Biofilm Formation, Motility, and Virulence of Listeria monocytogenes Are Reduced by Deletion of the Gene lmo0159, a Novel Listerial LPXTG Surface Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Primers
2.2. Cells and Cell Culture
2.3. Experimental Design
2.4. Construction of the ∆lmo0159 Mutants Strain
2.5. Growth Assay
2.6. Biofilm-Formation Assay
2.7. Motility Assay
2.7.1. Swimming Motility Assay
2.7.2. Plaque Forming Assay
2.8. Scratch Assay
2.9. Adhesion and Invasion Assays
2.10. Intracellular Proliferation Assay
2.11. The Mice Survival Rate Assay and the Detection of 50% Lethal Dose (LD50)
2.12. Bacterial Loads in Mice Livers, Spleens, and Brains
2.13. RT-PCR Assays
2.14. Statistical Analysis
3. Results
3.1. Construction of the lmo0159 Deletion Mutant (∆lmo0159)
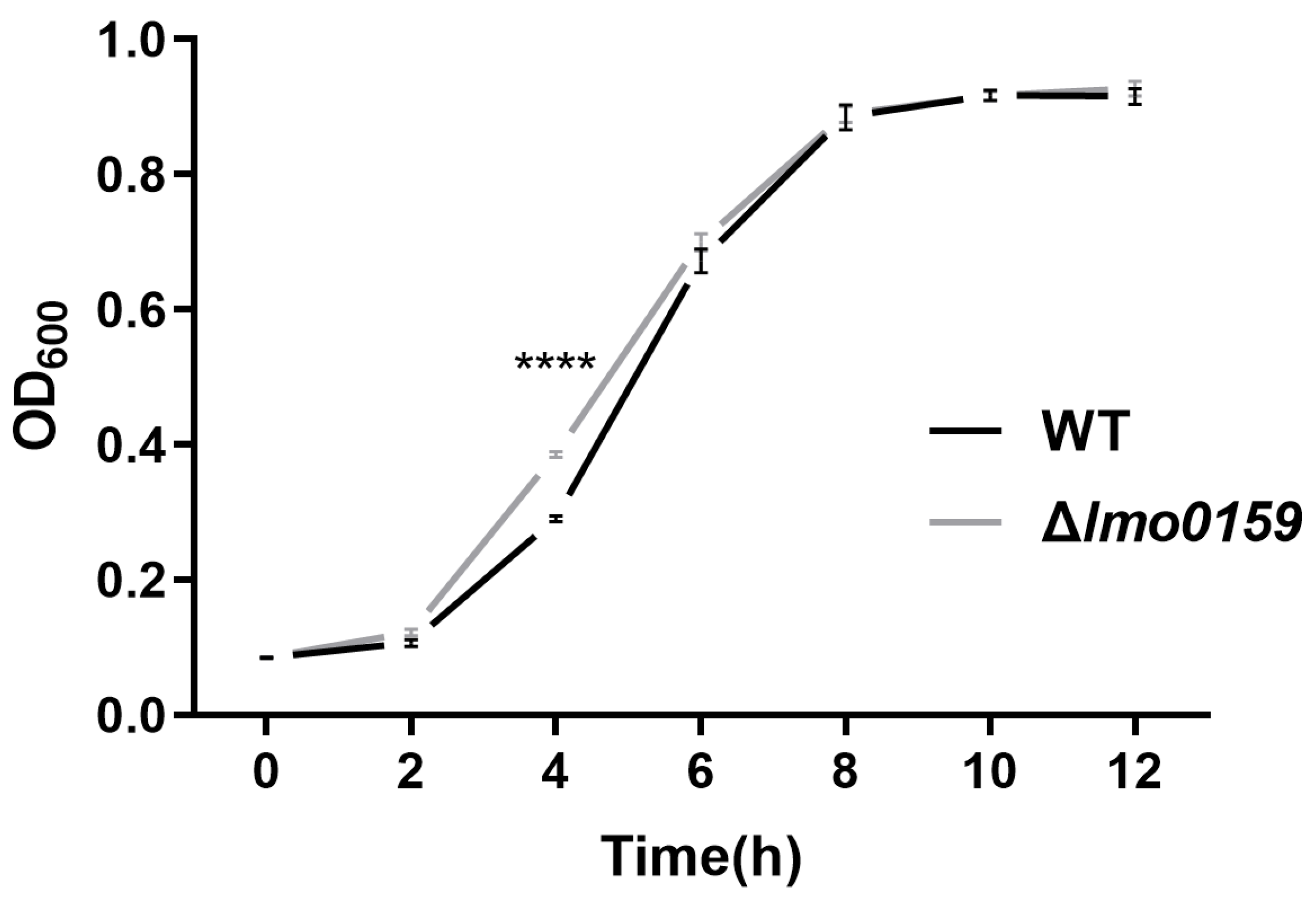
3.2. Impact of Lmo0159 Deletion on L. monocytogenes Growth
3.3. Lm0159 Mutant Is Defective in Biofilm Formation
3.4. Inhibiting Effects of Lmo0159 on the L. monocytogenes Motility
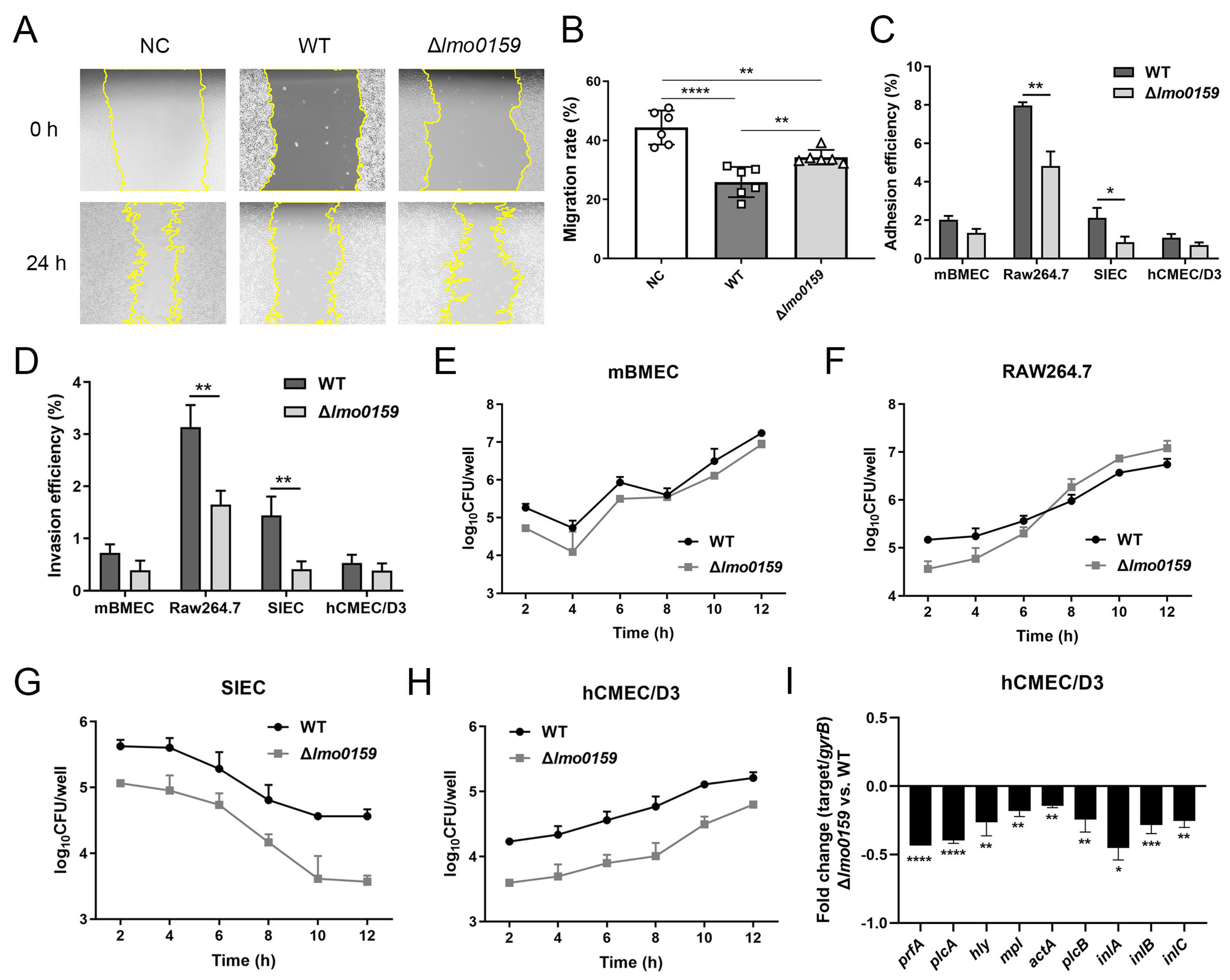
3.5. Effects of Lmo0159 on Cell Adhesion, Invasion, Intracellular Proliferation, and Virulence Factor Levels
3.6. Inactivation of Lmo0159 Impairs the Virulence of L. monocytogenes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cabanes, D.; Dehoux, P.; Dussurget, O.; Frangeul, L.; Cossart, P. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 2002, 10, 238–245. [Google Scholar] [CrossRef]
- Barbuddhe, S.B.; Chakraborty, T. Listeria as an enteroinvasive gastrointestinal pathogen. Curr. Top. Microbiol. Immunol. 2009, 337, 173–195. [Google Scholar] [CrossRef]
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef]
- Stavru, F.; Archambaud, C.; Cossart, P. Cell biology and immunology of Listeria monocytogenes infections: Novel insights. Immunol. Rev. 2011, 240, 160–184. [Google Scholar] [CrossRef]
- Carvalho, F.; Sousa, S.; Cabanes, D. How Listeria monocytogenes organizes its surface for virulence. Front. Cell. Infect. Microbiol. 2014, 4, 48. [Google Scholar] [CrossRef]
- Camejo, A.; Carvalho, F.; Reis, O.; Leitão, E.; Sousa, S.; Cabanes, D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2011, 2, 379–394. [Google Scholar] [CrossRef]
- Mariscotti, J.F.; Quereda, J.J.; Pucciarelli, M.G. Contribution of sortase A to the regulation of Listeria monocytogenes LPXTG surface proteins. Int. Microbiol. 2012, 15, 43–51. [Google Scholar] [CrossRef]
- Bonazzi, M.; Lecuit, M.; Cossart, P. Listeria monocytogenes internalin and E-cadherin: From bench to bedside. Cold Spring Harb. Perspect. Biol. 2009, 1, a003087. [Google Scholar] [CrossRef]
- Schubert, W.D.; Urbanke, C.; Ziehm, T.; Beier, V.; Machner, M.P.; Domann, E.; Wehland, J.; Chakraborty, T.; Heinz, D.W. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 2002, 111, 825–836. [Google Scholar] [CrossRef]
- Cabanes, D.; Sousa, S.; Cebriá, A.; Lecuit, M.; García-del Portillo, F.; Cossart, P. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. EMBO J. 2005, 24, 2827–2838. [Google Scholar] [CrossRef]
- Personnic, N.; Bruck, S.; Nahori, M.A.; Toledo-Arana, A.; Nikitas, G.; Lecuit, M.; Dussurget, O.; Cossart, P.; Bierne, H. The stress-induced virulence protein InlH controls interleukin-6 production during murine listeriosis. Infect. Immun. 2010, 78, 1979–1989. [Google Scholar] [CrossRef]
- Smith, K.; Youngman, P. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 1992, 74, 705–711. [Google Scholar] [CrossRef]
- Yundan, X.; Baotun, W.; Juan, F.; Wei, L.; Biao, J.; Chun, L.; Yanhua, H.; Youlu, S. The influence of enzyme EII of the cellobiose-phosphotransferase system on the virulence of Streptococcus agalactiae in Nile tilapia (Oreochromis niloticus). Aquaculture 2021, 535, 736340. [Google Scholar] [CrossRef]
- Ushijima, B.; Häse, C.C. Influence of Chemotaxis and Swimming Patterns on the Virulence of the Coral Pathogen Vibrio coralliilyticus. J. Bacteriol. 2018, 200, e00791-17. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Du, W.F.; Yang, J.; Liu, Z.G.; Yousef, A.E. Control of Listeria monocytogenes biofilm by paenibacterin, a natural antimicrobial lipopeptide. Food Control 2018, 84, 529–535. [Google Scholar] [CrossRef]
- Edouard, S.; Raoult, D. Use of the plaque assay for testing the antibiotic susceptibility of intracellular bacteria. Int. J. Dermatol. 2013, 8, 1301–1316. [Google Scholar] [CrossRef]
- Al-Shabib, N.A.; Husain, F.M.; Nadeem, M.; Khan, M.S.; Al-Qurainy, F.; Alyousef, A.A.; Arshad, M.; Khan, A.; Khan, J.M.; Alam, P.; et al. Bio-inspired facile fabrication of silver nanoparticles from in vitro grown shoots of Tamarix nilotica: Explication of its potential in impeding growth and biofilms of Listeria monocytogenes and assessment of wound healing ability. RSC Adv. 2020, 10, 30139–30149. [Google Scholar] [CrossRef]
- Li, H.; Qiao, Y.; Du, D.; Wang, J.; Ma, X. Deletion of the oligopeptide transporter Lmo2193 decreases the virulence of Listeria monocytogenes. J. Vet. Sci. 2020, 21, e88. [Google Scholar] [CrossRef]
- Liu, C.X.; Shi, W.D.; Kou, L.J.; Ma, X.; Wang, J.; Gao, S.J.; Lyu, S.F.; Chen, X.K.; Kong, C.L.; Ren, H.J.; et al. Permease IIC of Listeria monocytogenes pathogenicity island 4 regulates the transcription of key virulence factors. China Anim. Husb. Vet. Med. 2023, 50, 1340–1351. [Google Scholar] [CrossRef]
- Gandhi, M.; Chikindas, M.L. Listeria: A foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 2007, 113, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kolb-Mäurer, A.; Gentschev, I.; Fries, H.W.; Fiedler, F.; Bröcker, E.B.; Kämpgen, E.; Goebel, W. Listeria monocytogenes-infected human dendritic cells: Uptake and host cell response. Infect. Immun. 2000, 68, 3680–3688. [Google Scholar] [CrossRef]
- Lungu, B.; Ricke, S.C.; Johnson, M.G. Growth, survival, proliferation and pathogenesis of Listeria monocytogenes under low oxygen or anaerobic conditions: A review. Anaerobe 2009, 15, 7–17. [Google Scholar] [CrossRef]
- Bierne, H.; Cossart, P. Listeria monocytogenes surface proteins: From genome predictions to function. Microbiol. Mol. Biol. Rev. 2007, 71, 377–397. [Google Scholar] [CrossRef]
- Bierne, H.; Mazmanian, S.K.; Trost, M.; Pucciarelli, M.G.; Liu, G.; Dehoux, P.; Jänsch, L.; Garcia-del Portillo, F.; Schneewind, O.; Cossart, P. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 2002, 43, 869–881. [Google Scholar] [CrossRef]
- Doumith, M.; Cazalet, C.; Simoes, N.; Frangeul, L.; Jacquet, C.; Kunst, F.; Martin, P.; Cossart, P.; Glaser, P.; Buchrieser, C. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 2004, 72, 1072–1083. [Google Scholar] [CrossRef]
- Hain, T.; Ghai, R.; Billion, A.; Kuenne, C.T.; Steinweg, C.; Izar, B.; Mohamed, W.; Mraheil, M.A.; Domann, E.; Schaffrath, S.; et al. Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes. BMC Genom. 2012, 13, 144. [Google Scholar] [CrossRef]
- Calvo, E.; Pucciarelli, M.G.; Bierne, H.; Cossart, P.; Albar, J.P.; García-Del Portillo, F. Analysis of the Listeria cell wall proteome by two-dimensional nanoliquid chromatography coupled to mass spectrometry. Proteomics 2005, 5, 433–443. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Ling, C.; Ma, X.; Du, D.D.; Qian, L.X.; Li, H.H. Cloning and bioinformatics analysis of lmo0159 gene of Listeria monocytogenes isolated strain from Xinjiang. Genom. Appl. Biol. 2018, 37, 5287–5295. [Google Scholar] [CrossRef]
- Cacace, G.; Mazzeo, M.F.; Sorrentino, A.; Spada, V.; Malorni, A.; Siciliano, R.A. Proteomics for the elucidation of cold adaptation mechanisms in Listeria monocytogenes. J. Proteomics 2010, 73, 2021–2030. [Google Scholar] [CrossRef] [PubMed]
- Soni, K.A.; Nannapaneni, R.; Tasara, T. The contribution of transcriptomic and proteomic analysis in elucidating stress adaptation responses of Listeria monocytogenes. Foodborne Pathog. Dis. 2011, 8, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Kyoui, D.; Hirokawa, E.; Takahashi, H.; Kuda, T.; Kimura, B. Effect of glucose on Listeria monocytogenes biofilm formation, and assessment of the biofilm’s sanitation tolerance. Biofouling 2016, 32, 815–826. [Google Scholar] [CrossRef]
- Lemon, K.P.; Higgins, D.E.; Kolter, R. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 2007, 189, 4418–4424. [Google Scholar] [CrossRef]
- Kamp, H.D.; Higgins, D.E. A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog. 2011, 7, e1002153. [Google Scholar] [CrossRef]
- Mirzahosseini, H.K.; Najmeddin, F.; Najafi, A.; Ahmadi, A.; Sharifnia, H.; Khaledi, A.; Mojtahedzadeh, M. Correlation of biofilm formation, virulence factors, and phylogenetic groups among Escherichia coli strains causing urinary tract infection: A global systematic review and meta-analysis. J. Res. Med. Sci. 2023, 28, 66. [Google Scholar] [CrossRef]
- Lecuit, M.; Vandormael-Pournin, S.; Lefort, J.; Huerre, M.; Gounon, P.; Dupuy, C.; Babinet, C.; Cossart, P. A transgenic model for listeriosis: Role of internalin in crossing the intestinal barrier. Science 2001, 292, 1722–1725. [Google Scholar] [CrossRef]
- Lecuit, M.; Nelson, D.M.; Smith, S.D.; Khun, H.; Huerre, M.; Vacher-Lavenu, M.C.; Gordon, J.I.; Cossart, P. Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: Role of internalin interaction with trophoblast E-cadherin. Proc. Natl. Acad. Sci. USA 2004, 101, 6152–6157. [Google Scholar] [CrossRef]
- Disson, O.; Grayo, S.; Huillet, E.; Nikitas, G.; Langa-Vives, F.; Dussurget, O.; Ragon, M.; Le Monnier, A.; Babinet, C.; Cossart, P.; et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 2008, 455, 1114–1118. [Google Scholar] [CrossRef]
- Stachowiak, R.; Jagielski, T.; Roeske, K.; Osińska, O.; Gunerka, P.; Wiśniewski, J.; Bielecki, J. Lmo0171, a novel internalin-like protein, determines cell morphology of Listeria monocytogenes and its ability to invade human cell lines. Curr. Microbiol. 2015, 70, 267–274. [Google Scholar] [CrossRef]
- Chen, B.-Y.; Kim, T.-J.; Silva, J.L.; Jung, Y.-S. Positive Correlation Between the Expression of inlA and inlB Genes of Listeria monocytogenes and Its Attachment Strength on Glass Surface. Food Biophys. 2009, 4, 304–311. [Google Scholar] [CrossRef]
- Luo, Q.; Shang, J.; Feng, X.; Guo, X.; Zhang, L.; Zhou, Q. PrfA led to reduced biofilm formation and contributed to altered gene expression patterns in biofilm-forming Listeria monocytogenes. Curr. Microbiol. 2013, 67, 372–378. [Google Scholar] [CrossRef] [PubMed]
- de las Heras, A.; Cain, R.J.; Bielecka, M.K.; Vázquez-Boland, J.A. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 2011, 14, 118–127. [Google Scholar] [CrossRef]
- Wood, T.K.; González Barrios, A.F.; Herzberg, M.; Lee, J. Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 2006, 72, 361–367. [Google Scholar] [CrossRef]
- Disson, O.; Lecuit, M. Targeting of the central nervous system by Listeria monocytogenes. Virulence 2012, 3, 213–221. [Google Scholar] [CrossRef] [PubMed]

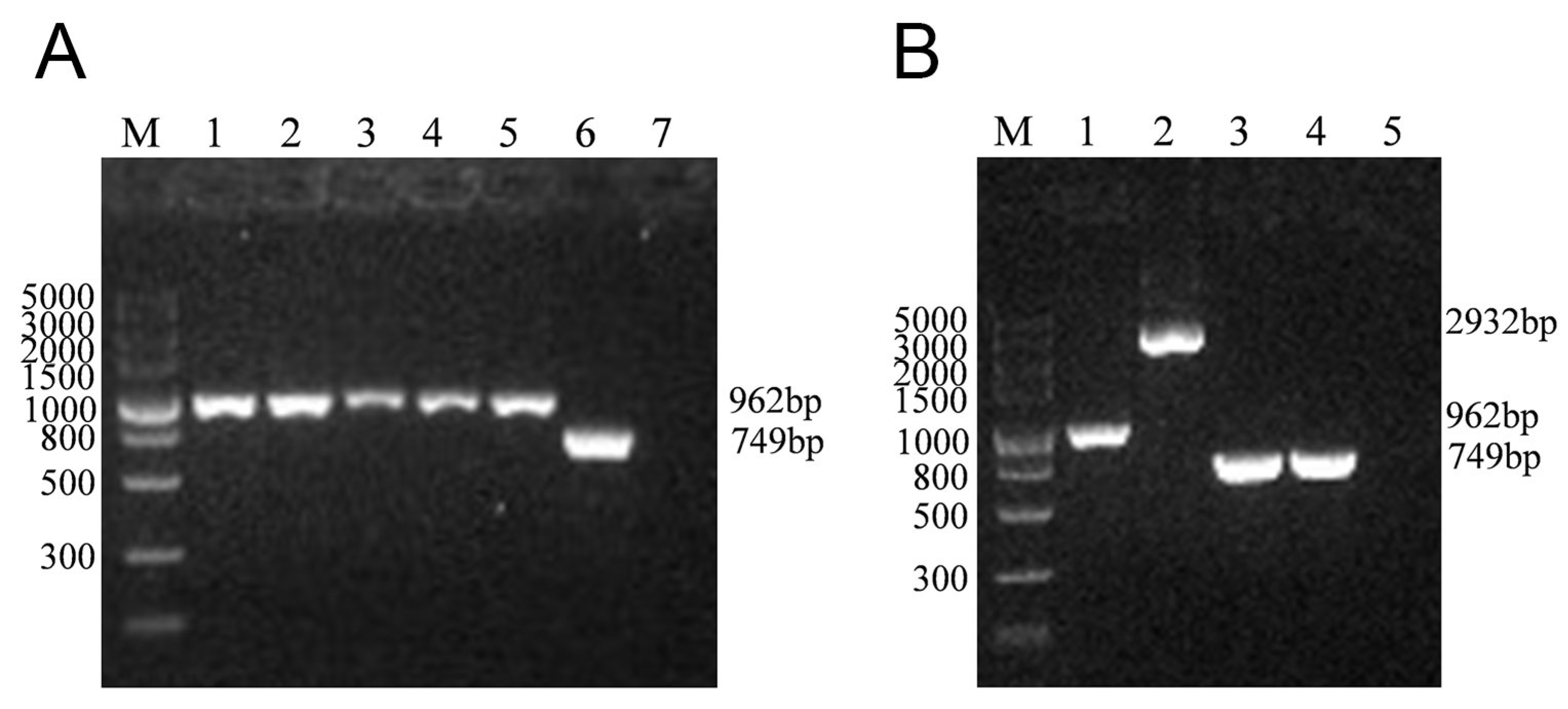



| Primer | Nucleotide Sequence (5′-3′) | Target Gene | Length (bp) |
|---|---|---|---|
| 0159-F1 | CGGGATCCCAAATGCTCATTCTGGACC (BamH I) | Upstream sequence of lmo0159 gene (A1) | 388 |
| 0159-R1 | TCGTTTCAGCAGAAGACCAGATTGTGCTAGCGACGATT | ||
| 0159-F2 | AATCGTCGCTAGCACAATCTGGTCTTCTGCTGAAACGA | Downstream sequence of lmo0159 gene (A2) | 409 |
| 0159-R2 | AACTGCAGTCGTAATAACCGAAGTCCG (Pst I) | ||
| 0159-A | TGAGTATGCTGGTATGGGA | Verify primers of lmo0159 deletion | 2932/962 |
| 0159-B | GTCGCCTTCCTTCACATTA | ||
| hly-F | CTGAATTCGGCTGTTACTAAAGAGCAGTTGC | hly | 749 |
| hly-R | ATGGATCCTTAGCCCCAGATGGAGATATTCTA |
| Gene | forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| degU | ACGCATAGAGAGTGCGAGGTATT | CCCAATTCCGCGGTTACTT |
| flaA | CTGGTATGAGTCGCCTTAG | CATTTGCGGTGTTTGGTTTG |
| mogR | AACTGCCGAAGAAATCTACCATTT | CGATTCCACCGTGTTCTTCA |
| fur | ACAGTTGCAAGGCCAGTGTCGGGTG | TGGAAGGTCGTATTGGACGCATTAA |
| fliP | TTGGCCGGGTGTGAATGT | CCATTTACACCAAGCGAATCC |
| Strain | Group | Does | No. of Animals | Death No. | Mortality | LD50 Median Lethal Dose |
|---|---|---|---|---|---|---|
| WT | 1 | 5.3 × 109 | 8 | 8 | 1 | 106.22 |
| 2 | 5.3 × 108 | 8 | 8 | 1 | ||
| 3 | 5.3 × 107 | 8 | 7 | 0.875 | ||
| 4 | 5.3 × 106 | 8 | 6 | 0.75 | ||
| 5 | 5.3 × 105 | 8 | 3 | 0.375 | ||
| Δlmo0159 | 1 | 3.7 × 109 | 8 | 8 | 1 | 107.19 |
| 2 | 3.7 × 108 | 8 | 7 | 0.875 | ||
| 3 | 3.7 × 107 | 8 | 5 | 0.625 | ||
| 4 | 3.7 × 106 | 8 | 3 | 0.375 | ||
| 5 | 3.7 × 105 | 8 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, W.; Zhang, Q.; Li, H.; Du, D.; Ma, X.; Wang, J.; Jiang, J.; Liu, C.; Kou, L.; Ren, J. Biofilm Formation, Motility, and Virulence of Listeria monocytogenes Are Reduced by Deletion of the Gene lmo0159, a Novel Listerial LPXTG Surface Protein. Microorganisms 2024, 12, 1354. https://doi.org/10.3390/microorganisms12071354
Shi W, Zhang Q, Li H, Du D, Ma X, Wang J, Jiang J, Liu C, Kou L, Ren J. Biofilm Formation, Motility, and Virulence of Listeria monocytogenes Are Reduced by Deletion of the Gene lmo0159, a Novel Listerial LPXTG Surface Protein. Microorganisms. 2024; 12(7):1354. https://doi.org/10.3390/microorganisms12071354
Chicago/Turabian StyleShi, Weidi, Qiwen Zhang, Honghuan Li, Dongdong Du, Xun Ma, Jing Wang, Jianjun Jiang, Caixia Liu, Lijun Kou, and Jingjing Ren. 2024. "Biofilm Formation, Motility, and Virulence of Listeria monocytogenes Are Reduced by Deletion of the Gene lmo0159, a Novel Listerial LPXTG Surface Protein" Microorganisms 12, no. 7: 1354. https://doi.org/10.3390/microorganisms12071354





