Universal Lineage-Independent Markers of Multidrug Resistance in Mycobacterium tuberculosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Read Mapping and Variant Calling
2.3. Statistical Analysis
3. Results
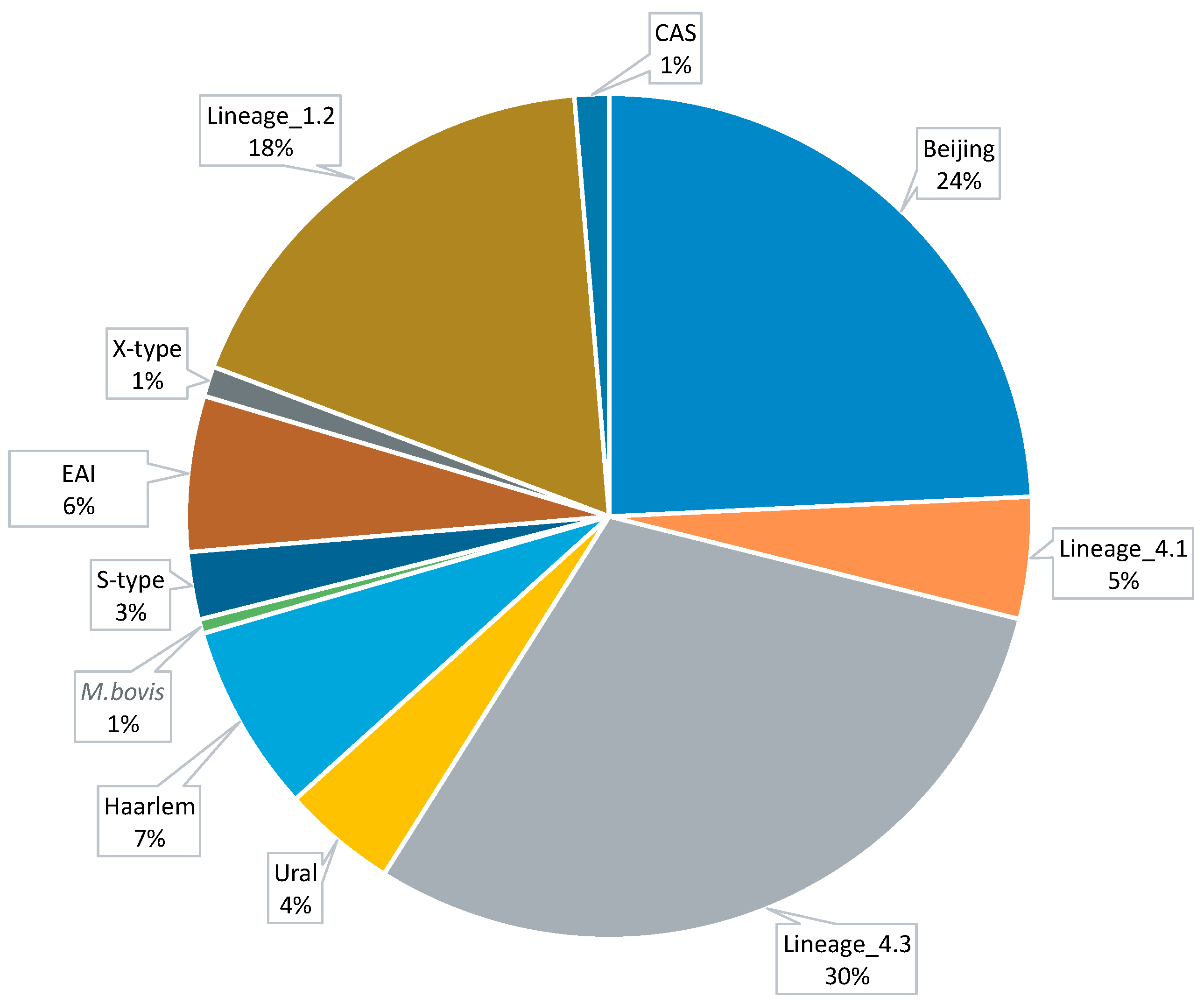
3.1. Data Collection and Computational Analysis
3.2. Universal Markers of Antibiotic Resistance
3.3. Attributable Risk Interactions between Mutations Associated with Multidrug Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kim, P.S.; Swaminathan, S. Ending TB: The world’s oldest pandemic. J. Int. AIDS Soc. 2021, 24, e25698. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.H.; Contamin, L.; Nguyen, T.V.A.; Bañuls, A.L. Insights into the processes that drive the evolution of drug resistance in Mycobacterium tuberculosis. Evol. Appl. 2018, 11, 1498–1511. [Google Scholar] [CrossRef] [PubMed]
- Trauner, A.; Borrell, S.; Reither, K.; Gagneux, S. Evolution of drug resistance in tuberculosis: Recent progress and implications for diagnosis and therapy. Drugs 2014, 74, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Gygli, S.M.; Borrell, S.; Trauner, A.; Gagneux, S. Antimicrobial resistance in Mycobacterium tuberculosis: Mechanistic and evolutionary perspectives. FEMS Microbiol. Rev. 2017, 41, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Vanino, E.; Granozzi, B.; Akkerman, O.W.; Munoz-Torrico, M.; Palmieri, F.; Seaworth, B.; Tiberi, S.; Tadolini, M. Update of drug-resistant tuberculosis treatment guidelines: A turning point. Int. J. Infect. Dis. 2023, 130, S12–S15. [Google Scholar] [CrossRef]
- Gill, C.M.; Dolan, L.; Piggott, L.M.; McLaughlin, A.M. New developments in tuberculosis diagnosis and treatment. Breathe 2022, 18, 210149. [Google Scholar] [CrossRef] [PubMed]
- Al-Saeedi, M.; Al-Hajoj, S. Diversity and evolution of drug resistance mechanisms in Mycobacterium tuberculosis. Infect. Drug Resist. 2017, 10, 333. [Google Scholar] [CrossRef]
- Perdigao, J.; Portugal, I. Genetics and roadblocks of drug resistant tuberculosis. Infect. Genet. Evol. 2019, 72, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Farhat, M.R.; Shapiro, B.J.; Kieser, K.J.; Sultana, R.; Jacobson, K.R.; Victor, T.C.; Warren, R.M.; Streicher, E.M.; Calver, A.; Sloutsky, A. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 2013, 45, 1183–1189. [Google Scholar] [CrossRef]
- Qin, L.; Wang, J.; Lu, J.; Yang, H.; Zheng, R.; Liu, Z.; Huang, X.; Feng, Y.; Hu, Z.; Ge, B. A deletion in the RD105 region confers resistance to multiple drugs in Mycobacterium tuberculosis. BMC Bio 2019, 17, 1–12. [Google Scholar] [CrossRef]
- Singh, P.; Jamal, S.; Ahmed, F.; Saqib, N.; Mehra, S.; Ali, W.; Roy, D.; Ehtesham, N.Z.; Hasnain, S.E. Computational modeling and bioinformatic analyses of functional mutations in drug target genes in Mycobacterium tuberculosis. Comput. Struct. Biotechnol. J. 2021, 19, 2423–2446. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.; Andersson, D.I. Evolutionary trajectories to antibiotic resistance. Ann. Rev. Microbiol. 2017, 71, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Martini, M.C.; Hicks, N.D.; Xiao, J.; Alonso, M.N.; Barbier, T.; Sixsmith, J.; Fortune, S.M.; Shell, S.S. Loss of RNase J leads to multi-drug tolerance and accumulation of highly structured mRNA fragments in Mycobacterium tuberculosis. PLoS Pathog. 2022, 18, e1010705. [Google Scholar] [CrossRef] [PubMed]
- Safi, H.; Lingaraju, S.; Amin, A.; Kim, S.; Jones, M.; Holmes, M.; McNeil, M.; Peterson, S.N.; Chatterjee, D.; Fleischmann, R. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat. Genet. 2013, 45, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Cohen, K.A.; Manson, A.L.; Desjardins, C.A.; Abeel, T.; Earl, A.M. Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: Progress, promise, and challenges. Genome Med. 2019, 11, 45. [Google Scholar] [CrossRef]
- Borrell, S.; Gagneux, S. Strain diversity, epistasis and the evolution of drug resistance in Mycobacterium tuberculosis. Clin. Microbiol. Infect. 2011, 17, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Borrell, S.; Teo, Y.; Giardina, F.; Streicher, E.M.; Klopper, M.; Feldmann, J.; Müller, B.; Victor, T.C.; Gagneux, S. Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evol. Med. Public Health 2013, 2013, 65–74. [Google Scholar] [CrossRef]
- Wong, A. Epistasis and the Evolution of Antimicrobial Resistance. Front. Microbiol. 2017, 8, 246. [Google Scholar] [CrossRef]
- Muzondiwa, D.; Hlanze, H.; Reva, O.N. The epistatic landscape of antibiotic resistance of different clades of Mycobacterium tuberculosis. Antibiotics 2021, 10, 857. [Google Scholar] [CrossRef]
- Boritsch, E.C.; Brosch, R. Evolution of Mycobacterium tuberculosis: New insights into pathogenicity and drug resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Naz, S.; Paritosh, K.; Sanyal, P.; Khan, S.; Singh, Y.; Varshney, U.; Nandicoori, V.K. GWAS and functional studies suggest a role for altered DNA repair in the evolution of drug resistance in Mycobacterium tuberculosis. eLife 2023, 12, e75860. [Google Scholar] [CrossRef] [PubMed]
- Vīksna, A.; Sadovska, D.; Berge, I.; Bogdanova, I.; Vaivode, A.; Freimane, L.; Norvaiša, I.; Ozere, I.; Ranka, R. Genotypic and phenotypic comparison of drug resistance profiles of clinical multidrug-resistant Mycobacterium tuberculosis isolates using whole genome sequencing in Latvia. BMC Infect. Dis. 2023, 23, 638. [Google Scholar] [CrossRef] [PubMed]
- Katale, B.Z.; Mbelele, P.M.; Lema, N.A.; Campino, S.; Mshana, S.E.; Rweyemamu, M.M.; Phelan, J.E.; Keyyu, J.D.; Majigo, M.; Mbugi, E.V.; et al. Whole genome sequencing of Mycobacterium tuberculosis isolates and clinical outcomes of patients treated for multidrug-resistant tuberculosis in Tanzania. BMC Genom. 2020, 21, 174. [Google Scholar] [CrossRef] [PubMed]
- Sriraman, K.; Nilgiriwala, K.; Saranath, D.; Chatterjee, A.; Mistry, N. Deregulation of genes associated with alternate drug resistance mechanisms in Mycobacterium tuberculosis. Curr. Microbiol. 2018, 75, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Torres Ortiz, A.; Coronel, J.; Vidal, J.R.; Bonilla, C.; Moore, D.A.; Gilman, R.H.; Balloux, F.; Kon, O.M.; Didelot, X.; Grandjean, L. Genomic signatures of pre-resistance in Mycobacterium tuberculosis. Nat. Commun. 2021, 12, 7312. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi-Rad, M.; Bifani, P.; Martin, C.; Kremer, K.; Samper, S.; Rauzier, J.; Kreiswirth, B.; Blazquez, J.; Jouan, M.; van Soolingen, D.; et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 2003, 9, 838–845. [Google Scholar] [CrossRef] [PubMed]
- CRyPTIC Consortium and the 100,000 Genomes Project. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N. Eng. J. Med. 2018, 379, 1403–1415. [CrossRef] [PubMed]
- Manson, A.L.; Cohen, K.A.; Abeel, T.; Desjardins, C.A.; Armstrong, D.T.; Barry III, C.E.; Brand, J.; Jureen, P.; Malinga, L.; Nordenberg, D. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat. Genet. 2017, 49, 395. [Google Scholar] [CrossRef]
- Gillespie, J.J.; Wattam, A.R.; Cammer, S.A.; Gabbard, J.L.; Shukla, M.P.; Dalay, O.; Driscoll, T.; Hix, D.; Mane, S.P.; Mao, C.; et al. PATRIC: The comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect. Immun. 2011, 79, 4286–4298. [Google Scholar] [CrossRef]
- Chernyaeva, E.N.; Shulgina, M.V.; Rotkevich, M.S.; Dobrynin, P.V.; Simonov, S.A.; Shitikov, E.A.; Ischenko, D.S.; Karpova, I.Y.; Kostryukova, E.S.; Ilina, E.N. Genome-wide Mycobacterium tuberculosis variation (GMTV) database: A new tool for integrating sequence variations and epidemiology. BMC Genom. 2014, 15, 308. [Google Scholar] [CrossRef]
- Riojas, M.A.; McGough, K.J.; Rider-Riojas, C.J.; Rastogi, N.; Hazbón, M.H. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic synonyms of Mycobacterium tuberculosis. Int. J. Syst. Evol. Microbiol. 2018, 68, 324–332. [Google Scholar] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Muzondiwa, D.; Mutshembele, A.; Pierneef, R.E.; Reva, O.N. Resistance sniffer: An online tool for prediction of drug resistance patterns of mycobacterium tuberculosis isolates using next generation sequencing data. Int. J. Med. Microbiol. 2020, 310, 151399. [Google Scholar] [CrossRef] [PubMed]
- Van Niekerk, K.; Pierneef, R.; Reva, O.N.; Korostetskiy, I.S.; Ilin, A.I.; Akhmetova, G.K. Clade-specific distribution of antibiotic resistance mutations in the population of Mycobacterium tuberculosis. Prospects for drug resistance reversion. Basic. Biol. Appl. Actinobacteria 2018, 7, 79–98. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Reva, O.N. Supplementary Materials on the Statistical Evaluation of Associations between Mutations in Protein-Coding Genes of M. tuberculosis and Multidrug Resistance; and Predicted Epistatic Interactions among Global Markers of Drug Resistance in the Overall M. tuberculosis Population. 2024. Available online: https://zenodo.org/records/11530797 (accessed on 26 June 2024).
- WHO-Reported List of Mtb Drug Resistance. Available online: https://www.who.int/publications/i/item/9789240028173 (accessed on 26 June 2024).
- Gómez-González, P.J.; Grabowska, A.D.; Tientcheu, L.D.; Tsolaki, A.G.; Hibberd, M.L.; Campino, S.; Phelan, J.E.; Clark, T.G. Functional genetic variation in pe/ppe genes contributes to diversity in Mycobacterium tuberculosis lineages and potential interactions with the human host. Front. Microbiol. 2023, 14, 1244319. [Google Scholar] [CrossRef]
- Cohen, T.; Sommers, B.; Murray, M. The effect of drug resistance on the fitness of Mycobacterium tuberculosis. Lancet Infect. Dis. 2003, 3, 13–21. [Google Scholar] [CrossRef]
- Ye, X.; Peng, T.; Li, Y.; Huang, T.; Wang, H.; Hu, Z. Identification of an important function of CYP123: Role in the monooxygenase activity in a novel estradiol degradation pathway in bacteria. J. Steroid Biochem. Mol. Biol. 2022, 215, 106025. [Google Scholar] [CrossRef] [PubMed]
- Gagneux, S.; Long, C.D.; Small, P.M.; Van, T.; Schoolnik, G.K.; Bohannan, B.J. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 2006, 312, 1944–1946. [Google Scholar] [CrossRef] [PubMed]
- Cohen, T.; Murray, M. Modeling epidemics of multidrug resistant M. tuberculosis of heterogeneous fitness. Nat. Med. 2004, 10, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Ilin, A.I.; Kulmanov, M.E.; Korotetskiy, I.S.; Lankina, M.V.; Akhmetova, G.K.; Shvidko, S.V.; Reva, O.N. Constraints of drug resistance in Mycobacterium tuberculosis—Prospects for pharmacological reversion of susceptibility to antibiotics. Open Conf. Proc. J. 2017, 8, 1. [Google Scholar] [CrossRef]
- Billows, N.; Phelan, J.; Xia, D.; Peng, Y.; Clark, T.G.; Chang, Y.M. Large-scale statistical analysis of Mycobacterium tuberculosis genome sequences identifies compensatory mutations associated with multi-drug resistance. Sci. Rep. 2024, 14, 12312. [Google Scholar] [CrossRef] [PubMed]
- Lagutkin, D.; Panova, A.; Vinokurov, A.; Gracheva, A.; Samoilova, A.; Vasilyeva, I. Genome-wide study of drug resistant Mycobacterium tuberculosis and its intra-host evolution during treatment. Microorganisms 2022, 10, 1440. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, J.; Yan, B.; Zhang, W.; Guddat, L.W.; Liu, X.; Rao, Z. Structural basis for the broad substrate specificity of two acyl-CoA dehydrogenases FadE5 from mycobacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 16324–16332. [Google Scholar] [CrossRef] [PubMed]
- Beites, T.; Jansen, R.S.; Wang, R.; Jinich, A.; Rhee, K.Y.; Schnappinger, D.; Ehrt, S. Multiple acyl-CoA dehydrogenase deficiency kills Mycobacterium tuberculosis in vitro and during infection. Nat. Commun. 2021, 12, 6593. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, Á.; Mares-Alejandre, R.E.; Muñoz-Muñoz, P.L.A.; Ruvalcaba-Ruiz, S.; González-Sánchez, R.A.; Bernáldez-Sarabia, J.; Meléndez-López, S.G.; Licea-Navarro, A.F.; Ramos-Ibarra, M.A. Molecular analysis of streptomycin resistance genes in clinical strains of Mycobacterium tuberculosis and biocomputational analysis of the MtGidB L101F variant. Antibiotics 2021, 10, 807. [Google Scholar] [CrossRef]
- Morlock, G.P.; Metchock, B.; Sikes, D.; Crawford, J.T.; Cooksey, R.C. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 2003, 47, 3799–3805. [Google Scholar] [CrossRef]
- Laborde, J.; Deraeve, C.; Bernardes-Génisson, V. Update of antitubercular prodrugs from a molecular perspective: Mechanisms of action, bioactivation pathways, and associated resistance. ChemMedChem 2017, 12, 1657–1676. [Google Scholar] [CrossRef] [PubMed]
- de Souza, V.C.; Antunes, D.; Santos, L.H.S.; Goliatt, P.V.Z.C.; Caffarena, E.R.; Guimarães, A.C.R.; Galvão, T.C. Insights into the mechanism of ethionamide resistance in Mycobacterium tuberculosis through an in silico structural evaluation of EthA and mutants identified in clinical isolates. Catalysts 2020, 10, 543. [Google Scholar] [CrossRef]
- Fernandez Do Porto, D.A.; Monteserin, J.; Campos, J.; Sosa, E.J.; Matteo, M.; Serral, F.; Yokobori, N.; Benevento, A.F.; Poklepovich, T.; Pardo, A.; et al. Five-year microevolution of a multidrug-resistant Mycobacterium tuberculosis strain within a patient with inadequate compliance to treatment. BMC Infect. Dis. 2021, 21, 394. [Google Scholar] [CrossRef] [PubMed]
- Bothra, A.; Arumugam, P.; Panchal, V.; Menon, D.; Srivastava, S.; Shankaran, D.; Nandy, A.; Jaisinghani, N.; Singh, A.; Gokhale, R.S.; et al. Phospholipid homeostasis, membrane tenacity and survival of Mtb in lipid rich conditions is determined by MmpL11 function. Sci. Rep. 2018, 8, 8317. [Google Scholar] [CrossRef] [PubMed]
- McNeil, M.B.; O’Malley, T.; Dennison, D.; Shelton, C.D.; Sunde, B.; Parish, T. Multiple mutations in Mycobacterium tuberculosis MmpL3 increase resistance to MmpL3 inhibitors. mSphere 2020, 5, e00985-20. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Nakata, N.; Mukai, T.; Kawagishi, I.; Ato, M. Coexpression of MmpS5 and MmpL5 contributes to both efflux transporter MmpL5 trimerization and drug resistance in Mycobacterium tuberculosis. mSphere 2021, 6, e00518-20. [Google Scholar] [CrossRef]
- Guo, J.; Zheng, X.; Xu, L.; Liu, Z.; Xu, K.; Li, S.; Wen, T.; Liu, S.; Pang, H. Characterization of a novel esterase Rv0045c from Mycobacterium tuberculosis. PLoS ONE 2010, 5, e13143. [Google Scholar] [CrossRef] [PubMed]
- Sherlin, D.; Anishetty, S. Mechanistic insights from molecular dynamic simulation of Rv0045c esterase in Mycobacterium tuberculosis. J. Mol. Model. 2015, 21, 1–8. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, W.; Gao, F.; Huang, Y.; Lv, C.; Wang, H. Comparison of the proteome of isoniazid-resistant and-susceptible strains of Mycobacterium tuberculosis. Microb. Drug Resist. 2006, 12, 231–238. [Google Scholar] [CrossRef]
- Jhingan, G.D.; Kumari, S.; Jamwal, S.V.; Kalam, H.; Arora, D.; Jain, N.; Kumaar, L.K.; Samal, A.; Rao, K.V.; Kumar, D. Comparative proteomic analyses of avirulent, virulent, and clinical strains of Mycobacterium tuberculosis identify strain-specific patterns. J. Biol. Chem. 2016, 291, 14257–14273. [Google Scholar] [CrossRef]
- Diamond, S.; Rubin, B.E.; Shultzaberger, R.K.; Chen, Y.; Barber, C.D.; Golden, S.S. Redox crisis underlies conditional light–dark lethality in cyanobacterial mutants that lack the circadian regulator, RpaA. Proc. Natl. Acad. Sci. USA 2017, 114, E580–E589. [Google Scholar] [CrossRef] [PubMed]
- Furió, V.; Moreno-Molina, M.; Chiner-Oms, Á.; Villamayor, L.M.; Torres-Puente, M.; Comas, I. An evolutionary functional genomics approach identifies novel candidate regions involved in isoniazid resistance in Mycobacterium tuberculosis. Commun. Biol. 2021, 4, 1322. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Hu, P.; Zhang, L.; Wang, Z.-X.; Fleming, J.; Ni, B.; Luo, J.; Guan, C.-X.; Bai, L.; Tan, Y. Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene. Commun. Biol. 2023, 6, 156. [Google Scholar] [CrossRef] [PubMed]
- Tudó, G.; Laing, K.; Mitchison, D.A.; Butcher, P.D.; Waddell, S.J. Examining the basis of isoniazid tolerance in nonreplicating Mycobacterium tuberculosis using transcriptional profiling. Future Med. Chem. 2010, 2, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Shekar, S.; Yeo, Z.X.; Wong, J.C.; Chan, M.K.; Ong, D.C.; Tongyoo, P.; Wong, S.-Y.; Lee, A.S. Detecting novel genetic variants associated with isoniazid-resistant Mycobacterium tuberculosis. PLoS ONE 2014, 9, e102383. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Chen, L.; Zhang, H. Serum proteomic analysis of Mycobacterium tuberculosis antigens for discriminating active tuberculosis from latent infection. J. Int. Med. Res. 2020, 48, 0300060520910042. [Google Scholar] [CrossRef] [PubMed]
- Raman, K.; Chandra, N. Mycobacterium tuberculosis interactome analysis unravels potential pathways to drug resistance. BMC Microbiol. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Qian, J.; Chen, R.; Wang, H.; Zhang, X. Role of the PE/PPE family in host-pathogen interactions and prospects for anti-tuberculosis vaccine and diagnostic tool design. Front. Cell Infect. Microbiol. 2020, 10, 594288. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.; Alam, A.; Ehtram, A.; Rani, A.; Grover, S.; Ehtesham, N.Z.; Hasnain, S.E. The Mycobacterium tuberculosis PE_PGRS protein family acts as an immunological decoy to subvert host immune response. Int. J. Mol. Sci. 2022, 23, 525. [Google Scholar] [CrossRef]
- Allue Guardia, A.; Garcia, J.I.; Torrelles, J.B. Evolution of drug-resistant Mycobacterium tuberculosis strains and their adaptation to the human lung environment. Front. Microbiol. 2021, 12, 137. [Google Scholar] [CrossRef]
- Yew, W.W.; Chan, D.P.; Chang, K.C.; Zhang, Y. Does oxidative stress contribute to antituberculosis drug resistance? J. Thorac. Dis. 2019, 11, E100–E102. [Google Scholar] [CrossRef] [PubMed]
- Shastri, M.D.; Shukla, S.D.; Chong, W.C.; Dua, K.; Peterson, G.M.; Patel, R.P.; Hansbro, P.M.; Eri, R.; O’Toole, R.F. Role of oxidative stress in the pathology and management of human tuberculosis. Oxid. Med. Cell Longev. 2018, 7695364. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Balaji, K.N. The PE and PPE proteins of Mycobacterium tuberculosis. Tuberculosis 2011, 91, 41–447. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sharma, M. Proline-Glutamate/Proline-Proline-Glutamate (PE/PPE) proteins of Mycobacterium tuberculosis: The multifaceted immune-modulators. Acta Trop. 2021, 222, 106035. [Google Scholar]
- Flores, J.; Espitia, C. Differential expression of PE and PE_PGRS genes in Mycobacterium tuberculosis strains. Gene 2003, 318, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Ates, L.S. New insights into the mycobacterial PE and PPE proteins provide a framework for future research. Mol. Microbiol. 2020, 113, 4–21. [Google Scholar] [CrossRef]
- Arora, G.; Bothra, A.; Prosser, G.; Arora, K.; Sajid, A. Role of post-translational modifications in the acquisition of drug resistance in Mycobacterium tuberculosis. FEBS J. 2021, 288, 3375–3393. [Google Scholar] [CrossRef]
- Ilin, A.I.; Kulmanov, M.E.; Korotetskiy, I.S.; Islamov, R.A.; Akhmetova, G.K.; Lankina, M.V.; Reva, O.N. Genomic insight into mechanisms of reversion of antibiotic resistance in multidrug resistant Mycobacterium tuberculosis induced by a nanomolecular iodine-containing complex FS-1. Front. Cell Infect. Microbiol. 2017, 7, 151. [Google Scholar] [CrossRef]
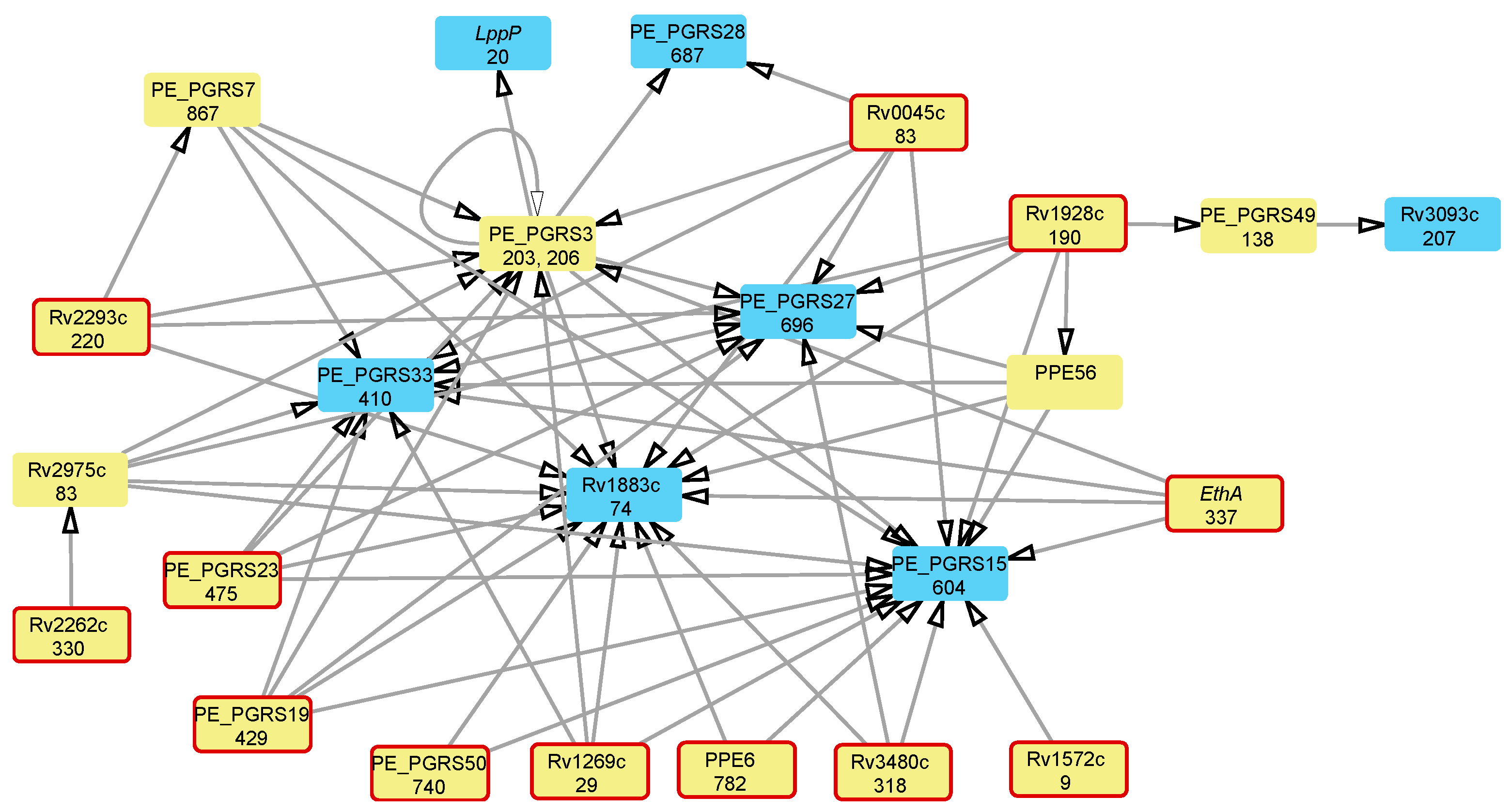
| Locus Tag | Gene Product | Polymorphic Codon | Associated Antibiotic Resistance * |
|---|---|---|---|
| Rv1269c | Conserved secreted protein | 29 | AM, CM, EMB, INH, MFX, OFX, PTO, PZA, RIF, and SM |
| Rv0766c | Cytochrome P450 Cyp123 | 278, 279 | AM, CM, EMB, INH, MFX, OFX, PTO, PZA, RIF, and SM |
| Rv3480c | Triacylglycerol synthase, diacylglycerol acyltransferase | 318 | AM, CM, EMB, MFX, OFX, PTO, PZA, and RIF |
| Rv1928c | Short-chain dehydrogenase/reductase | 190 | CM, EMB, MFX, OFX, PTO, PZA, and SM |
| Rv0045c | Possible hydrolase | 83 | AM, CM, EMB, MFX, OFX, PTO, and PZA |
| Rv3854c | Monooxygenase EthA | 337 † | AM, CM, EMB, OFX, PTO, PZA, and SM |
| Rv0823c | Transcriptional regulatory protein | 322 | AM, CM, EMB, MFX, OFX, PTO, and PZA |
| Rv3919c | Glucose-inhibited division protein GidB | 65 † | AM, EMB, OFX, PTO, PZA, and SM |
| Rv3346c | Conserved transmembrane protein | 19 | EMB, INH, PZA, RIF, and SM |
| Rv1522c | Transmembrane transport protein MmpL12 | 549 | EMB, INH, PZA, RIF, and SM |
| Rv3093c | Oxidoreductase | 207 | EMB, INH, PZA, RIF, and SM |
| Rv3761c | Acyl-CoA dehydrogenase FadE36 | 303 | INH, PZA, RIF, and SM |
| Rv2330c | Lipoprotein LppP | 20 | EMB, INH, PZA, RIF, and SM |
| Rv2434c | Conserved transmembrane protein | 317 | AM, CM, MFX, OFX, and PTO |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hlanze, H.; Mutshembele, A.; Reva, O.N. Universal Lineage-Independent Markers of Multidrug Resistance in Mycobacterium tuberculosis. Microorganisms 2024, 12, 1340. https://doi.org/10.3390/microorganisms12071340
Hlanze H, Mutshembele A, Reva ON. Universal Lineage-Independent Markers of Multidrug Resistance in Mycobacterium tuberculosis. Microorganisms. 2024; 12(7):1340. https://doi.org/10.3390/microorganisms12071340
Chicago/Turabian StyleHlanze, Hleliwe, Awelani Mutshembele, and Oleg N. Reva. 2024. "Universal Lineage-Independent Markers of Multidrug Resistance in Mycobacterium tuberculosis" Microorganisms 12, no. 7: 1340. https://doi.org/10.3390/microorganisms12071340
APA StyleHlanze, H., Mutshembele, A., & Reva, O. N. (2024). Universal Lineage-Independent Markers of Multidrug Resistance in Mycobacterium tuberculosis. Microorganisms, 12(7), 1340. https://doi.org/10.3390/microorganisms12071340





