Pathogenicity of Multidrug-Resistant Salmonella typhimurium Isolated from Ducks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Main Reagents
2.2. Sampling Strategy and Isolation and Identification of Salmonella
2.3. Drug Susceptibility Test
2.4. Detection of Salmonella Serotype
2.5. Detection of Virulence Genes
2.6. Biofilm Formation Assay
2.7. Motility Assay
2.8. Animal Experiment
2.9. Real-Time Quantitative PCR
2.10. Data Analysis
3. Results
3.1. Isolation, Serotype, and Antibiotic Resistance of Salmonella Strains
3.2. Virulence Genes of S. typhimurium
3.3. Analysis of the Biofilm Formation Ability and Motility of S. typhimurium
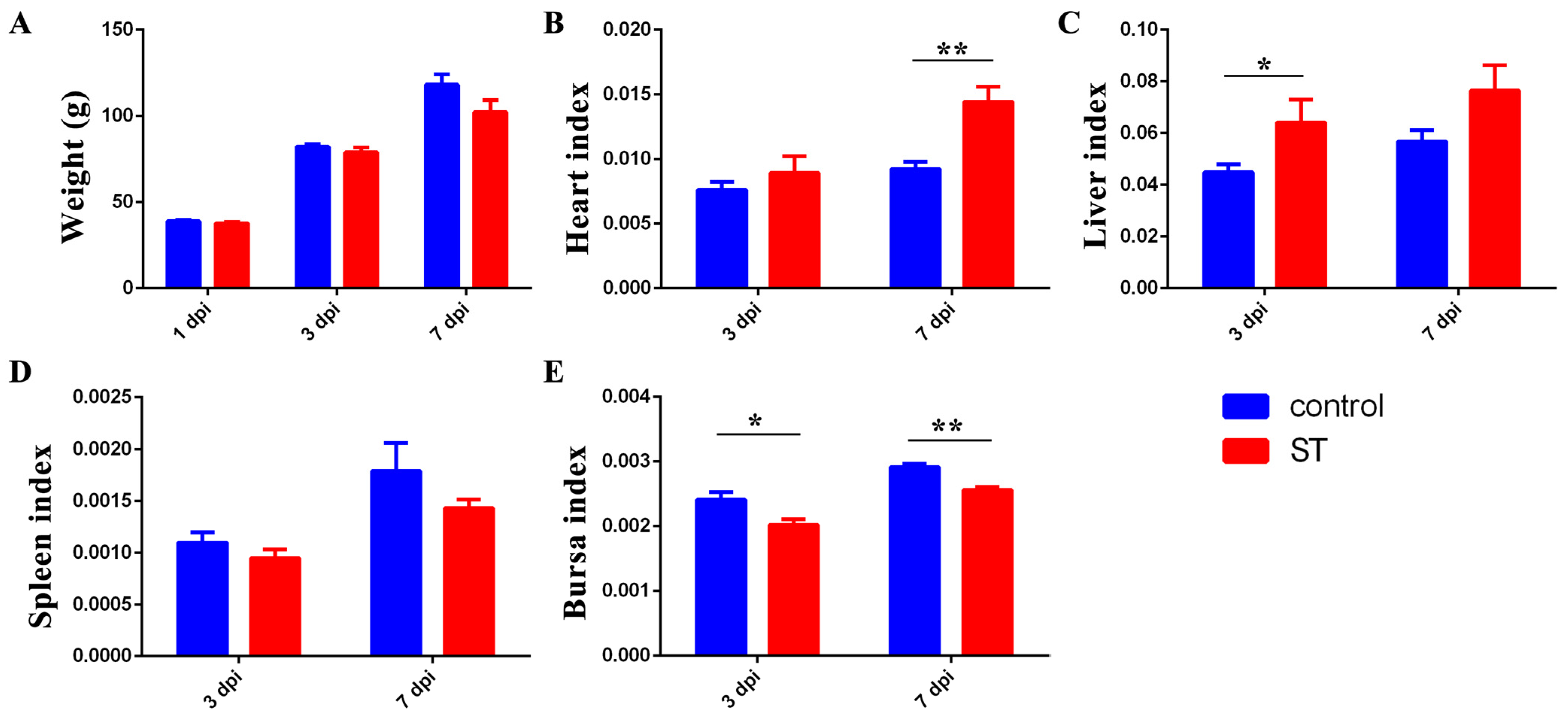
3.4. Infection with 22SD07 Significantly Reduces Body Weight and Affects Organ Indexes
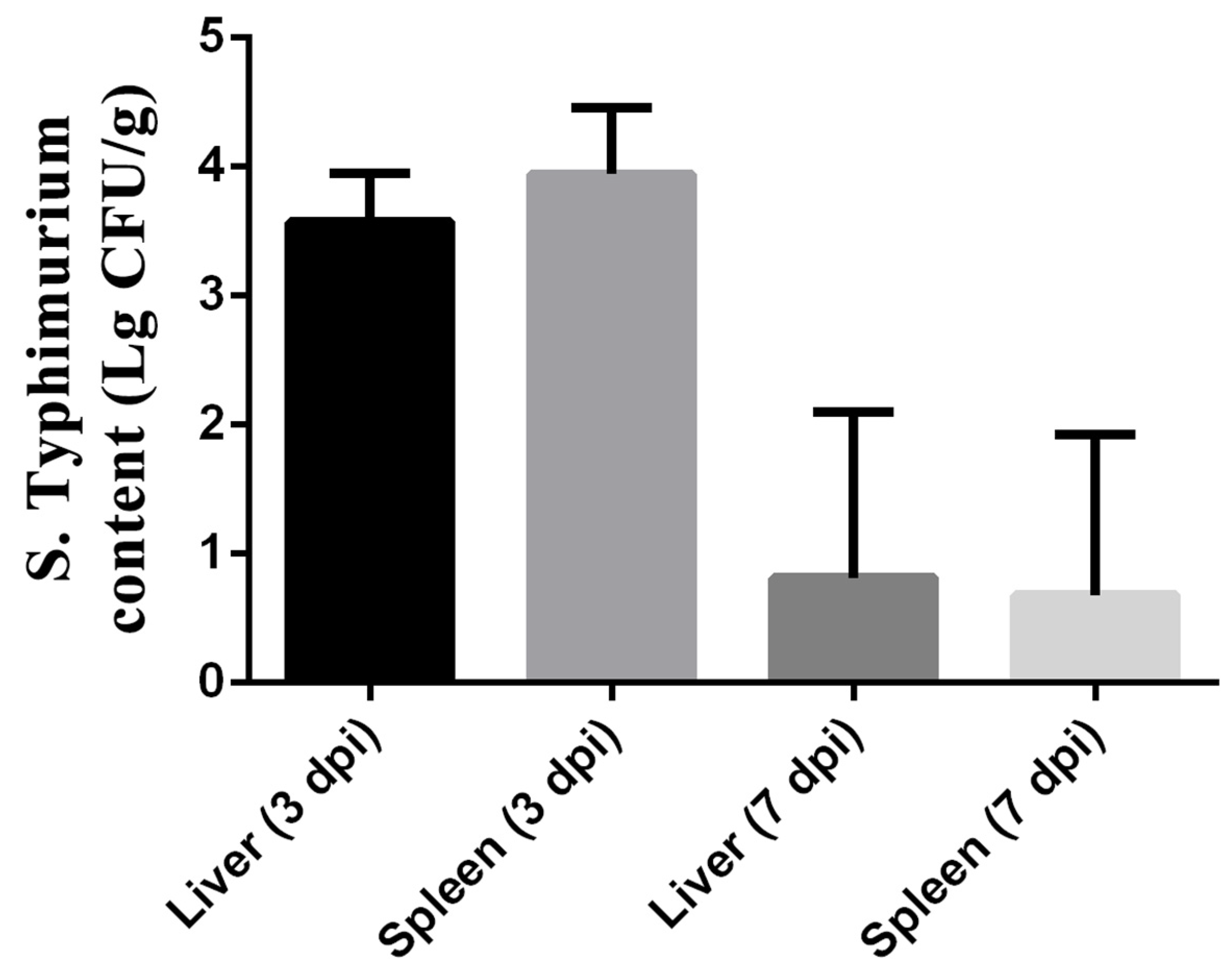
3.5. S. typhimurium Translocation
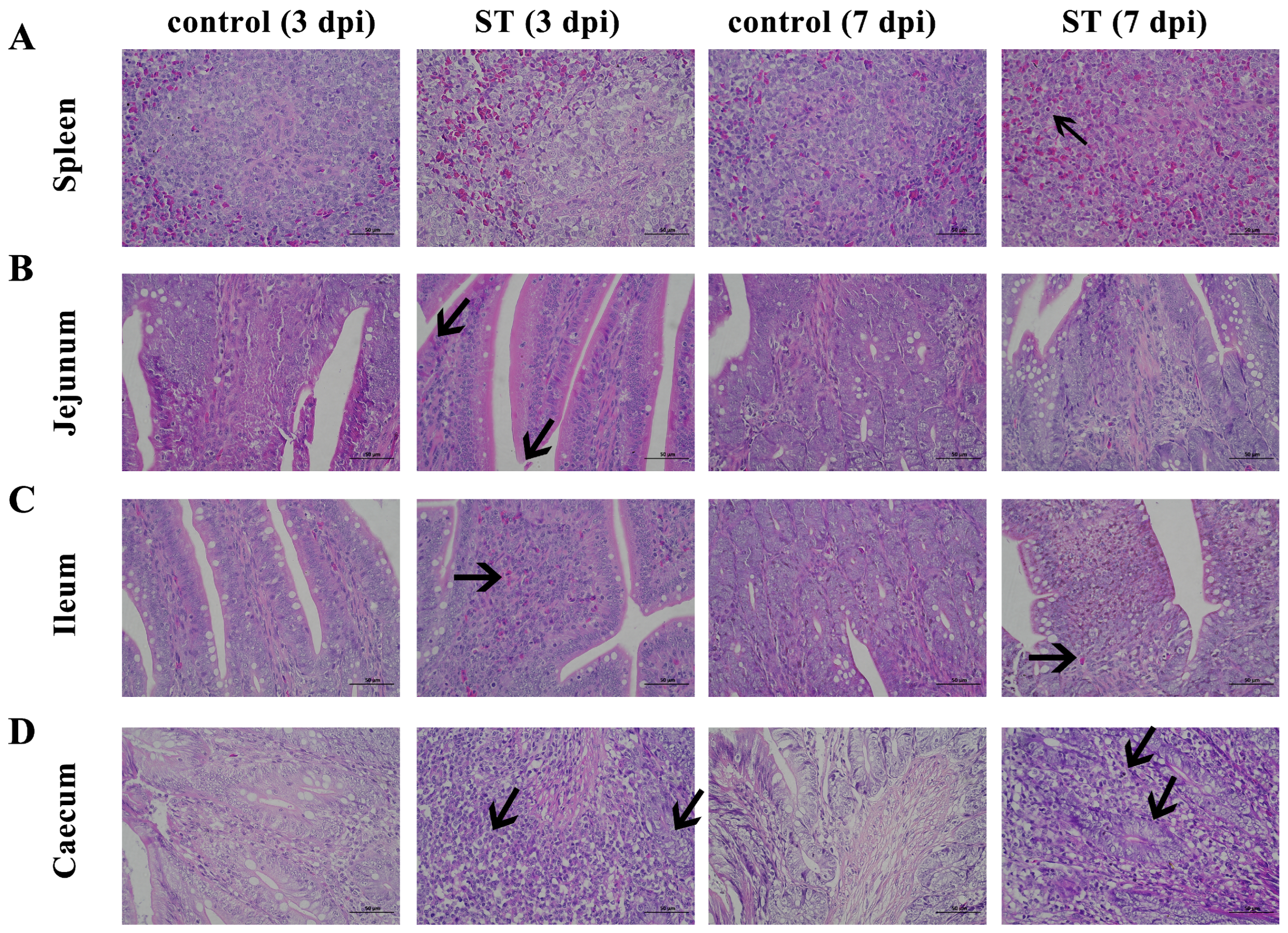
3.6. Histopathological Analysis
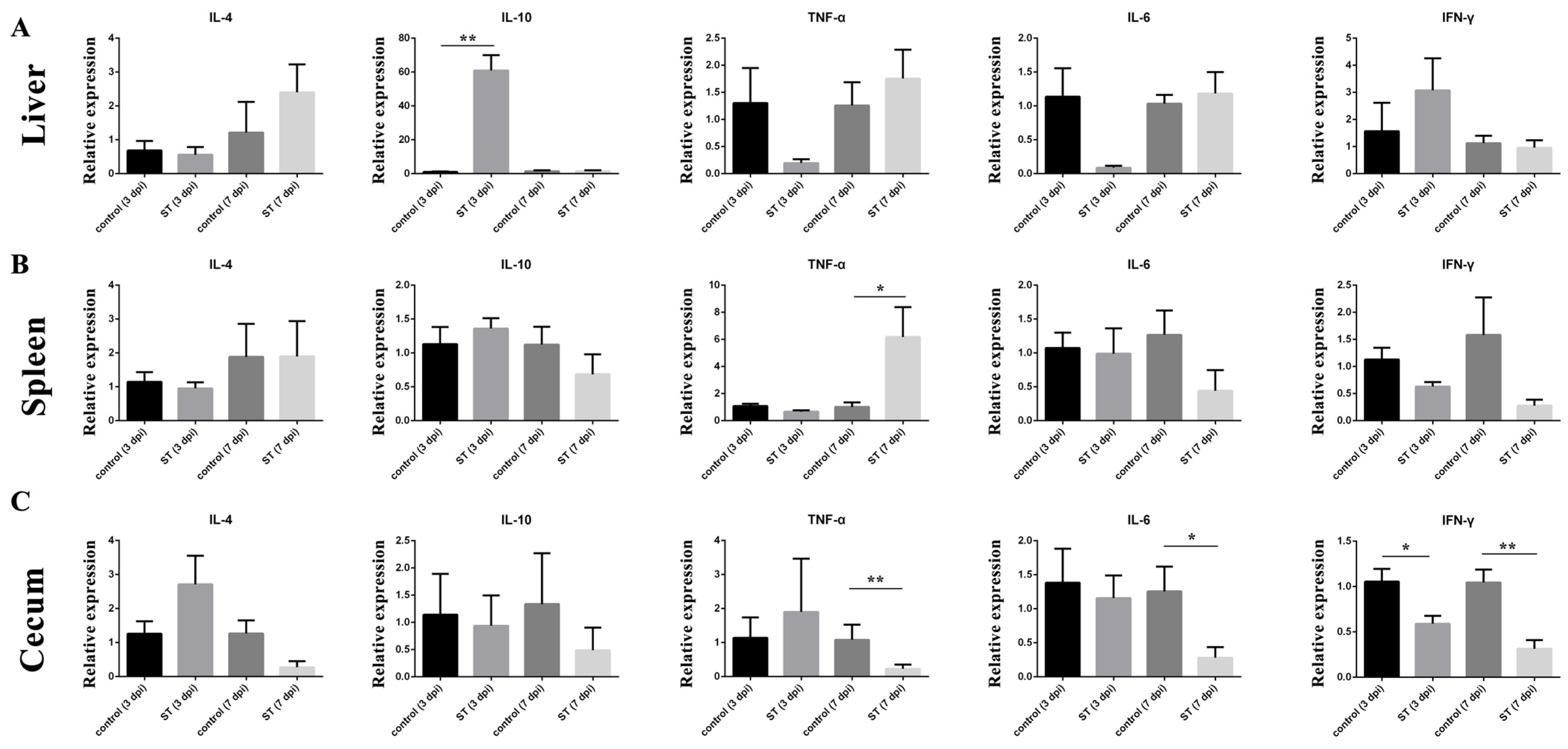
3.7. Cytokine Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bakkeren, E.; Huisman, J.S.; Fattinger, S.A.; Hausmann, A.; Furter, M.; Egli, A.; Slack, E.; Sellin, M.E.; Bonhoeffer, S.; Regoes, R.R.; et al. Salmonella persisters promote the spread of antibiotic resistance plasmids in the gut. Nature 2019, 573, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, G.; Tang, W.; Song, X.; Zhao, X.; Wang, C.; Li, Y.; Zou, M. Antimicrobial resistance and genomic characteristics of Salmonella from broilers in Shandong Province. Front. Vet. Sci. 2023, 10, 1292401. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, M.; Zhou, C.; Gu, G.; Liang, J.; Hou, X.; Wang, M.; Wei, P. Prevalence and antimicrobial resistance of retail-meat-borne Salmonella in southern China during the years 2009–2016: The diversity of contamination and the resistance evolution of multidrug-resistant isolates. Int. J. Food Microbiol. 2020, 333, 108790. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Wang, D.; Hao, W.; Sun, R.; Sun, J.; Liu, Y.; Liao, X. Prevalence, antibiotic resistance, virulence genes and molecular characteristics of Salmonella isolated from ducks and wild geese in China. Food Microbiol. 2024, 118, 104423. [Google Scholar] [CrossRef] [PubMed]
- Zucker, B.A.; Trojan, S.; Muller, W. Airborne gram-negative bacterial flora in animal houses. J. Vet. Med. B Infect. Dis. Vet. Public Health 2000, 47, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Habib, I.; Elbediwi, M.; Mohteshamuddin, K.; Mohamed, M.I.; Lakshmi, G.B.; Abdalla, A.; Anes, F.; Ghazawi, A.; Khan, M.; Khalifa, H. Genomic profiling of extended-spectrum beta-lactamase-producing Escherichia coli from Pets in the United Arab Emirates: Unveiling colistin resistance mediated by mcr-1.1 and its probable transmission from chicken meat—A One Health perspective. J. Infect. Public Health 2023, 16, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Foster, N.; Tang, Y.; Berchieri, A.; Geng, S.; Jiao, X.; Barrow, P. Revisiting Persistent Salmonella Infection and the Carrier State: What Do We Know? Pathogens 2021, 10, 1299. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.M.; Jusoh, M.B.; Rose, F.Z.C.; Azami, M.M.; Roslee, R. Salmonella serovars trend in poultry Malaysia from 2011 to 2020. Vet. Res. Commun. 2024, 48, 1791–1802. [Google Scholar] [CrossRef] [PubMed]
- Hazards, E.P.o.B.; Koutsoumanis, K.; Allende, A.; Alvarez-Ordonez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Salmonella control in poultry flocks and its public health impact. EFSA J. 2019, 17, e05596. [Google Scholar] [CrossRef]
- Chen, Z.; Bai, J.; Wang, S.; Zhang, X.; Zhan, Z.; Shen, H.; Zhang, H.; Wen, J.; Gao, Y.; Liao, M.; et al. Prevalence, Antimicrobial Resistance, Virulence Genes and Genetic Diversity of Salmonella Isolated from Retail Duck Meat in Southern China. Microorganisms 2020, 8, 444. [Google Scholar] [CrossRef]
- Kang, X.; Wang, M.; Meng, C.; Li, A.; Jiao, X.; Pan, Z. Prevalence and whole-genome sequencing analysis of Salmonella reveal its spread along the duck production chain. Poult. Sci. 2022, 101, 101993. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, W.; Hou, S.; Wang, Y.; Wang, S.; Gao, J.; Zhang, R.; Jiang, S.; Zhu, Y. Epidemiological investigation on drug resistance of Salmonella isolates from duck breeding farms in Shandong Province and surrounding areas, China. Poult. Sci. 2022, 101, 101961. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xiao, T.; Wu, L.; Li, Y.; Duan, X.; Liu, W.; Liu, K.; Jin, W.; Ren, H.; Sun, J.; et al. Comprehensive profiling of serotypes, antimicrobial resistance and virulence of Salmonella isolates from food animals in China, 2015–2021. Front. Microbiol. 2023, 14, 1133241. [Google Scholar] [CrossRef] [PubMed]
- Landini, P.; Antoniani, D.; Burgess, J.G.; Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 2010, 86, 813–823. [Google Scholar] [CrossRef]
- Lin, Q.; Sheng, M.; Kang, Z.; Xu, J.; Gao, Y.; Ma, S.; Xin, B.; Tan, Y. Synergistic and antibiofilm activity of DNase I and glucose oxidase loaded chitosan nanoparticles against dual-species biofilms of Listeria monocytogenes and Salmonella. Int. J. Biol. Macromol. 2024, 269, 131943. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.M.; Smithson, A.; Horcajada, J.P.; Martinez, J.A.; Mensa, J.P.; Vila, J. Implication of biofilm formation in the persistence of urinary tract infection caused by uropathogenic Escherichia coli. Clin. Microbiol. Infect. 2006, 12, 1034–1036. [Google Scholar] [CrossRef] [PubMed]
- Gilbertie, J.M.; Levent, G.; Norman, K.N.; Vinasco, J.; Scott, H.M.; Jacob, M.E. Comprehensive phenotypic and genotypic characterization and comparison of virulence, biofilm, and antimicrobial resistance in urinary Escherichia coli isolated from canines. Vet. Microbiol. 2020, 249, 108822. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; International Collaboration on Enteric Disease “Burden of Illness” Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef]
- Chen, Z.; Bai, J.; Zhang, X.; Wang, S.; Chen, K.; Lin, Q.; Xu, C.; Qu, X.; Zhang, H.; Liao, M.; et al. Highly prevalent multidrug resistance and QRDR mutations in Salmonella isolated from chicken, pork and duck meat in Southern China, 2018–2019. Int. J. Food Microbiol. 2021, 340, 109055. [Google Scholar] [CrossRef]
- Shi, W.; Tang, W.; Li, Y.; Han, Y.; Cui, L.; Sun, S. Comparative Analysis between Salmonella enterica Isolated from Imported and Chinese Native Chicken Breeds. Microorganisms 2023, 11, 390. [Google Scholar] [CrossRef]
- Zheng, Y.; Pan, X.; Hou, J.; Shi, W.; Sun, S.; Song, M.; Gao, Z. Effects of Salmonella typhimurium Infection on the Gut Microbiota of Cherry Valley Meat Ducks. Microorganisms 2024, 12, 602. [Google Scholar] [CrossRef]
- Song, Y.; Wang, F.; Liu, Y.; Song, Y.; Zhang, L.; Zhang, F.; Gu, X.; Sun, S. Occurrence and Characterization of Salmonella Isolated From Chicken Breeder Flocks in Nine Chinese Provinces. Front. Vet. Sci. 2020, 7, 479. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e0021321. [Google Scholar] [CrossRef] [PubMed]
- Dawan, J.; Ahn, J. Variability in Adaptive Resistance of Salmonella typhimurium to Sublethal Levels of Antibiotics. Antibiotics 2022, 11, 1725. [Google Scholar] [CrossRef]
- Cui, L.; Zhao, X.; Li, R.; Han, Y.; Hao, G.; Wang, G.; Sun, S. Companion Animals as Potential Reservoirs of Antibiotic Resistant Diarrheagenic Escherichia coli in Shandong, China. Antibiotics 2022, 11, 828. [Google Scholar] [CrossRef]
- Fardsanei, F.; Soltan Dallal, M.M.; Douraghi, M.; Zahraei Salehi, T.; Mahmoodi, M.; Memariani, H.; Nikkhahi, F. Genetic diversity and virulence genes of Salmonella enterica subspecies enterica serotype Enteritidis isolated from meats and eggs. Microb. Pathog. 2017, 107, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Koutsoudis, M.D.; Tsaltas, D.; Minogue, T.D.; von Bodman, S.B. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. USA 2006, 103, 5983–5988. [Google Scholar] [CrossRef]
- Wu, S.; Cui, L.; Han, Y.; Lin, F.; Huang, J.; Song, M.; Lan, Z.; Sun, S. Characteristics, Whole-Genome Sequencing and Pathogenicity Analysis of Escherichia coli from a White Feather Broiler Farm. Microorganisms 2023, 11, 2939. [Google Scholar] [CrossRef] [PubMed]
- Raufu, I.A.; Ahmed, O.A.; Aremu, A.; Ameh, J.A.; Timme, R.E.; Hendriksen, R.S.; Ambali, A.G. Occurrence, antimicrobial resistance and whole genome sequence analysis of Salmonella serovars from pig farms in Ilorin, North-central Nigeria. Int. J. Food Microbiol. 2021, 350, 109245. [Google Scholar] [CrossRef]
- Jiang, Z.; Anwar, T.M.; Peng, X.; Biswas, S.; Elbediwi, M.; Li, Y.; Fang, W.; Yue, M. Prevalence and antimicrobial resistance of Salmonella recovered from pig-borne food products in Henan, China. Food Control 2021, 121, 107535. [Google Scholar] [CrossRef]
- Liu, K.; Wang, M.; Zhang, Y.; Fang, C.; Zhang, R.; Fang, L.; Sun, J.; Liu, Y.; Liao, X. Distribution of antibiotic resistance genes and their pathogen hosts in duck farm environments in south-east coastal China. Appl. Microbiol. Biotechnol. 2024, 108, 136. [Google Scholar] [CrossRef] [PubMed]
- Angulo, F.J.; Nargund, V.N.; Chiller, T.C. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J. Vet. Med. B Infect. Dis. Vet. Public Health 2004, 51, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.; Aqib, A.I.; Muzammil, I.; Majeed, N.; Bhutta, Z.A.; Kulyar, M.F.; Fatima, M.; Zaheer, C.F.; Muneer, A.; Murtaza, M.; et al. MRSA compendium of epidemiology, transmission, pathophysiology, treatment, and prevention within one health framework. Front. Microbiol. 2022, 13, 1067284. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The Complex Relationship between Virulence and Antibiotic Resistance. Genes 2017, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Sydor, A.M.; Boddy, K.C.; Coyaud, E.; Laurent, E.M.N.; Au, A.; Tan, J.M.J.; Yan, B.R.; Moffat, J.; Muise, A.M.; et al. Salmonella exploits membrane reservoirs for invasion of host cells. Nat. Commun. 2024, 15, 3120. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Scholz, J.; Wald, J.; Thuenauer, R.; Hennell James, R.; Ellenberg, I.; Windhorst, S.; Faix, J.; Marlovits, T.C. Structural basis for subversion of host cell actin cytoskeleton during Salmonella infection. Sci. Adv. 2023, 9, eadj5777. [Google Scholar] [CrossRef]
- Barilli, E.; Bacci, C.; StellaVilla, Z.; Merialdi, G.; D’Incau, M.; Brindani, F.; Vismarra, A. Antimicrobial resistance, biofilm synthesis and virulence genes in Salmonella isolated from pigs bred on intensive farms. Ital. J. Food Saf. 2018, 7, 7223. [Google Scholar] [CrossRef]
- Chakroun, I.; Mahdhi, A.; Morcillo, P.; Cordero, H.; Cuesta, A.; Bakhrouf, A.; Mahdouani, K.; Esteban, M.A. Motility, biofilm formation, apoptotic effect and virulence gene expression of atypical Salmonella typhimurium outside and inside Caco-2 cells. Microb. Pathog. 2018, 114, 153–162. [Google Scholar] [CrossRef]
- Fabrega, A.; Vila, J. Salmonella enterica serovar Typhimurium skills to succeed in the host: Virulence and regulation. Clin. Microbiol. Rev. 2013, 26, 308–341. [Google Scholar] [CrossRef]
- Kroger, C.; Colgan, A.; Srikumar, S.; Handler, K.; Sivasankaran, S.K.; Hammarlof, D.L.; Canals, R.; Grissom, J.E.; Conway, T.; Hokamp, K.; et al. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 2013, 14, 683–695. [Google Scholar] [CrossRef]
- Fass, E.; Groisman, E.A. Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 2009, 12, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Merino, L.; Procura, F.; Trejo, F.M.; Bueno, D.J.; Golowczyc, M.A. Biofilm formation by Salmonella sp. in the poultry industry: Detection, control and eradication strategies. Food Res. Int. 2019, 119, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Kroupitski, Y.; Golberg, D.; Belausov, E.; Pinto, R.; Swartzberg, D.; Granot, D.; Sela, S. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 2009, 75, 6076–6086. [Google Scholar] [CrossRef] [PubMed]
- Kogut, M.H.; He, H.; Kaiser, P. Lipopolysaccharide binding protein/CD14/TLR4-dependent recognition of Salmonella LPS induces the functional activation of chicken heterophils and up-regulation of pro-inflammatory cytokine and chemokine gene expression in these cells. Anim. Biotechnol. 2005, 16, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, D.; Abdelfattah-Hassan, A.; Badawi, M.; Ismail, T.A.; Bendary, M.M.; Abdelaziz, A.M.; Mosbah, R.A.; Mohamed, D.I.; Arisha, A.H.; El-Hamid, M.I.A. Thymol nanoemulsion promoted broiler chicken’s growth, gastrointestinal barrier and bacterial community and conferred protection against Salmonella typhimurium. Sci. Rep. 2021, 11, 7742. [Google Scholar] [CrossRef] [PubMed]
- Laptev, G.Y.; Filippova, V.A.; Kochish, I.I.; Yildirim, E.A.; Ilina, L.A.; Dubrovin, A.V.; Brazhnik, E.A.; Novikova, N.I.; Novikova, O.B.; Dmitrieva, M.E.; et al. Examination of the Expression of Immunity Genes and Bacterial Profiles in the Caecum of Growing Chickens Infected with Salmonella Enteritidis and Fed a Phytobiotic. Animals 2019, 9, 615. [Google Scholar] [CrossRef] [PubMed]
- Fasina, Y.O.; Hoerr, F.J.; McKee, S.R.; Conner, D.E. Influence of Salmonella enterica serovar Typhimurium infection on intestinal goblet cells and villous morphology in broiler chicks. Avian Dis. 2010, 54, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.A.; Ponnuraj, N.P.; Lee, J.J.; Utterback, P.; Gaskins, H.R.; Dilger, R.N.; Pettigrew, J.E. Effects of dietary clays on performance and intestinal mucus barrier of broiler chicks challenged with Salmonella enterica serovar Typhimurium and on goblet cell function in vitro. Poult. Sci. 2014, 93, 839–847. [Google Scholar] [CrossRef]
- Shao, Y.; Lei, Z.; Yuan, J.; Yang, Y.; Guo, Y.; Zhang, B. Effect of zinc on growth performance, gut morphometry, and cecal microbial community in broilers challenged with Salmonella enterica serovar typhimurium. J. Microbiol. 2014, 52, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.D.; Bayir, H.O.; Cosby, D.E.; Cox, N.A.; Williams, S.M.; Fowler, J. Evaluation of encapsulated sodium butyrate on growth performance, energy digestibility, gut development, and Salmonella colonization in broilers. Poult. Sci. 2017, 96, 3638–3644. [Google Scholar] [CrossRef]

| Primer | Sequence | Genebank |
|---|---|---|
| DuIFN-γ-F | GCTGATGGCAATCCTGTTTT | KF746069.1 |
| DuIFN-γ-R | GGATTTTCAAGCCAGTCAGC | |
| DuIL-10-F | GGGGAGAGGAAACTGAGAGATG | NM_001310368.1 |
| DuIL-10-R | TCACTGGAGGGTAAAATGCAGA | |
| DuIL-6-F | TTCGACGAGGAGAAATGCTT | JF437643.1 |
| DuIL-6-R | CCTTATCGTCGTTGCCAGAT | |
| DuTNF-α-F | ATCAGCTGGCTAAGACCGTG | XM_005506221.3 |
| DuTNF-α-R | GGGATTGTACAAGGCAGCCA | |
| DuIL-4-F | ATCCTCTCCACGCAGGTTTC | MF346730.1 |
| DuIL-4-R | TGGTGCTCTTTGTCACGATG | |
| 18S-F | TCAGATACCGTCGTAGTTCC | |
| 18S-R | TTCCGTCAATTCCTTTAAGTT |
| No. | Serotype | Drug Resistance | Virulence Genes |
|---|---|---|---|
| 22SD01 | S. typhimurium | AMK-TMP | invA, sipC, sipA, ssaR, ssrA, stnp1, hilA |
| 22SD02 | S. typhimurium | AMK-TE-DOX | invA, sipC, sipA, ssaR, ssrA, stnp1, hilA |
| 22SD03 | S. typhimurium | AMK | invA, sipC, sipA, ssaR, ssrA, stnp1, hilA |
| 22SD04 | S. typhimurium | AMK-DOX | invA, sipC, sipA, ssaR, ssrA, stnp1 |
| 22SD05 | S. typhimurium | DOX | invA, sipC, sipA, ssaR, ssrA, stnp1, hilA |
| 22SD06 | S. typhimurium | DOX | invA, sipC, sipA, ssaR, ssrA, stnp1, hilA |
| 22SD07 | S. typhimurium | CTX-DOX-CTR-TE-AMX-AMP-CAZ | invA, sipC, sipA, ssaR, ssrA, stnp1, hilA |
| 22SD08 | S. typhimurium | AMK-DOX-ENR | invA, sipC, sipA, ssaR, ssrA, stnp1, hilA |
| Drug Resistance | Proportion (%) |
|---|---|
| DOX | 75 (6/8) |
| AMK | 62.5 (5/8) |
| TE | 25 (2/8) |
| TMP | 12.5 (1/8) |
| CTX | 12.5 (1/8) |
| CTR | 12.5 (1/8) |
| AMX | 12.5 (1/8) |
| AMP | 12.5 (1/8) |
| CAZ | 12.5 (1/8) |
| ENR | 12.5 (1/8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Yu, Z.; Wu, S.; Song, M.; Cui, L.; Sun, S.; Wu, J. Pathogenicity of Multidrug-Resistant Salmonella typhimurium Isolated from Ducks. Microorganisms 2024, 12, 1359. https://doi.org/10.3390/microorganisms12071359
Xu Y, Yu Z, Wu S, Song M, Cui L, Sun S, Wu J. Pathogenicity of Multidrug-Resistant Salmonella typhimurium Isolated from Ducks. Microorganisms. 2024; 12(7):1359. https://doi.org/10.3390/microorganisms12071359
Chicago/Turabian StyleXu, Yulin, Zhitong Yu, Shaopeng Wu, Mengze Song, Lulu Cui, Shuhong Sun, and Jiaqiang Wu. 2024. "Pathogenicity of Multidrug-Resistant Salmonella typhimurium Isolated from Ducks" Microorganisms 12, no. 7: 1359. https://doi.org/10.3390/microorganisms12071359
APA StyleXu, Y., Yu, Z., Wu, S., Song, M., Cui, L., Sun, S., & Wu, J. (2024). Pathogenicity of Multidrug-Resistant Salmonella typhimurium Isolated from Ducks. Microorganisms, 12(7), 1359. https://doi.org/10.3390/microorganisms12071359





