato, C.; Bonch-Osmolovskaya, E.A.; Robb, F.T. Carboxydobrachium pacificum gen. nov., sp. nov., a new anaerobic, thermophilic, CO-utilizing marine bacterium from Okinawa Trough. Int. J. Syst. Evol. Microbiol. 2001, 51 Pt 1, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Xu, Y.; Liu, Y.; Ma, Y.; Zhou, P. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int. J. Syst. Evol. Microbiol. 2001, 51 Pt 4, 1335–1341. [Google Scholar] [CrossRef]
- Kim, B.C.; Grote, R.; Lee, D.W.; Antranikian, G.; Pyun, Y.R. Thermoanaerobacter yonseiensis sp. nov., a novel extremely thermophilic, xylose-utilizing bacterium that grows at up to 85 degrees C. Int. J. Syst. Evol. Microbiol. 2001, 51 Pt 4, 1539–1548. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, Y.J.; Park, G.S.; Kim, B.C.; Lee, S.J.; Shin, J.H.; Lee, D.W. Draft Genome Sequence of an Anaerobic and Extremophilic Bacterium, Caldanaerobacter yonseiensis, Isolated from a Geothermal Hot Stream. Genome Announc. 2013, 7, e00923-13. [Google Scholar] [CrossRef]
- Javier-Lopez, R.; Mandolini, E.; Dzhuraeva, M.; Bobodzhanova, K.; Birkeland, N.K. Fervidobacterium pennivorans subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile. Microorganisms 2022, 11, 22. [Google Scholar]
- Gousterova, A.; Braikova, D.; Goshev, I.; Christov, P.; Tishinov, K.; Vasileva-Tonkova, E.; Haertlé, T.; Nedkov, P. Degradation of keratin and collagen containing wastes by newly isolated thermoactinomycetes or by alkaline hydrolysis. Lett. Appl. Microbiol. 2005, 40, 335–340. [Google Scholar] [CrossRef]
- Rouse, J.G.; Van Dyke, M.E. A Review of Keratin-Based Biomaterials for Biomedical Applications. Materials 2010, 3, 999–1014. [Google Scholar] [CrossRef]
- Lai, Y.; Wu, X.; Zheng, X.; Li, W.; Wang, L. Insights into the keratin efficient degradation mechanism mediated by Bacillus sp. CN2 based on integrating functional degradomics. Biotechnol. Biofuels. Bioprod. 2023, 16, 59. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.M.; Grimes, J.L.; Mikkelsen, R.L. The use of poultry litter as co-substrate and source of inorganic nutrients and microorganisms for the ex situ biodegradation of petroleum compounds. Poult. Sci. 1999, 78, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Nam, G.W.; Lee, D.W.; Lee, H.S.; Lee, N.J.; Kim, B.C.; Choe, E.A.; Hwang, J.K.; Suhartono, M.T.; Pyun, Y.R. Native-feather degradation by Fervidobacterium islandicum AW-1, a newly isolated keratinase-producing thermophilic anaerobe. Arch. Microbiol. 2002, 178, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Elmayergi, H.H.; Smith, R.E. Influence of growth of Streptomyces fradiae on pepsin-HCl digestibility and methionine content of feather meal. Can. J. Microbiol. 1971, 17, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Lange, L.; Huang, Y.; Busk, P.K. Microbial decomposition of keratin in nature-a new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 2016, 100, 2083–2096. [Google Scholar] [CrossRef] [PubMed]
- Sharma, I.; Pranaw, K.; Soni, H.; Rawat, H.K.; Kango, N. Parametrically optimized feather degradation by Bacillus velezensis NCIM 5802 and delineation of keratin hydrolysis by multi-scale analysis for poultry waste management. Sci. Rep. 2022, 12, 17118. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Jeong, H.; Park, G.S.; Kwak, Y.; Lee, S.J.; Lee, S.J.; Park, M.K.; Kim, J.Y.; Kang, H.K.; Shin, J.H.; et al. Genome sequence of a native-feather degrading extremely thermophilic Eubacterium, Fervidobacterium islandicum AW-1. Stand. Genomic. Sci. 2015, 29, 71. [Google Scholar] [CrossRef] [PubMed]
- Mashzhan, A.; Javier-López, R.; Kistaubayeva, A.; Savitskaya, I.; Birkeland, N.K. Analysis and Characteristics of Thermal Springs in Kazakhstan. In Microbial Communities and their Interactions in the Extreme Environment; Egamberdieva, D., Birkeland, N.K., Li, W.-J., Panosyan, H., Eds.; Springer: Singapore, 2021; pp. 97–114. [Google Scholar]
- Miller, T.L.; Wolin, M.J. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 1974, 27, 985–987. [Google Scholar] [CrossRef]
- Mashzhan, A.; Javier-López, R.; Kistaubayeva, A.; Savitskaya, I.; Birkeland, N.K. Metagenomics and Culture-Based Diversity Analysis of the Bacterial Community in the Zharkent Geothermal Spring in Kazakhstan. Curr. Microbiol. 2021, 78, 2926–2934. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends. Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic. Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 8, 402. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Sant’Anna, F.H.; Lebedinsky, A.V.; Sokolova, T.G.; Robb, F.T.; Gonzalez, J.M. Analysis of three genomes within the thermophilic bacterial species Caldanaerobacter subterraneus with a focus on carbon monoxide dehydrogenase evolution and hydrolase diversity. BMC Genom. 2015, 16, 757. [Google Scholar] [CrossRef]
- Jang, H.J.; Kim, B.C.; Pyun, Y.R.; Kim, Y.S. A novel subtilisin-like serine protease from Thermoanaerobacter yonseiensis KB-1: Its cloning, expression, and biochemical properties. Extremophiles 2002, 6, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and FastDistance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S. Estimating Phylogenetic Trees from Distance Matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Kreft, L.; Botzki, A.; Coppens, F.; Vandepoele, K.; Van Bel, M. PhyD3: A phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 2017, 33, 2946–2947. [Google Scholar] [CrossRef] [PubMed]



| Feature | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Origin | Geothermal hot spring | Terrestrial hot spring | Oilfield water | Geothermal hot stream | Pacific Ocean hot vents | Terrestrial hot spring |
| Cell size (µm) | 0.2–0.7 × 1–20 | 0.5–0.6 × 1–10 | 0.5–0.7 × 2–8 | 0.4–0.8 × 1–3 | 0.3 × 4–10 | 0.3–0.5 × 1.5–15 |
| Spore | + | - | - | + | - | + |
| Motility | - | - | - | + | - | - |
| Flagella | - | - | Peritrichous | - | - | - |
| Temperature (°C) | ||||||
| Range | 55–80 | 50–80 | 40–75 | 50–85 | 50–80 | 50–75 |
| Optimum | 70 | 75 | 65 | 75 | 70 | 68–70 |
| pH | ||||||
| Range | 4.0–9.0 | 5.5–9.0 | 6.0–8.5 | 4.5–9.0 | 5.8–7.6 | 4.8–8.0 |
| Optimum | 6.8 | 7.0–7.5 | 7.5 | 6.5 | 6.8–7.1 | 6.8 |
| NaCl (%) | ||||||
| Range | 0–3 | 0–2.5 | 0–3 | 0–4 | ND | 0–2 |
| Optimum | 0.4 | 0.2 | 0 | 0 | 2–2.5 | 0.5 |
| Starch hydrolysis | slow | + | + | + | + | + |
| Xylan degradation | + | - | + | - | ND | ND |
| Utilization of: | ||||||
| Glucose | + | + | + | + | + | + |
| Sucrose | + | - | - | + | - | + |
| Xylose | + | - | + | + | ND | + |
| Arabinose | + | - | - | - | ND | + |
| Pyruvate | slow | - | + | ND | + | + |
| Galactose | + | + | + | + | + | + |
| Peptone | + | + | + | + | + | + |
| DNA G+C content (mol%) | 37.6 | 33 | 41 | 36.6 | 33 ± 1 | 34.2 ± 0.5 |
| Scientific Name | C. subterraneus KAk | C. subterraneus subsp. subterraneus DSM 13054T | C. subterraneus subsp. pacificus JMT | C. subterraneus subsp. tengcongensis MB4T | C. subterraneus subsp. younseiensis KB-1T |
|---|---|---|---|---|---|
| Genome size (Mb) | 2.4 | 2.6 | 2.4 | 2.7 | 2.7 |
| G+C content (%) | 37.6 | 37.5 | 37.5 | 37.5 | 37.5 |
| N50 (kb) | 108.9 | 82.6 | 56.3 | 2.7 | 62.5 |
| L50 | 8 | 11 | 13 | 1 | 14 |
| Number of contigs | 56 | 83 | 135 | 1 | 102 |
| Number of coding sequences | 2361 | 2613 | 2511 | 2588 | 2711 |
| Number of tRNAs | 52 | 56 | 49 | 55 | 59 |
| 5S rRNA | 3 | 3 | 4 | 4 | 4 |
| 16S rRNA | 2 | 5 | 4 | 4 | 5 |
| 23S rRNA | 3 | 5 | 3 | 4 | 9 |
| NCBI Accession no. | JAULJF000000000 | SLWU00000000 | ABXP00000000 | AE008691 | AXDC01000000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mashzhan, A.; Kistaubayeva, A.; Javier-López, R.; Bissenbay, A.; Birkeland, N.-K. Caldanaerobacter subterraneus subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile. Microorganisms 2024, 12, 1277. https://doi.org/10.3390/microorganisms12071277
Mashzhan A, Kistaubayeva A, Javier-López R, Bissenbay A, Birkeland N-K. Caldanaerobacter subterraneus subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile. Microorganisms. 2024; 12(7):1277. https://doi.org/10.3390/microorganisms12071277
Chicago/Turabian StyleMashzhan, Akzhigit, Aida Kistaubayeva, Rubén Javier-López, Akerke Bissenbay, and Nils-Kåre Birkeland. 2024. "Caldanaerobacter subterraneus subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile" Microorganisms 12, no. 7: 1277. https://doi.org/10.3390/microorganisms12071277
APA StyleMashzhan, A., Kistaubayeva, A., Javier-López, R., Bissenbay, A., & Birkeland, N.-K. (2024). Caldanaerobacter subterraneus subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile. Microorganisms, 12(7), 1277. https://doi.org/10.3390/microorganisms12071277






