Utilizing Feline Lentiviral Infection to Establish a Translational Model for COVID-19 in People with Human Immunodeficiency Virus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Source and Care and Feline Immunodeficiency Viral Infection
2.2. SARS-CoV-2 Viral Source, Propagation, and Inoculation
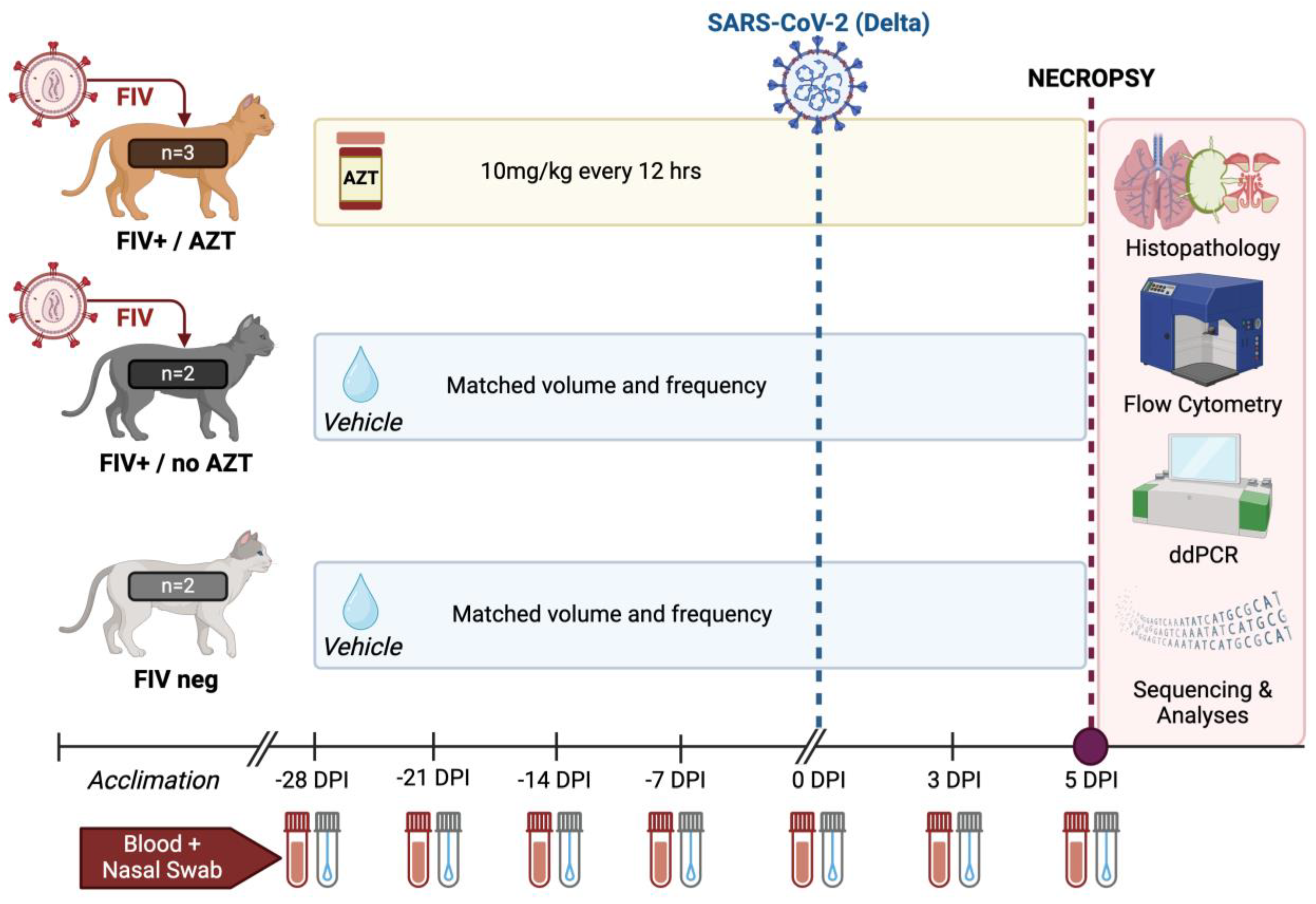
2.3. Experimental Design and Viral Challenge
2.4. Clinical Evaluation
2.5. Sample Collection
2.6. FIV and SARS-CoV-2 Viral RNA Analyses
2.7. Flow Cytometry
2.8. Histopathology
2.9. Next-Generation Sequencing and Analyses
2.10. Statistical Analyses
3. Results
3.1. FIV Viral RNA Expression Is Confirmed in Plasma of Previously Infected FIV+ Cats
3.2. AZT-Treated FIV-Infected Cats Have Significant Increases in CD4+ Cells Post SARS-CoV-2 Infection
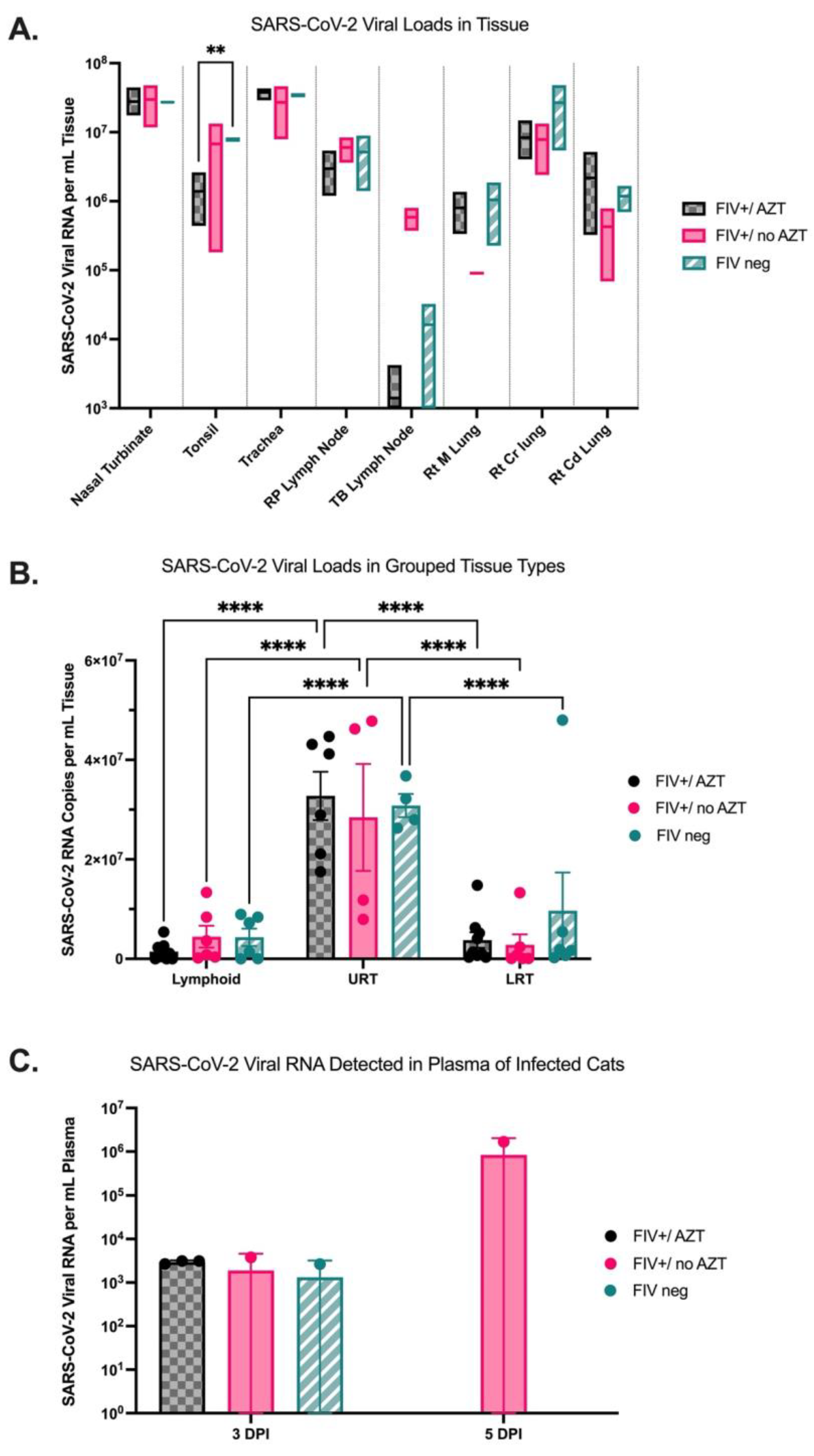
3.3. Quantification of SARS-CoV-2 Viral RNA in Tissue Indicates Higher Viral Loads in Upper Respiratory Tract in All Infected Cats and Detectable Viral RNA in Plasma of FIV+/no AZT Cat at 5 DPI
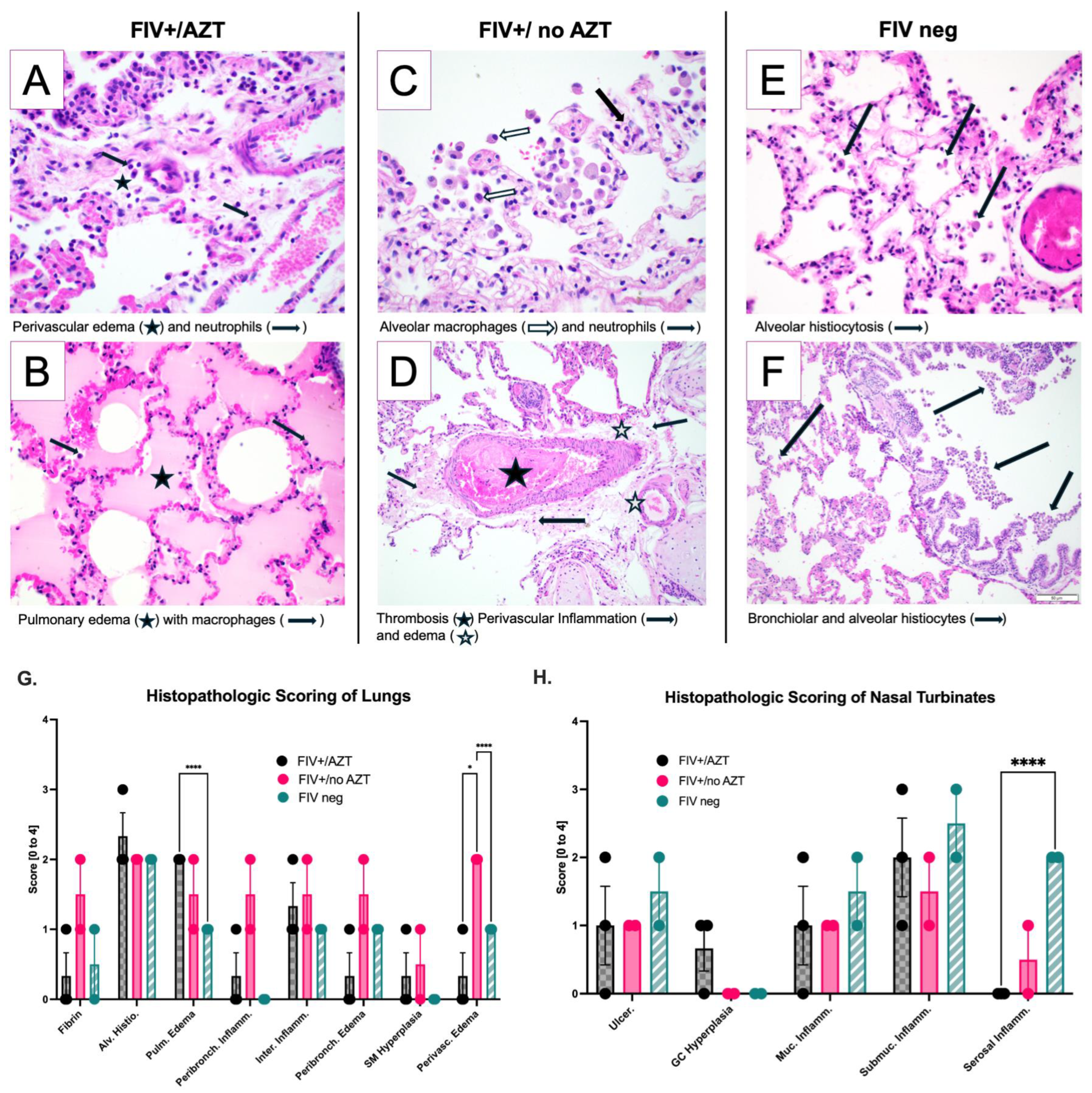
3.4. Significant Histopathologic Differences between Groups after SARS-CoV-2 Infection
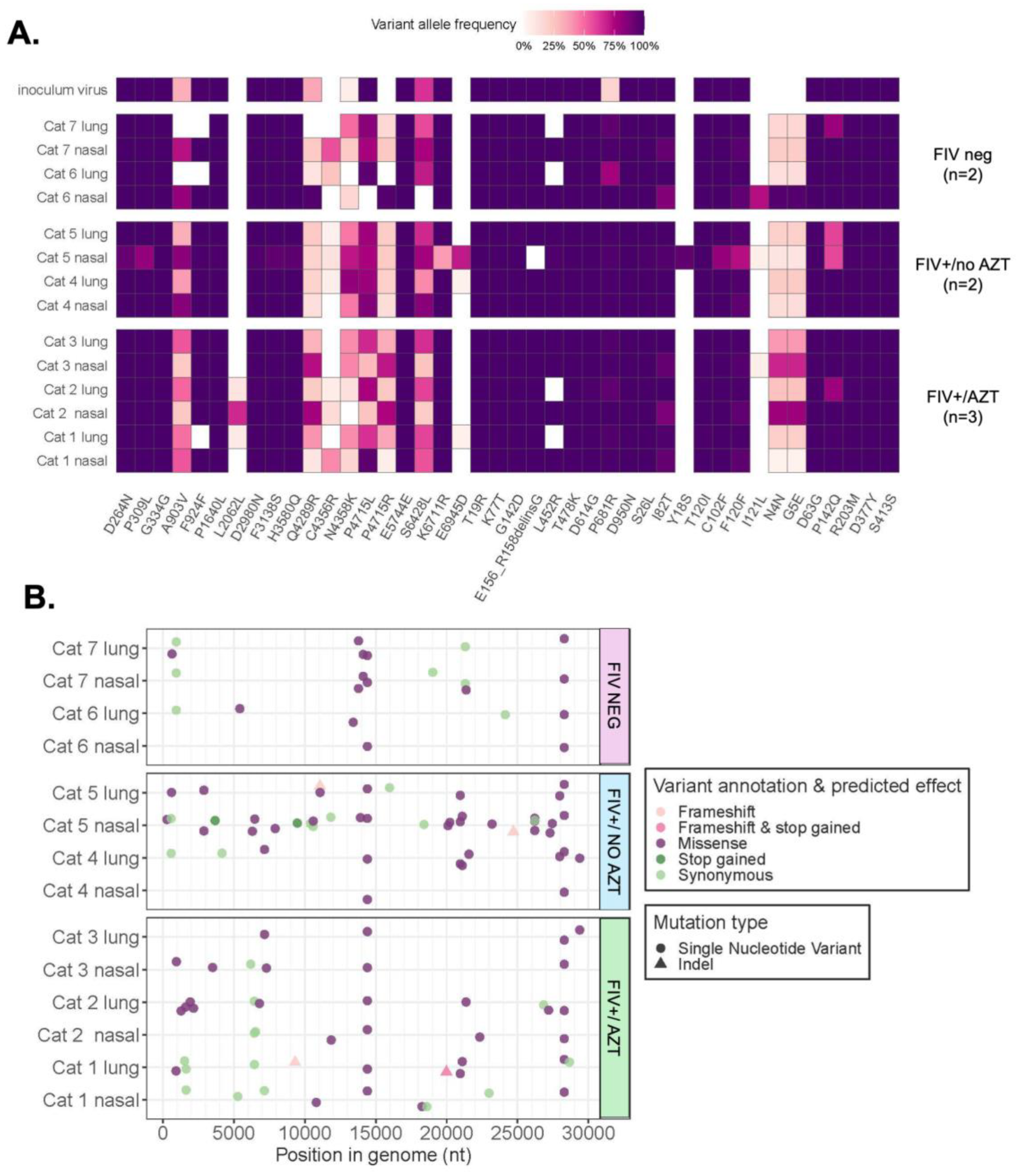
3.5. Sequencing and Mutation Analyses Reveal Positive Selection Signatures and Mutations after SARS-CoV-2’s Passage through Feline Host
3.5.1. Assessment of Variant Numbers in Tissue and Treatment Type
3.5.2. Signature of Selection
3.5.3. Variants Observed in SARS-CoV-2 Genomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global HIV Statistics. 2022. Available online: https://www.unaids.org/sites/default/files/media_asset (accessed on 4 April 2022).
- HIV and AIDS Resources. Available online: https://www.hiv.gov (accessed on 11 April 2024).
- WHO Coronavirus, Dashboard. COVID-19 Who Int. Available online: https://data.who.int/dashboards/covid19/cases (accessed on 14 May 2022).
- CDC People with Certain Medical Conditions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html (accessed on 18 February 2024).
- Danwang, C.; Noubiap, J.J.; Robert, A.; Yombi, J.C. Outcomes of patients with HIV and COVID-19 co-infection: A systematic review and meta-analysis. AIDS Res. Ther. 2022, 19, 3. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, Z.; Ding, S.; Liu, S.; Tang, Z.; Jia, L.; Liu, J.; Liu, Y. HIV infection and risk of COVID-19 mortality: A meta-analysis. Medicine 2021, 100, e26573. [Google Scholar] [CrossRef]
- Bhaskaran, K.; Rentsch, C.T.; MacKenna, B.; Schultze, A.; Mehrkar, A.; Bates, C.J.; Eggo, R.M.; Morton, C.E.; Bacon, S.C.J.; Inglesby, P.; et al. HIV infection and COVID-19 death: A population-based cohort analysis of UK primary care data and linked national death registrations within the OpenSAFELY platform. Lancet HIV 2021, 8, e24–e32. [Google Scholar] [CrossRef]
- Russell, C.D.; Lone, N.I.; Baillie, J.K. Comorbidities, multimorbidity and COVID-19. Nat. Med. 2023, 29, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.M.; Frieman, M.B. Growth and Quantification of MERS-CoV Infection. Curr. Protoc. Microbiol. 2015, 37, 15E.2.1-9. [Google Scholar] [CrossRef] [PubMed]
- Mengato, D.; Mazzitelli, M.; Francavilla, A.; Bettio, M.; Sasset, L.; Presa, N.; Pivato, L.; Lo Menzo, S.; Trevenzoli, M.; Venturini, F.; et al. Changing patterns and clinical outcomes of hospitalized patients with COVID-19 severe pneumonia treated with remdesivir according to vaccination status: Results from a real-world retrospective study. Clin. Exp. Med. 2023, 23, 2749–2756. [Google Scholar] [CrossRef] [PubMed]
- Mazzitelli, M.; Trunfio, M.; Sasset, L.; Scaglione, V.; Ferrari, A.; Mengato, D.; Gardin, S.; Bonadiman, N.; Calandrino, L.; Agostini, E.; et al. Risk of Hospitalization and Sequelae in Patients with COVID-19 Treated with 3-Day Early Remdesivir vs. Controls in the Vaccine and Omicron era: A Real-Life Cohort Study. J. Med. Virol. 2023, 95, e28660. [Google Scholar] [CrossRef] [PubMed]
- Falcone, M.; Suardi, L.R.; Tiseo, G.; Barbieri, C.; Giusti, L.; Galfo, V.; Forniti, A.; Caroselli, C.; Della Sala, L.; Tempini, S.; et al. Early Use of Remdesivir and Risk of Disease Progression in Hospitalized Patients with Mild to Moderate COVID-19. Clin. Ther. 2022, 44, 364–373. [Google Scholar] [CrossRef]
- Patel, R.H.; Pella, P.M. COVID-19 in a patient with HIV infection. J. Med. Virol. 2020, 92, 2356–2357. [Google Scholar] [CrossRef]
- Ridgway, J.P.; Farley, B.; Benoit, J.-L.; Frohne, C.; Hazra, A.; Pettit, N.; Pho, M.; Pursell, K.; Saltzman, J.; Schmitt, J.; et al. A Case Series of Five People Living with HIV Hospitalized with COVID-19 in Chicago, Illinois. AIDS Patient Care STDs 2020, 34, 331–335. [Google Scholar] [CrossRef]
- Höft, M.A.; Burgers, W.A.; Riou, C. The immune response to SARS-CoV-2 in people with HIV. Cell. Mol. Immunol. 2024, 21, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Rudd, J.M.; Tamil Selvan, M.; Cowan, S.; Kao, Y.-F.; Midkiff, C.C.; Narayanan, S.; Ramachandran, A.; Ritchey, J.W.; Miller, C.A. Clinical and Histopathologic Features of a Feline SARS-CoV-2 Infection Model Are Analogous to Acute COVID-19 in Humans. Viruses 2021, 13, 1550. [Google Scholar] [CrossRef]
- Hosie, M.J.; Lutz, H. Feline Immunodeficiency Virus. In Schalm’s Veterinary Hematology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2022; pp. 424–430. ISBN 978-1-119-50053-7. [Google Scholar]
- Kenyon, J.C.; Lever, A.M.L. The Molecular Biology of Feline Immunodeficiency Virus (FIV). Viruses 2011, 3, 2192–2213. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Ho, E.W.; Brown, M.L.; Yamamoto, J.K. Isolation of a T-Lymphotropic Virus from Domestic Cats with an Immunodeficiency-Like Syndrome. Science 1987, 235, 790–793. [Google Scholar] [CrossRef]
- Elder, J.H.; Lin, Y.-C.; Fink, E.; Grant, C.K. Feline Immunodeficiency Virus (FIV) as A Model for Study of Lentivirus Infections: Parallels with HIV. Curr. HIV Res. 2010, 8, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Burkhard, M.; Dean, G.A. Transmission and Immunopathogenesis of FIV in Cats as a Model for HIV. Curr. HIV Res. 2003, 1, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Sparger, E.E. FIV as a Model for HIV: An Overview. In In Vivo Models of HIV Disease and Control; Friedman, H., Specter, S., Bendinelli, M., Eds.; Infectious Diseases and Pathogenesis; Springer: Boston, MA, USA, 2006; pp. 149–237. ISBN 978-0-387-25741-9. [Google Scholar]
- Barlough, J.E.; Ackley, C.D.; George, J.W.; Levy, N.; Acevedo, R.; Moore, P.F.; Rideout, B.A.; Cooper, M.D.; Pedersen, N.C. Acquired Immune Dysfunction in Cats with Experimentally Induced Feline Immunodeficiency Virus Infection: Comparison of Short-Term and Long-Term Infections. JAIDS J. Acquir. Immune Defic. Syndr. 1991, 4, 219–227. Available online: https://journals.lww.com/jaids/abstract/1991/03000/acquired_immune_dysfunction_in_cats_with.2.aspx (accessed on 18 February 2024).
- Miller, C.; Abdo, Z.; Ericsson, A.; Elder, J.; VandeWoude, S. Applications of the FIV Model to Study HIV Pathogenesis. Viruses 2018, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- Kornya, M.R.; Little, S.E.; Scherk, M.A.; Sears, W.C.; Bienzle, D. Association between Oral Health Status and Retrovirus Test Results in Cats. J. Am. Vet. Med. Assoc. 2014, 245, 8. Available online: https://avmajournals.avma.org/view/journals/javma/245/8/javma.245.8.916.xml (accessed on 18 February 2024). [CrossRef]
- Laitinen, T.; Meili, T.; Koyioni, M.; Koutentis, P.A.; Poso, A.; Hofmann-Lehmann, R.; Asquith, C.R.M. Synthesis and evaluation of 1,2,3-dithiazole inhibitors of the nucleocapsid protein of feline immunodeficiency virus (FIV) as a model for HIV infection. Bioorg. Med. Chem. 2022, 68, 116834. [Google Scholar] [CrossRef]
- Mohammadi, H.; Bienzle, D. Pharmacological Inhibition of Feline Immunodeficiency Virus (FIV). Viruses 2012, 4, 708–724. [Google Scholar] [CrossRef]
- Nastri, B.M.; Pagliano, P.; Zannella, C.; Folliero, V.; Masullo, A.; Rinaldi, L.; Galdiero, M.; Franci, G. HIV and Drug-Resistant Subtypes. Microorganisms 2023, 11, 221. [Google Scholar] [CrossRef] [PubMed]
- North, T.W.; North, G.L.; Pedersen, N.C. Feline immunodeficiency virus, a model for reverse transcriptase-targeted chemotherapy for acquired immune deficiency syndrome. Antimicrob. Agents Chemother. 1989, 33, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Auwerx, J.; Esnouf, R.; Clercq, E.D.; Balzarini, J. Susceptibility of Feline Immunodeficiency Virus/Human Immunodeficiency Virus Type 1 Reverse Transcriptase Chimeras to Non-Nucleoside RT Inhibitors. Mol. Pharmacol. 2004, 65, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Earl, D.D.; Yamamoto, J.K. Is AZT/3TC therapy effective against FIV infection or immunopathogenesis? Vet. Immunol. Immunopathol. 2002, 85, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Antony, J.M.; Martinez, J.A.; Glerum, D.M.; Brussee, V.; Hoke, A.; Zochodne, D.; Power, C. Didanosine causes sensory neuropathy in an HIV/AIDS animal model: Impaired mitochondrial and neurotrophic factor gene expression. Brain 2007, 130, 2011–2023. [Google Scholar] [CrossRef]
- Schwartz, A.M.; McCrackin, M.A.; Schinazi, R.F.; Hill, P.B.; Vahlenkamp, T.W.; Tompkins, M.B.; Hartmann, K. Antiviral efficacy of nine nucleoside reverse transcriptase inhibitors against feline immunodeficiency virus in feline peripheral blood mononuclear cells. Am. J. Vet. Res. 2014, 75, 273–281. [Google Scholar] [CrossRef]
- Bisset, L.R.; Lutz, H.; Böni, J.; Hofmann-Lehmann, R.; Lüthy, R.; Schüpbach, J. Combined effect of zidovudine (ZDV), lamivudine (3TC) and abacavir (ABC) antiretroviral therapy in suppressing in vitro FIV replication. Antivir. Res. 2002, 53, 35–45. [Google Scholar] [CrossRef]
- Hartmann, K. Feline immunodeficiency virus infection: An overview. Vet. J. 1998, 155, 123–137. [Google Scholar] [CrossRef]
- Diehl, L.J.; Mathiason-Dubard, C.K.; O’Neil, L.L.; Obert, L.A.; Hoover, E.A. Induction of accelerated feline immunodeficiency virus disease by acute-phase virus passage. J. Virol. 1995, 69, 6149–6157. [Google Scholar] [CrossRef]
- Kim, J.; Behzadi, E.S.; Nehring, M.; Carver, S.; Cowan, S.R.; Conry, M.K.; Rawlinson, J.E.; VandeWoude, S.; Miller, C.A. Combination Antiretroviral Therapy and Immunophenotype of Feline Immunodeficiency Virus. Viruses 2023, 15, 822. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Miller, C.; Powers, J.; Musselman, E.; Mackie, R.; Elder, J.; VandeWoude, S. Immunopathologic Effects of Prednisolone and Cyclosporine A on Feline Immunodeficiency Virus Replication and Persistence. Viruses 2019, 11, 805. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.-F.; Peake, B.; Madden, R.; Cowan, S.R.; Scimeca, R.C.; Thomas, J.E.; Reichard, M.V.; Ramachandran, A.; Miller, C.A. A probe-based droplet digital polymerase chain reaction assay for early detection of feline acute cytauxzoonosis. Vet. Parasitol. 2021, 292, 109413. [Google Scholar] [CrossRef] [PubMed]
- Braun, K.M.; Moreno, G.K.; Halfmann, P.J.; Hodcroft, E.B.; Baker, D.A.; Boehm, E.C.; Weiler, A.M.; Haj, A.K.; Hatta, M.; Chiba, S.; et al. Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. PLoS Pathog. 2021, 17, e1009373. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Polak, S.B.; Van Gool, I.C.; Cohen, D.; von der Thüsen, J.H.; van Paassen, J. A systematic review of pathological findings in COVID-19: A pathophysiological timeline and possible mechanisms of disease progression. Mod. Pathol. 2020, 33, 2128–2138. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Xiong, Y.; Liu, H.; Niu, L.; Guo, J.; Liao, M.; Xiao, S.-Y. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod. Pathol. 2020, 33, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- von der Thüsen, J.; van der Eerden, M. Histopathology and genetic susceptibility in COVID-19 pneumonia. Eur. J. Clin. Investig. 2020, 50, e13259. [Google Scholar] [CrossRef]
- Tamil Selvan, M.; Gunasekara, S.; Xiao, P.; Griffin, K.; Cowan, S.R.; Narayanan, S.; Ramachandran, A.; Hagen, D.E.; Ritchey, J.W.; Rudd, J.M.; et al. SARS CoV-2 (Delta Variant) Infection Kinetics and Immunopathogenesis in Domestic Cats. Viruses 2022, 14, 1207. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.; Boegler, K.; Carver, S.; MacMillan, M.; Bielefeldt-Ohmann, H.; VandeWoude, S. Pathogenesis of oral FIV infection. PLoS ONE 2017, 12, e0185138. [Google Scholar] [CrossRef] [PubMed]
- Artic Network. Available online: https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html (accessed on 1 April 2024).
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.W.; Moncla, L.H.; Hughes, A.L. SNPGenie: Estimating evolutionary parameters to detect natural selection using pooled next-generation sequencing data. Bioinformatics 2015, 31, 3709–3711. [Google Scholar] [CrossRef] [PubMed]
- R Core Team, R. R Core Team R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Collado, V.M.; Domenech, A.; Miró, G.; Martin, S.; Escolar, E.; Gomez-Lucia, E. Epidemiological Aspects and Clinicopathological Findings in Cats Naturally Infected with Feline Leukemia Virus (FeLV) and/or Feline Immunodeficiency Virus (FIV). Sci. Res. Publ. 2012, 2012, 13–20. [Google Scholar] [CrossRef]
- Murphy, B.; Vapniarsky, N.; Hillman, C.; Castillo, D.; McDonnel, S.; Moore, P.; Luciw, P.A.; Sparger, E.E. FIV establishes a latent infection in feline peripheral blood CD4+ T lymphocytes in vivo during the asymptomatic phase of infection. Retrovirology 2012, 9, 12. [Google Scholar] [CrossRef]
- Kohmoto, M.; Uetsuka, K.; Ikeda, Y.; Inoshima, Y.; Shimojima, M.; Sato, E.; Inada, G.; Toyosaki, T.; Miyazawa, T.; Doi, K.; et al. Eight-year observation and comparative study of specific pathogen-free cats experimentally infected with feline immunodeficiency virus (FIV) subtypes A and B: Terminal acquired immunodeficiency syndrome in a cat infected with FIV petaluma strain. J. Vet. Med. Sci. 1998, 60, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sun, J.; Patel, R.C.; Zhang, J.; Guo, S.; Zheng, Q.; Olex, A.L.; Olatosi, B.; Weissman, S.B.; Islam, J.Y.; et al. Associations between HIV infection and clinical spectrum of COVID-19: A population level analysis based on US National COVID Cohort Collaborative (N3C) data. Lancet HIV 2021, 8, e690–e700. [Google Scholar] [CrossRef]
- Mondi, A.; Cimini, E.; Colavita, F.; Cicalini, S.; Pinnetti, C.; Matusali, G.; Casetti, R.; Maeurer, M.; Vergori, A.; Mazzotta, V.; et al. COVID-19 in people living with HIV: Clinical implications of dynamics of the immune response to SARS-CoV-2. J. Med. Virol. 2021, 93, 1796–1804. [Google Scholar] [CrossRef]
- Nomah, D.K.; Reyes-Urueña, J.; Díaz, Y.; Moreno, S.; Aceiton, J.; Bruguera, A.; Vivanco-Hidalgo, R.M.; Llibre, J.M.; Domingo, P.; Falcó, V.; et al. Sociodemographic, clinical, and immunological factors associated with SARS-CoV-2 diagnosis and severe COVID-19 outcomes in people living with HIV: A retrospective cohort study. Lancet HIV 2021, 8, e701–e710. [Google Scholar] [CrossRef]
- Western Cape Department of Health in collaboration with the National Institute for Communicable Diseases, South Africa. Risk Factors for Coronavirus Disease 2019 (COVID-19) Death in a Population Cohort Study from the Western Cape Province, South Africa. Clin. Infect. Dis. 2021, 73, e2005–e2015. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, T.; Zhang, H. Recovery from the coronavirus disease-2019 (COVID-19) in two patients with coexisted (HIV) infection. J. Med. Virol. 2020, 92, 2325–2327. [Google Scholar] [CrossRef]
- Härter, G.; Spinner, C.D.; Roider, J.; Bickel, M.; Krznaric, I.; Grunwald, S.; Schabaz, F.; Gillor, D.; Postel, N.; Mueller, M.C.; et al. COVID-19 in people living with human immunodeficiency virus: A case series of 33 patients. Infection 2020, 48, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Shalev, N.; Scherer, M.; LaSota, E.D.; Antoniou, P.; Yin, M.T.; Zucker, J.; Sobieszczyk, M.E. Clinical Characteristics and Outcomes in People Living with Human Immunodeficiency Virus Hospitalized for Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2294–2297. [Google Scholar] [CrossRef]
- Mirzaei, H.; McFarland, W.; Karamouzian, M.; Sharifi, H. COVID-19 Among People Living with HIV: A Systematic Review. AIDS Behav. 2021, 25, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, J.; Díez, C.; Martín-Vicente, M.; Micán, R.; Pérez-Elías, M.J.; García-Fraile, L.J.; Vidal, F.; Suárez-García, I.; Podzamczer, D.; Del Romero, J.; et al. Prevalence and factors associated with SARS-CoV-2 seropositivity in the Spanish HIV Research Network Cohort. Clin. Microbiol. Infect. 2021, 27, 1678–1684. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, K. Efficacy of antiviral chemotherapy for retrovirus-infected cats: What does the current literature tell us? J. Feline Med. Surg. 2015, 17, 925–939. [Google Scholar] [CrossRef]
- Ferreira, I.; Datir, R.; Papa, G.; Kemp, S.; Meng, B.; Rakshit, P.; Singh, S.; Pandey, R.; Ponnusamy, K.; Radhakrishnan, V.S.; et al. SARS-CoV-2 B.1.617 emergence and sensitivity to vaccine-elicited antibodies. BioRxiv 2021. [Google Scholar] [CrossRef]
- Winger, A.; Caspari, T. The Spike of Concern—The Novel Variants of SARS-CoV-2. Viruses 2021, 13, 1002. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, T.; Fang, Y.; Liu, J.; Ye, Q.; Ding, L. SARS-CoV-2 spike L452R mutation increases Omicron variant fusogenicity and infectivity as well as host glycolysis. Signal Transduct. Target. Ther. 2022, 7, 1–3. [Google Scholar] [CrossRef]
- Yadav, P.D.; Sapkal, G.N.; Abraham, P.; Ella, R.; Deshpande, G.; Patil, D.Y.; Nyayanit, D.A.; Gupta, N.; Sahay, R.R.; Shete, A.M.; et al. Neutralization of Variant under Investigation B.1.617.1 with Sera of BBV152 Vaccinees. Clin. Infect. Dis. 2022, 74, 366–368. [Google Scholar] [CrossRef]



| Clinical Score | |||||
|---|---|---|---|---|---|
| 0 (Healthy) | 1 (Mild) | 2 (Moderate) | 3 (Marked) | ||
| Assessed/Observed Clinical Parameter | Body Weight | No weight loss | <5% weight loss | 5 to 10% weight loss | >10% weight loss |
| Temperature | 37.2 to 39.0 °C | 39.1 to 39.4 °C | 39.5 to 39.7 °C | >39.7 °C | |
| Pulse Oximetry | 98 to 100% | 96 to 97% | 93 to 95% | <93% | |
| Activity | Normal | Mild reduction when disturbed * (mild lethargy) | Moderate reduction when disturbed * (moderate lethargy) | Little to no activity disturbed * and reduced activity when stimulated ** | |
| Behavior | Normal | Noticeable but minimal reduction in interest in food and/or attention | Moderate reduction in interest in food and/or attention | Anorexia and/or complete lack of interest | |
| Respiratory Effort | Normal resting respiratory effort and rate | Mild tachypnea (>35 breaths per minute at rest) with no overt increase in respiratory effort | Moderate tachypnea (>40 breaths per minute at rest) with moderate increase in effort | Marked tachypnea (>45 breaths per minute at rest) with marked dyspnea or effort | |
| Ocular and/or Nasal Discharge | None | Mild discharge observed from nares or eyes | Moderate discharge observed from either nares or eyes or from both nares and eyes | Marked or purulent discharge noted from nares and/or eyes | |
| Coughing | None | Occasional, rare cough observed | Intermittent coughing (at least one episode per 30 min) | Marked, persistent coughing (2+ episodes per 30 min) | |
| Wheezing | None | Occasional, rare wheeze observed | Intermittent wheezing (at least one episode per 30 min) | Marked, persistent wheezing (2+ episodes per 30 min) | |
| Mutation | Emergent (Absent → Present) | Absence (Present → Absent) | Amplified (Present → More Present) | Weakened (Present → Less Present) | Protein |
|---|---|---|---|---|---|
| C4356R | ●● ◊◊ ΔΔ | ORF1ab | |||
| P4715R | ●●● ◊◊ ΔΔ | ||||
| E6945D | ● ◊◊ | ||||
| A903V | ΔΔ | ||||
| L2062L | ●● | ||||
| F924F | ● ΔΔ | ||||
| N4358K | ◊◊ | ||||
| L452R | ●● ΔΔ | Spike | |||
| P681R | ●●● ◊◊ ΔΔ | ||||
| E156 | ◊ | ||||
| N4N | ●●● ◊◊ ΔΔ | N | |||
| G5E | ●●● ◊◊ ΔΔ | ||||
| P142Q | ● | ||||
| Y18S | ◊ | ORF7 | |||
| I121L | ● Δ | ORF8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shatnawi, S.; Gunasekara, S.; Bashor, L.; Tamil Selvan, M.; Nehring, M.; Cowan, S.; Ritchey, J.; VandeWoude, S.; Taylor, B.; Miller, C.; et al. Utilizing Feline Lentiviral Infection to Establish a Translational Model for COVID-19 in People with Human Immunodeficiency Virus Infection. Microorganisms 2024, 12, 1289. https://doi.org/10.3390/microorganisms12071289
Shatnawi S, Gunasekara S, Bashor L, Tamil Selvan M, Nehring M, Cowan S, Ritchey J, VandeWoude S, Taylor B, Miller C, et al. Utilizing Feline Lentiviral Infection to Establish a Translational Model for COVID-19 in People with Human Immunodeficiency Virus Infection. Microorganisms. 2024; 12(7):1289. https://doi.org/10.3390/microorganisms12071289
Chicago/Turabian StyleShatnawi, Shoroq, Sachithra Gunasekara, Laura Bashor, Miruthula Tamil Selvan, Mary Nehring, Shannon Cowan, Jerry Ritchey, Susan VandeWoude, Brianne Taylor, Craig Miller, and et al. 2024. "Utilizing Feline Lentiviral Infection to Establish a Translational Model for COVID-19 in People with Human Immunodeficiency Virus Infection" Microorganisms 12, no. 7: 1289. https://doi.org/10.3390/microorganisms12071289







